Abstract
What determines the nuclear organization within a cell and whether this organization itself can impose cellular function within a tissue remains unknown. To explore the relationship between nuclear organization and tissue architecture and function, we used a model of human mammary epithelial cell acinar morphogenesis. When cultured within a reconstituted basement membrane (rBM), HMT-3522 cells form polarized and growth-arrested tissue-like acini with a central lumen and deposit an endogenous BM. We show that rBM-induced morphogenesis is accompanied by relocalization of the nuclear matrix proteins NuMA, splicing factor SRm160, and cell cycle regulator Rb. These proteins had distinct distribution patterns specific for proliferation, growth arrest, and acini formation, whereas the distribution of the nuclear lamina protein, lamin B, remained unchanged. NuMA relocalized to foci, which coalesced into larger assemblies as morphogenesis progressed. Perturbation of histone acetylation in the acini by trichostatin A treatment altered chromatin structure, disrupted NuMA foci, and induced cell proliferation. Moreover, treatment of transiently permeabilized acini with a NuMA antibody led to the disruption of NuMA foci, alteration of histone acetylation, activation of metalloproteases, and breakdown of the endogenous BM. These results experimentally demonstrate a dynamic interaction between the extracellular matrix, nuclear organization, and tissue phenotype. They further show that rather than passively reflecting changes in gene expression, nuclear organization itself can modulate the cellular and tissue phenotype.
The cell nucleus is organized by a nonchromatin internal structure referred to as the nuclear matrix (NM; refs. 1–3). Identified NM components include coiled-coil proteins (4), cell cycle regulators (5), tissue-specific transcription factors (6, 7), and RNA splicing factors (for review see ref. 2). Although splicing factors have been shown to redistribute during cellular differentiation (8, 9) and following the induction of gene expression (10), spatial distribution of nuclear components are thought to be the consequence of changes in gene expression (8, 10, 11). However, whether NM composition and structure may themselves affect gene expression and cellular function has not been examined.
To systematically study the effect of cell growth and tissue differentiation on nuclear organization, we used a reconstituted basement membrane (rBM)-directed model of mammary gland morphogenesis (12). The HMT-3522 human mammary epithelial cells (HMECs) were isolated from reduction mammoplasty and became immortalized in culture (13). When embedded within a rBM, these cells arrest growth, organize an endogenous BM, and form polarized acinus-like structures with vectorial secretion of sialomucin into a central lumen (12). We used this model to compare the nuclear organization of HMECs cultured on a plastic surface [two-dimensional (2D) monolayer] vs. a three-dimensional (3D) rBM. Nuclear organization was assessed by examining the distribution of the coiled-coil NM proteins lamin B (14) and NuMA (15), the cell cycle regulator Rb (p110Rb; ref. 5), and the splicing factor SRm160 (formerly known as B1C8; ref. 16). These proteins had distinct spatial distribution patterns specific for proliferation, growth arrest, and acini formation. Moreover, disruption of nuclear organization in acini by either perturbing histone acetylation or directly modifying the distribution of NM proteins altered the acinar phenotype.
We previously hypothesized (17) and thereafter provided evidence that the extracellular matrix (ECM) directs morphogenesis and gene expression in mammary epithelial cells (12, 18, 19). Here we show that a reciprocal relationship exists between the ECM and nuclear organization. These findings underscore a role for nuclear organization in regulation of gene expression and provide a possible framework for how cell–ECM interactions determine cell and tissue phenotype.
MATERIALS AND METHODS
Cell Culture.
HMT-3522 HMECs (S-1 passage-50 cells; ref. 13) were propagated in 2D cultures in chemically defined medium (12), and growth arrest was induced by removing epidermal growth factor (EGF) for 48 hr. Cultures were prepared by embedding single cells (8.5 × 105 cells per ml of matrix) in rBM (Matrigel, Collaborative Research) or collagen-I matrix (Cellagen AC-5, ICN) in 4-well chamber slides (Nalge). These cultures were grown for 5–10 days. Growth arrest and morphogenesis were routinely observed by days 7–9.
Antibodies and Inhibitors.
For Western blots and/or immunostaining, we used mAbs against type IV collagen (clone CIV, Dako), β-catenin (clone 14, Transduction Laboratories, Lexington, KY), SRm160 splicing factor (clone B1C8, 16), lamin B (clone 101-B7, Matritech, Cambridge, MA), NuMA (clone 204–41, Matritech, and clone B1C11, a gift from S. Penman, Massachusetts Institute of Technology, Cambridge, MA), and polyclonal antibodies (pAbs) against Ki-67 (NovoCastra, Newcastle, U.K.), acetylated histone H4 (Upstate Biotechnology, Lake Placid, NY), and p110Rb (Santa Cruz Biotechnology). For bioperturbation assays, we used mAbs against lamins A/C (clone 636, Novocastra, Newcastle, U.K.) and NuMA (clone 22, Transduction Laboratories, Lexington, KY), in addition to B1C11 and 101-B7. Trichostatin A (Wako Chemicals, Richmond, VA) was used as an inhibitor of histone deacetylase (40 nM).
Indirect Immunofluorescence.
Cells were permeabilized in situ (0.5% Triton X-100 in 100 mM NaCl/300 mM sucrose/10 mM Pipes, pH 6.8/5 mM MgCl2 containing 1 mM Pefabloc Sc (AEBSF) (Boehringer Manheim)/10 μg/ml leupeptin/10 μg/ml aprotinin/10 μg/ml trypsin inhibitor type II/250 μM NaF), fixed in 2% paraformaldehyde, and immunostained as described (18). Human mammary tissue was snap-frozen in n-hexane and embedded in Tissue-Tek O.C.T. compound (Sakura Firetek, Torrance, CA); 5-μm sections were fixed in methanol and immunostained in accordance with human protocol (KF) 01–216/93 in the laboratory of O.W.P.
Image Acquisition, Processing, and Data Analysis.
Samples were analyzed by using a Bio-Rad MRC 1024 laser scanning confocal microscope attached to a Nikon Diaphot 200 microscope. Fluorescence specificity was verified by sequential fluorophore excitation. NuMA foci were analyzed by using image space-3d analysis program (Molecular Probes) and normalized to 3D rBM cluster-cell number by highlighting and counting each nucleus using image space-measure 2d. The voxel threshold was set at 0.2 μm.
Immunoblot Analysis.
Total cell extracts (2% SDS in phosphate-buffered saline, pH 7.4, containing 1 mM Pefabloc/10 μg/ml leupeptin/10 μg/ml aprotinin/10 μg/ml trypsin inhibitor type II/250 μM NaF) were prepared in situ for 2D cultures or from acini isolated from 3D cultures by dispase treatment (5,000 units per ml caseinolytic activity, Collaborative Research). Equal amounts of protein were separated and immunoblotted as described (18).
In Situ NM Preparation.
In situ NM preparation was as previously described (20), except that 0.05% Triton X-100 and micrococcal nuclease (5 units per ml; Sigma) were used.
Antibody-Mediated Perturbation of Nuclear Organization.
rBM-induced acini (day 10) were permeabilized for less than 2 min in situ (0.01% digitonin in 25 mM Hepes, pH 7.2/78 mM potassium acetate/3 mM magnesium acetate/1 mM EGTA/300 mM sucrose/1% RIA-grade BSA), rinsed twice in digitonin-free buffer, and incubated in medium containing dialyzed specific or mock mAbs (15 μg/ml) for 48 hr, after which the cells were incubated with fresh medium for an additional 48 hr. Antibody concentrations and incubation times were determined empirically. Trypan blue dye-exclusion tests and apoptosis studies verified the absence of digitonin toxicity.
RESULTS
Internal Nuclear Organization Is Remodeled When HMECs Are Cultured Within a Basement Membrane.
HMT-3522 HMECs, like primary HMECs, undergo morphogenesis to form tissue-like acini when cultured in a 3D rBM (12, 18). Neither cell type undergoes acinar differentiation when cultured as 2D monolayers. In proliferating 2D cultures, NuMA was diffusely distributed in the nucleus (Fig. 1b) except when localized to the spindle poles in mitotic cells (15), and splicing factor SRm160 was distributed into numerous speckles of heterogenous sizes (Fig. 1c; ref. 16). In rBM-induced acini, NuMA was redistributed into an average of eight nuclear foci (ranging from 1 to 1.6 μm in diameter) surrounded by diffusely localized NuMA protein (Fig. 1e), and SRm160 was distributed into an average of seven large speckles (Fig. 1f). In contrast, lamin B maintained a peripheral ring-like distribution around the nucleus, with some internal localization, regardless of culture conditions (Fig. 1 a and d). The distribution pattern of these proteins was conserved in NM preparations in situ, where chromatin was removed before immunolocalization (staining is shown for 3D rBM cultures only (Fig. 1 g–i).
Figure 1.
NM protein redistribution in HMECs after 3D rBM-induced acinar morphogenesis. Confocal fluorescence images (0.2-μm optical sections) of lamin B, NuMA, and splicing factor SRm160 in cells grown as 2D monolayers (a–c) and within 3D rBMs (d–i). NuMA was diffusely distributed in the nuclei of cells grown as monolayers (b), but reorganized into large nuclear foci in cells induced to undergo morphogenesis (acini formation) in response to a rBM (e). SRm160 was distributed as multiple nuclear speckles in cells cultured as monolayer (c), whereas it was concentrated into fewer and larger speckles in the acini (f). Lamin B, in contrast, consistently localized to the nuclear periphery and within intranuclear patches (a and d). The distribution of lamin B (g), NuMA (h), and SRm160 (i) after in situ NM preparation of cells cultured in 3D rBM was similar to that observed in intact cells (d–f). Arrows indicate nuclei found within the plane of the section. (Bar = 10 μm.)
We next examined NuMA and SRm160 distribution at different stages of 3D rBM-induced morphogenesis. After embedment in rBM, cells proliferated to form small clusters by days 3–5 but lacked β-catenin at cell–cell junctions, and collagen IV staining was discontinuous (Fig. 2 Aa–Ac). After growth arrest (days 6–10), cells assembled a continuous endogenous BM and formed polarized acinus-like structures with organized adherens junctions (Fig. 2 Ad–Af). NuMA was uniformly distributed in the nuclei of proliferating cells (Fig. 2Ba), but became concentrated into distinct foci of differing sizes after growth arrest (day 7; Fig. 2Bb), and into larger and fewer foci on completion of morphogenesis (day 10, Fig. 2Bc). NuMA and the splicing factor SRm160 were not colocalized in proliferating cells (Fig. 2 Ba′ and Ba′′), but NuMA foci and SRm160 speckles were closer together after growth arrest (Fig. 2 Bb′ and Bb′′) and were completely colocalized in large assemblies after the completion of morphogenesis (Fig. 2B, c′ and c′′). These spatial changes in NuMA arrangement occurred without significant modifications in the level of NuMA expression or molecular weight, as determined by using Western blot analysis (Fig. 2Be). These experiments demonstrate that specific NM proteins undergo spatial rearrangement during rBM-induced acinar morphogenesis. Because the existence of NuMA in differentiated tissue has been questioned (21), we studied NuMA in the normal resting human mammary gland. Intense staining was observed in the epithelial cells of acini and ducts, where NuMA was distributed in foci of different sizes and resembled the acinar stages recapitulated in 3D rBM cultures (Fig. 2Bd).
Figure 2.
(A) Distribution of structural proteins during rBM-induced acinar morphogenesis. Confocal fluorescence images (0.2-μm optical sections) of collagen IV, β-catenin, and Ki-67 in HMECs embedded within a rBM for 3–4 days (proliferating cells; a–c), and for 7–10 days (growth-arrested acini; d–f). Coincident with growth arrest and acinar morphogenesis, HMECs deposited an organized endogenous collagen IV-rich BM (a vs. d), whereas β-catenin relocalized from the cytosol and basal plasma membrane to sites of cell–cell adhesion (b vs. e). Acinar morphogenesis was associated with cell cycle exit, as indicated by the loss of Ki-67 staining (c vs. f). (B). Spatial analysis of NuMA and splicing factor SRm160 redistribution during rBM-induced acinar morphogenesis. Confocal Texas red fluorescence images (0.2-μm optical sections) of NuMA (a–c) and double-labeled NuMA (Texas red), and fluorescein isothiocyanate (FITC) green-stained SRm160 (a′, a′′, b′, b′′, c′, and c′′) in HMT-3522 cells proliferating (a, a′, and a′′) and undergoing morphogenesis (b, b′, b′′, c, c′, and c′′) in response to a rBM. In proliferating cells, NuMA was diffusely distributed (a) and did not colocalize with SRm160 (a′ and a′′). After growth arrest, NuMA coalesced into foci of increasing size (0.2–2 μm; f) in association with the establishment of mature tissue-like structures (acini; b and c) Nine nuclei are shown in b. Only the larger NuMA foci observed in late morphogenesis fully colocalized with SRm160 (b′, b′′, c′, and c′′). (d) In the ductal and acinar HMECs of the mammary gland, in vivo, NuMA was localized in foci with a size distribution comparable to that observed in most of the HMEC nuclei of differentiating rBM cultures shown in b. (e) Western blot analysis of NuMA and Lamin B showed no difference in protein expression or size between proliferating and growth-arrested HMECs grown within rBMs. Arrows indicate nuclei. (Bars = 10 μm.)
Growth Arrest Is Associated With Changes in NuMA and Rb Distribution.
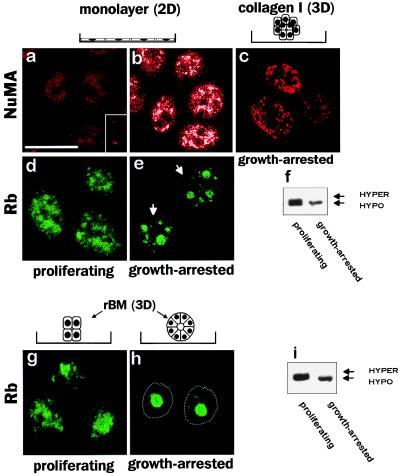
ECM-directed growth arrest is an early and critical step in mammary epithelial cell morphogenesis (12). To distinguish between the effect of ECM-directed growth arrest and changes caused by tissue structure and polarity, the localization of NuMA and SRm160 was compared between growth-arrested and proliferating cells cultured in monolayers. Less than 5% of the cells remained in the cell cycle after growth arrest induced by EGF removal, as indicated by the absence of detectable Ki-67 immunostaining (data not shown). NuMA was uniformly distributed in the nuclei of proliferating cells but coalesced into denser areas on growth arrest (Fig. 3 a and b). The irregular geometric quality of these dense areas was distinct from the circular foci pattern observed in growth-arrested 3D rBM-grown cells. In contrast, no significant change in the multispeckled distribution of SRm160 was detected under these conditions (data not shown). The relationship between nuclear organization and growth status was further investigated by examining the distribution of the cell cycle regulator Rb. Rb redistributed from a diffuse nuclear pattern in proliferating HMECs into a few large foci in growth-arrested cells (Fig. 3 d and e). Strikingly, the distribution of Rb in the growth-arrested 2D cultures was distinct from that observed in the growth-arrested 3D cultures (compare Fig. 3 e and h), which may reflect differences in the state of growth arrest between 2D monolayer and 3D rBM cultures. The monofocal pattern of Rb observed in 3D culture coincided with growth arrest. Western blot analysis showed that hypophosphorylated Rb was associated with the NM in 3D cultures (data not shown) as was previously reported for 2D cultures (5). Moreover, the diffuse distribution observed in proliferating cells was associated with the hyperphosphorylated form of the protein (Fig. 3 f and i).
Figure 3.
Effect of growth status on the distribution of NM proteins. Confocal fluorescence images (0.2-μm optical sections) of Texas red-stained NuMA (a–c) and fluorescein isothiocyanate (FITC) green-stained Rb (d, e, g, and h) in cells proliferating as 2D monolayers (a and d) and within 3D rBMs (g) and cells growth-arrested in monolayer (b and e) and within collagen-I (c) or a rBM (h). NuMA was diffusely distributed in the nucleus of proliferating HMECs grown as monolayers (a) and reorganized into random aggregates on growth arrest induced by EGF removal (b). The settings for image recording were the same as for a. Aggregates appear white because of saturation of the signal. NuMA was distributed in random aggregates or in small foci in growth-arrested and BM-free cell colonies obtained after 10 days of culture within collagen-I (c). Rb was diffusely distributed in the nucleus of proliferating cells grown either in monolayer (d) or in 3D rBM (g); however, on growth arrest, the protein redistributed into several foci in the monolayer propagated cells (e) but coalesced into a central, single nuclear focus in the rBM-induced acini (h); the dotted line indicates outer nuclear limit. Western blot analysis of Rb in proliferating and growth-arrested cells grown as monolayers (f) or within a 3D rBM (i) shows that the hyperphosphorylated isoform was present only in proliferating cells. Arrows indicate nuclei. (Bar = 10 μm.)
Because growth arrest in 3D rBM precedes the final stages of acinar morphogenesis (12), we examined the relationship between the large NuMA foci and the formation of a polarized endogenous BM. HMECs cultured in a 3D collagen-I matrix form growth-arrested organized colonies but do not assemble a polarized, endogenous BM (22). Therefore, we compared NuMA distribution in cells grown in rBM to those grown in type I collagen. After 12 days in collagen I, NuMA was distributed as small foci or irregular dense aggregates (Fig. 3c), similar to the pattern observed in growth-arrested cells in 2D cultures. Thus, NuMA redistribution into dense areas and small foci is induced by growth arrest, but the coalescence of the foci into larger and distinct structures requires the presence of a BM.
Cross-Modulation Between NuMA Distribution, Chromatin Structure, and the Acinar Phenotype.
The degree of histone acetylation has been shown to regulate chromatin structure and gene expression (19, 23). Histone acetylation was altered in the acini by using the histone deacetylase inhibitor trichostatin A. After 2 hr of treatment, NuMA foci began to disperse, and several cells entered the cell cycle, as measured by an increase in the Ki-67 labeling index. After 24 hr of treatment, NuMA was diffusely distributed in all nuclei (Fig. 4 e vs. a), and the acinar phenotype was altered as shown by loss of the endogenous BM (Fig. 4 f vs. b), redistribution of β-catenin (Fig. 4 g vs. c), and the presence of mitotic cells, as shown by mitotic spindle-pole staining of NuMA (Fig. 4e, arrow). In contrast, trichostatin A did not alter the cell phenotype or the distribution of NuMA (data not shown).
Figure 4.
Cross-modulation between chromatin structure, NM organization, and the acinar phenotype. Confocal fluorescence images (0.2-μm optical sections) of NuMA (a, e, and i), collagen IV (b, f, and j), β-catenin (c, g, and k), and acetylated histone H4 (d, h, and l) in control, trichostatin A (TSA)-treated, and NuMA mAb-incubated acini (day 10 of 3D rBM culture). (a–d) Nuclear organization and acinar phenotype in controls. Acini exhibit NuMA foci (a), an organized endogenous collagen IV-rich BM (b), cell–cell localized β-catenin (c), and dispersed acetylated H4 histone (d). (e–h) Effects of TSA on nuclear architecture and acinar phenotype. After 24 hr of TSA treatment (40 nM), >55% of the cells entered the cell cycle, as indicated by an increase in Ki-67 labeling index (m) and the appearance of mitotic cells (arrow in e). NuMA was uniformly distributed in the nuclei (e), collagen IV disappeared (f), β-catenin was released from the cell–cell interface (g), and the pattern of histone H4 acetylation was altered (h). (i–l) Effects of mAb-induced NuMA foci disruption on nuclear organization and acinar phenotype. Introduction of a NuMA mAb into the nuclei of the acini by using reversible digitonin permeabilization led to the disruption of NuMA foci (i), degradation of the collagen IV-rich BM (arrows in j), and the nuclear marginalization of acetylated H4 histone (l). There was no consistent alteration observed for β-catenin other than increased basal labeling (k). These effects were not observed with mock IgGs or mAbs to lamins A/C or B. (n) BM degradation after mAb-induced NuMA disruption in acini. Analysis of the percentage of acini with intact collagen IV-rich BMs in relation to control/digitonin-permeabilized (DP) acini (a), mock-IgG mAb-treated/DP acini (b), NuMA mAb-treated/nonpermeabilized acini (c), NuMA mAb-treated/DP acini (d), NuMA mAb-treated/DP acini + the metalloproteinase inhibitor GM6001 (e), NuMA mAb-treated/DP acini + the inactive metalloproteinase inhibitor GM1210 (f), NuMA mAb-treated/DP acini + the uPA inhibitor, aprotinin (g), and Lamin B mAb-treated/DP acini (h). Acini (>35%) degraded their endogenous BMs in response to disruption of NuMA (d). The BM loss could be rescued by treatment with the metalloproteinase inhibitor GM6001 (e), but not its inactive analogue (f) or a uPA protease inhibitor (g). (Bar = 10 μm.)
Because NuMA is essential for postmitotic nuclear assembly and participates in the loss of nuclear integrity during apoptosis (24, 25), we asked whether disruption of NuMA foci in the acini could globally influence nuclear organization and affect the acinar phenotype. Rapid and reversible digitonin permeabilization was used to load cells with either anti-NuMA mAbs or with an IgG1 mock mAb. The NuMA mAb B1C11, but not an N-terminal-specific mAb (clone 22; data not shown), disrupted NuMA organization, causing the protein to become diffusely redistributed within the nucleus as revealed by the secondary Ab (Fig. 4i). Chromatin structure was altered, as shown by the rearrangement of acetylated histone H4 distribution (Fig. 4 l vs. d). More dramatically, disruption of NuMA organization altered the acinar phenotype, as indicated by loss of the endogenously deposited BM (Fig. 4j). Because the loss could be prevented by treatment with GM6001, a potent metalloprotease inhibitor (Fig. 4n; ref. 26), we conclude that NuMA disruption led to induction and/or activation of a metalloprotease. Similar treatment of the acini with mAbs against lamins A/C or lamin B did not induce any change in histone H4 acetylation, BM integrity, or lamin distribution, even though these Abs reached their nuclear targets, as shown by secondary Ab staining (Fig. 4n and data not shown).
DISCUSSION
By modifying the cellular microenvironment, we have demonstrated that nuclear organization rearranges dramatically in HMECs after growth arrest and tissue-like acinar morphogenesis (Scheme S1). The use of the 3D-rBM culture assay has enabled us also to show that alterations of nuclear organization can modify the cellular and tissue phenotype.
Scheme 1.
Previously documented changes in nuclear organization have been broadly descriptive. By systematically analyzing the distribution of three NM proteins in 2D and 3D cultures, we have determined that precise nuclear rearrangements occur with growth arrest and after rBM-induced morphogenesis. In 3D rBM cultures, both NuMA and Rb were diffusely distributed in the nucleus of proliferating cells. After growth arrest, NuMA was relocalized into discrete foci, whereas Rb redistributed into a central nuclear mass. These patterns of distribution were different from those observed in growth-arrested cells in monolayer 2D cultures, suggesting that there may be different states of growth arrest in 2D and 3D rBM cultures (27). Because NuMA distribution in 3D collagen I cultures was comparable to that observed in growth-arrested 2D cultures, our results suggest that 3D organization of cells per se cannot explain the differences seen between monolayer and 3D rBM cultures. This finding implies that BM signaling is necessary for the ultimate nuclear organization within the acini. Indeed, the presence of large and distinct NuMA foci was observed only in mature 3D rBM cultures and in adult resting mammary gland in vivo, where the acini were surrounded by a continuous endogenous BM. The mammary gland undergoes developmental cycles of growth and differentiation even in adults; this may account for the heterogeneity of foci size observed in vivo and may further explain the absence of the very large NuMA foci in subpopulations of differentiated acini (Fig. 2Bc). Whether the pattern of NuMA distribution indeed corresponds to different levels of differentiation in vivo requires further analysis.
The antibody-directed disruption of NuMA foci in the acini induced changes in the distribution pattern of acetylated histone H4, the activation of metalloprotease(s), and the loss of BM integrity. These results, as well as our observation that NuMA progressively coalesces and eventually colocalizes with enlarged splicing-factor speckles during acini differentiation, suggests that some nuclear proteins may contain the molecular information necessary for the development and/or maintenance of the acinar phenotype. Interestingly, trichostatin-induced alteration of histone acetylation in acini also led to the disruption of NuMA foci and was associated with the loss of BM and the induction of cell proliferation. Although we do not know the molecular mechanisms responsible for phenotypic alterations induced by nuclear reorganization, our experiments demonstrate also the existence of reciprocal interactions between nuclear organization, chromatin structure, and the acinar phenotype. The BM has been shown previously to be necessary for the formation and maintenance of the functional acinus (12, 28, 29). We report here that BM-induced acinar formation is associated with the distinct spatial organization of a repertoire of NM proteins and that, conversely, perturbation of nuclear organization alters the BM and influences the acinar phenotype. These results illustrate the dynamic reciprocity between the ECM and the structural organization of the nucleus, and underscore the importance of ECM–NM communication (17) in phenotypic plasticity.
Acknowledgments
We thank K. Schmeichel for critical reading of the manuscript and J. Campisi, S. Penman, J. Lawrence, M. Simian, and P. Pujuguet for helpful comments. This work was supported by the U.S. Department of Energy, Office of Biological and Environmental Research (Grant DE-AC03-76SF00098), the National Institutes of Health (Grant CA-64786) to M.J.B.; the World Health Organization/International Agency for Research on Cancer and Department Of Defense/Breast Cancer Research Program fellowships to S.A.L., a University of California/Breast Cancer Research Program fellowship to V.M.W., the American Cancer Society (Grant IRG-93-033-05) to J.A.N., and a grant from the Danish Medical Research Council to O.W.P.
ABBREVIATIONS
- NM
nuclear matrix
- BM
basement membrane
- rBM
reconstituted BM
- HMEC
human mammary epithelial cells
- 2D and 3D
two and three dimensional
- Rb
retinoblastoma protein
- ECM
extracellular matrix
- EGF
epidermal growth factor
References
- 1.Berezney R, Coffey D S. Biochem Biophys Res Commun. 1974;60:1410–1417. doi: 10.1016/0006-291x(74)90355-6. [DOI] [PubMed] [Google Scholar]
- 2.Nickerson J A, Blencowe B J, Penman S. Int Rev Cytol. 1995;162A:67–123. doi: 10.1016/s0074-7696(08)61229-2. [DOI] [PubMed] [Google Scholar]
- 3.Nickerson J A, Krockmalnic G, Van K M, Penman S. Proc Natl Acad Sci USA. 1997;94:4446–4450. doi: 10.1073/pnas.94.9.4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Odgren P R, Harvie L W, Fey E G. Proteins. 1996;24:467–484. doi: 10.1002/(SICI)1097-0134(199604)24:4<467::AID-PROT6>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 5.Mancini M A, Shan B, Nickerson J A, Penman S, Lee W-H. Proc Natl Acad Sci USA. 1994;91:418–422. doi: 10.1073/pnas.91.1.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Wijnen A J, Bidwell J P, Fey E G, Penman S, Lian J B, Stein J, Stein G S. Biochemistry. 1993;32:8397–8402. doi: 10.1021/bi00084a003. [DOI] [PubMed] [Google Scholar]
- 7.Nardozza T A, Quigley M M, Getzenberg R H. J Cell Biochem. 1996;61:467–477. doi: 10.1002/(SICI)1097-4644(19960601)61:3%3C467::AID-JCB14%3E3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 8.Antoniou M, Carmo-Fonseca M, Ferreira J, Lamond A I. J Cell Biol. 1993;123:1055–1068. doi: 10.1083/jcb.123.5.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sahlas D J, Milankov K, Park P C, De Boni U. J Cell Sci. 1993;105:347–357. doi: 10.1242/jcs.105.2.347. [DOI] [PubMed] [Google Scholar]
- 10.Misteli T, Caceres J F, Spector D L. Nature (London) 1997;387:523–527. doi: 10.1038/387523a0. [DOI] [PubMed] [Google Scholar]
- 11.Singer R H, Green M R. Cell. 1997;91:291–294. doi: 10.1016/s0092-8674(00)80411-0. [DOI] [PubMed] [Google Scholar]
- 12.Petersen O W, Rønnov-Jessen L, Howlett A R, Bissell M J. Proc Natl Acad Sci USA. 1992;89:9064–9068. doi: 10.1073/pnas.89.19.9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Briand P, Petersen O W, Van Deurs B. In Vitro Cell Dev Biol. 1987;23:181–188. doi: 10.1007/BF02623578. [DOI] [PubMed] [Google Scholar]
- 14.Gerace L, Comeau C, Benson M. J Cell Sci Suppl. 1984;1:137–160. doi: 10.1242/jcs.1984.supplement_1.10. [DOI] [PubMed] [Google Scholar]
- 15.Lydersen B, Pettijohn D. Cell. 1980;22:489–499. doi: 10.1016/0092-8674(80)90359-1. [DOI] [PubMed] [Google Scholar]
- 16.Blencowe B J, Issner R, Nickerson J A, Sharp P A. Genes Dev. 1998;12:996–1009. doi: 10.1101/gad.12.7.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bissell M J, Hall H G, Parry G. J Theor Biol. 1982;99:31–68. doi: 10.1016/0022-5193(82)90388-5. [DOI] [PubMed] [Google Scholar]
- 18.Weaver V M, Petersen O W, Wang F, Larabell C A, Briand P, Damsky C, Bissell M J. J Cell Biol. 1997;137:231–245. doi: 10.1083/jcb.137.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Myers C A, Schmidhauser C, Mellentin-Michelotti J, Fragoso G, Roskelley C D, Casperson G, Mossi R, Pujuguet P, Hager G, Bissell M J. Mol Cell Biol. 1998;18:2184–2195. doi: 10.1128/mcb.18.4.2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He D, Nickerson J A, Penman S. J Cell Biol. 1990;110:569–580. doi: 10.1083/jcb.110.3.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Merdes A, Cleveland D W. J Cell Sci. 1998;111:71–79. doi: 10.1242/jcs.111.1.71. [DOI] [PubMed] [Google Scholar]
- 22.Howlett A R, Bailey N, Damsky C, Petersen O W, Bissell M J. J Cell Sci. 1995;108:1945–1957. doi: 10.1242/jcs.108.5.1945. [DOI] [PubMed] [Google Scholar]
- 23.Pazin M J, Kadonaga J T. Cell. 1997;89:325–328. doi: 10.1016/s0092-8674(00)80211-1. [DOI] [PubMed] [Google Scholar]
- 24.Compton D A, Cleveland D W. Curr Opin Cell Biol. 1994;6:343–346. doi: 10.1016/0955-0674(94)90024-8. [DOI] [PubMed] [Google Scholar]
- 25.Weaver V M, Carson C E, Walker P R, Chaly N, Lach B, Raymond Y, Brown D L, Sikorska M. J Cell Sci. 1996;109:45–56. doi: 10.1242/jcs.109.1.45. [DOI] [PubMed] [Google Scholar]
- 26.Grobelny D, Poncz L, Galardy R E. Biochemistry. 1992;31:7152–7154. doi: 10.1021/bi00146a017. [DOI] [PubMed] [Google Scholar]
- 27.Dhawan J, Farmer S R. J Biol Chem. 1990;266:8470–8475. [PubMed] [Google Scholar]
- 28.Streuli C H, Bailey N, Bissell M J. J Cell Biol. 1991;115:1383–1395. doi: 10.1083/jcb.115.5.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boudreau N, Sympson C J, Werb Z, Bissell M J. Science. 1995;267:891–893. doi: 10.1126/science.7531366. [DOI] [PMC free article] [PubMed] [Google Scholar]