Abstract
Wnt family members are critical to many developmental processes, and components of the Wnt signaling pathway have been linked to tumorigenesis in familial and sporadic colon carcinomas. Here we report the identification of two genes, WISP-1 and WISP-2, that are up-regulated in the mouse mammary epithelial cell line C57MG transformed by Wnt-1, but not by Wnt-4. Together with a third related gene, WISP-3, these proteins define a subfamily of the connective tissue growth factor family. Two distinct systems demonstrated WISP induction to be associated with the expression of Wnt-1. These included (i) C57MG cells infected with a Wnt-1 retroviral vector or expressing Wnt-1 under the control of a tetracyline repressible promoter, and (ii) Wnt-1 transgenic mice. The WISP-1 gene was localized to human chromosome 8q24.1–8q24.3. WISP-1 genomic DNA was amplified in colon cancer cell lines and in human colon tumors and its RNA overexpressed (2- to >30-fold) in 84% of the tumors examined compared with patient-matched normal mucosa. WISP-3 mapped to chromosome 6q22–6q23 and also was overexpressed (4- to >40-fold) in 63% of the colon tumors analyzed. In contrast, WISP-2 mapped to human chromosome 20q12–20q13 and its DNA was amplified, but RNA expression was reduced (2- to >30-fold) in 79% of the tumors. These results suggest that the WISP genes may be downstream of Wnt-1 signaling and that aberrant levels of WISP expression in colon cancer may play a role in colon tumorigenesis.
Wnt-1 is a member of an expanding family of cysteine-rich, glycosylated signaling proteins that mediate diverse developmental processes such as the control of cell proliferation, adhesion, cell polarity, and the establishment of cell fates (1, 2). Wnt-1 originally was identified as an oncogene activated by the insertion of mouse mammary tumor virus in virus-induced mammary adenocarcinomas (3, 4). Although Wnt-1 is not expressed in the normal mammary gland, expression of Wnt-1 in transgenic mice causes mammary tumors (5).
In mammalian cells, Wnt family members initiate signaling by binding to the seven-transmembrane spanning Frizzled receptors and recruiting the cytoplasmic protein Dishevelled (Dsh) to the cell membrane (1, 2, 6). Dsh then inhibits the kinase activity of the normally constitutively active glycogen synthase kinase-3β (GSK-3β) resulting in an increase in β-catenin levels. Stabilized β-catenin interacts with the transcription factor TCF/Lef1, forming a complex that appears in the nucleus and binds TCF/Lef1 target DNA elements to activate transcription (7, 8). Other experiments suggest that the adenomatous polyposis coli (APC) tumor suppressor gene also plays an important role in Wnt signaling by regulating β-catenin levels (9). APC is phosphorylated by GSK-3β, binds to β-catenin, and facilitates its degradation. Mutations in either APC or β-catenin have been associated with colon carcinomas and melanomas, suggesting these mutations contribute to the development of these types of cancer, implicating the Wnt pathway in tumorigenesis (1).
Although much has been learned about the Wnt signaling pathway over the past several years, only a few of the transcriptionally activated downstream components activated by Wnt have been characterized. Those that have been described cannot account for all of the diverse functions attributed to Wnt signaling. Among the candidate Wnt target genes are those encoding the nodal-related 3 gene, Xnr3, a member of the transforming growth factor (TGF)-β superfamily, and the homeobox genes, engrailed, goosecoid, twin (Xtwn), and siamois (2). A recent report also identifies c-myc as a target gene of the Wnt signaling pathway (10).
To identify additional downstream genes in the Wnt signaling pathway that are relevant to the transformed cell phenotype, we used a PCR-based cDNA subtraction strategy, suppression subtractive hybridization (SSH) (11), using RNA isolated from C57MG mouse mammary epithelial cells and C57MG cells stably transformed by a Wnt-1 retrovirus. Overexpression of Wnt-1 in this cell line is sufficient to induce a partially transformed phenotype, characterized by elongated and refractile cells that lose contact inhibition and form a multilayered array (12, 13). We reasoned that genes differentially expressed between these two cell lines might contribute to the transformed phenotype.
In this paper, we describe the cloning and characterization of two genes up-regulated in Wnt-1 transformed cells, WISP-1 and WISP-2, and a third related gene, WISP-3. The WISP genes are members of the CCN family of growth factors, which includes connective tissue growth factor (CTGF), Cyr61, and nov, a family not previously linked to Wnt signaling.
MATERIALS AND METHODS
SSH.
SSH was performed by using the PCR-Select cDNA Subtraction Kit (CLONTECH). Tester double-stranded cDNA was synthesized from 2 μg of poly(A)+ RNA isolated from the C57MG/Wnt-1 cell line and driver cDNA from 2 μg of poly(A)+ RNA from the parent C57MG cells. The subtracted cDNA library was subcloned into a pGEM-T vector for further analysis.
cDNA Library Screening.
Clones encoding full-length mouse WISP-1 were isolated by screening a λgt10 mouse embryo cDNA library (CLONTECH) with a 70-bp probe from the original partial clone 568 sequence corresponding to amino acids 128–169. Clones encoding full-length human WISP-1 were isolated by screening λgt10 lung and fetal kidney cDNA libraries with the same probe at low stringency. Clones encoding full-length mouse and human WISP-2 were isolated by screening a C57MG/Wnt-1 or human fetal lung cDNA library with a probe corresponding to nucleotides 1463–1512. Full-length cDNAs encoding WISP-3 were cloned from human bone marrow and fetal kidney libraries.
Expression of Human WISP RNA.
PCR amplification of first-strand cDNA was performed with human Multiple Tissue cDNA panels (CLONTECH) and 300 μM of each dNTP at 94°C for 1 sec, 62°C for 30 sec, 72°C for 1 min, for 22–32 cycles. WISP and glyceraldehyde-3-phosphate dehydrogenase primer sequences are available on request.
In Situ Hybridization.
33P-labeled sense and antisense riboprobes were transcribed from an 897-bp PCR product corresponding to nucleotides 601–1440 of mouse WISP-1 or a 294-bp PCR product corresponding to nucleotides 82–375 of mouse WISP-2. All tissues were processed as described (40).
Radiation Hybrid Mapping.
Genomic DNA from each hybrid in the Stanford G3 and Genebridge4 Radiation Hybrid Panels (Research Genetics, Huntsville, AL) and human and hamster control DNAs were PCR-amplified, and the results were submitted to the Stanford or Massachusetts Institute of Technology web servers.
Cell Lines, Tumors, and Mucosa Specimens.
Tissue specimens were obtained from the Department of Pathology (University of Pittsburgh) for patients undergoing colon resection and from the University of Leeds, United Kingdom. Genomic DNA was isolated (Qiagen) from the pooled blood of 10 normal human donors, surgical specimens, and the following ATCC human cell lines: SW480, COLO 320DM, HT-29, WiDr, and SW403 (colon adenocarcinomas), SW620 (lymph node metastasis, colon adenocarcinoma), HCT 116 (colon carcinoma), SK-CO-1 (colon adenocarcinoma, ascites), and HM7 (a variant of ATCC colon adenocarcinoma cell line LS 174T). DNA concentration was determined by using Hoechst dye 33258 intercalation fluorimetry. Total RNA was prepared by homogenization in 7 M GuSCN followed by centrifugation over CsCl cushions or prepared by using RNAzol.
Gene Amplification and RNA Expression Analysis.
Relative gene amplification and RNA expression of WISPs and c-myc in the cell lines, colorectal tumors, and normal mucosa were determined by quantitative PCR. Gene-specific primers and fluorogenic probes (sequences available on request) were designed and used to amplify and quantitate the genes. The relative gene copy number was derived by using the formula 2(Δct) where ΔCt represents the difference in amplification cycles required to detect the WISP genes in peripheral blood lymphocyte DNA compared with colon tumor DNA or colon tumor RNA compared with normal mucosal RNA. The ∂-method was used for calculation of the SE of the gene copy number or RNA expression level. The WISP-specific signal was normalized to that of the glyceraldehyde-3-phosphate dehydrogenase housekeeping gene. All TaqMan assay reagents were obtained from Perkin–Elmer Applied Biosystems.
RESULTS
Isolation of WISP-1 and WISP-2 by SSH.
To identify Wnt-1-inducible genes, we used the technique of SSH using the mouse mammary epithelial cell line C57MG and C57MG cells that stably express Wnt-1 (11). Candidate differentially expressed cDNAs (1,384 total) were sequenced. Thirty-nine percent of the sequences matched known genes or homologues, 32% matched expressed sequence tags, and 29% had no match. To confirm that the transcript was differentially expressed, semiquantitative reverse transcription–PCR and Northern analysis were performed by using mRNA from the C57MG and C57MG/Wnt-1 cells.
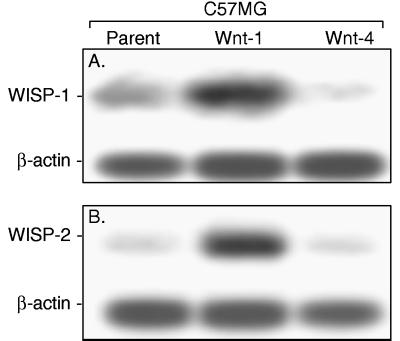
Two of the cDNAs, WISP-1 and WISP-2, were differentially expressed, being induced in the C57MG/Wnt-1 cell line, but not in the parent C57MG cells or C57MG cells overexpressing Wnt-4 (Fig. 1 A and B). Wnt-4, unlike Wnt-1, does not induce the morphological transformation of C57MG cells and has no effect on β-catenin levels (13, 14). Expression of WISP-1 was up-regulated approximately 3-fold in the C57MG/Wnt-1 cell line and WISP-2 by approximately 5-fold by both Northern analysis and reverse transcription–PCR.
Figure 1.
WISP-1 and WISP-2 are induced by Wnt-1, but not Wnt-4, expression in C57MG cells. Northern analysis of WISP-1 (A) and WISP-2 (B) expression in C57MG, C57MG/Wnt-1, and C57MG/Wnt-4 cells. Poly(A)+ RNA (2 μg) was subjected to Northern blot analysis and hybridized with a 70-bp mouse WISP-1-specific probe (amino acids 278–300) or a 190-bp WISP-2-specific probe (nucleotides 1438–1627) in the 3′ untranslated region. Blots were rehybridized with human β-actin probe.
An independent, but similar, system was used to examine WISP expression after Wnt-1 induction. C57MG cells expressing the Wnt-1 gene under the control of a tetracycline-repressible promoter produce low amounts of Wnt-1 in the repressed state but show a strong induction of Wnt-1 mRNA and protein within 24 hr after tetracycline removal (8). The levels of Wnt-1 and WISP RNA isolated from these cells at various times after tetracycline removal were assessed by quantitative PCR. Strong induction of Wnt-1 mRNA was seen as early as 10 hr after tetracycline removal. Induction of WISP mRNA (2- to 6-fold) was seen at 48 and 72 hr (data not shown). These data support our previous observations that show that WISP induction is correlated with Wnt-1 expression. Because the induction is slow, occurring after approximately 48 hr, the induction of WISPs may be an indirect response to Wnt-1 signaling.
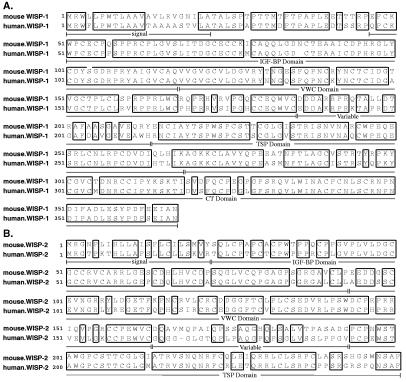
cDNA clones of human WISP-1 were isolated and the sequence compared with mouse WISP-1. The cDNA sequences of mouse and human WISP-1 were 1,766 and 2,830 bp in length, respectively, and encode proteins of 367 aa, with predicted relative molecular masses of ≈40,000 (Mr 40 K). Both have hydrophobic N-terminal signal sequences, 38 conserved cysteine residues, and four potential N-linked glycosylation sites and are 84% identical (Fig. 2A).
Figure 2.
Encoded amino acid sequence alignment of mouse and human WISP-1 (A) and mouse and human WISP-2 (B). The potential signal sequence, insulin-like growth factor-binding protein (IGF-BP), VWC, thrombospondin (TSP), and C-terminal (CT) domains are underlined.
Full-length cDNA clones of mouse and human WISP-2 were 1,734 and 1,293 bp in length, respectively, and encode proteins of 251 and 250 aa, respectively, with predicted relative molecular masses of ≈27,000 (Mr 27 K) (Fig. 2B). Mouse and human WISP-2 are 73% identical. Human WISP-2 has no potential N-linked glycosylation sites, and mouse WISP-2 has one at position 197. WISP-2 has 28 cysteine residues that are conserved among the 38 cysteines found in WISP-1.
Identification of WISP-3.
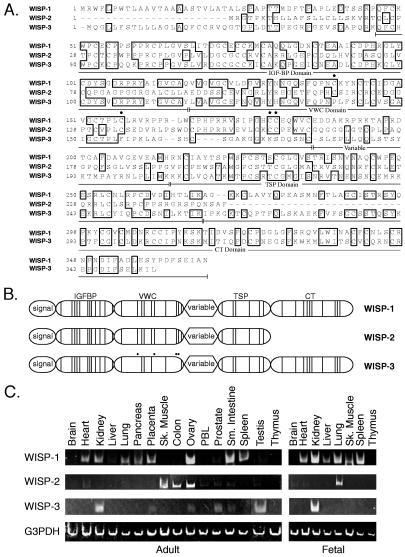
To search for related proteins, we screened expressed sequence tag (EST) databases with the WISP-1 protein sequence and identified several ESTs as potentially related sequences. We identified a homologous protein that we have called WISP-3. A full-length human WISP-3 cDNA of 1,371 bp was isolated corresponding to those ESTs that encode a 354-aa protein with a predicted molecular mass of 39,293. WISP-3 has two potential N-linked glycosylation sites and 36 cysteine residues. An alignment of the three human WISP proteins shows that WISP-1 and WISP-3 are the most similar (42% identity), whereas WISP-2 has 37% identity with WISP-1 and 32% identity with WISP-3 (Fig. 3A).
Figure 3.
(A) Encoded amino acid sequence alignment of human WISPs. The cysteine residues of WISP-1 and WISP-2 that are not present in WISP-3 are indicated with a dot. (B) Schematic representation of the WISP proteins showing the domain structure and cysteine residues (vertical lines). The four cysteine residues in the VWC domain that are absent in WISP-3 are indicated with a dot. (C) Expression of WISP mRNA in human tissues. PCR was performed on human multiple-tissue cDNA panels (CLONTECH) from the indicated adult and fetal tissues.
WISPs Are Homologous to the CTGF Family of Proteins.
Human WISP-1, WISP-2, and WISP-3 are novel sequences; however, mouse WISP-1 is the same as the recently identified Elm1 gene. Elm1 is expressed in low, but not high, metastatic mouse melanoma cells, and suppresses the in vivo growth and metastatic potential of K-1735 mouse melanoma cells (15). Human and mouse WISP-2 are homologous to the recently described rat gene, rCop-1 (16). Significant homology (36–44%) was seen to the CCN family of growth factors. This family includes three members, CTGF, Cyr61, and the protooncogene nov. CTGF is a chemotactic and mitogenic factor for fibroblasts that is implicated in wound healing and fibrotic disorders and is induced by TGF-β (17). Cyr61 is an extracellular matrix signaling molecule that promotes cell adhesion, proliferation, migration, angiogenesis, and tumor growth (18, 19). nov (nephroblastoma overexpressed) is an immediate early gene associated with quiescence and found altered in Wilms tumors (20). The proteins of the CCN family share functional, but not sequence, similarity to Wnt-1. All are secreted, cysteine-rich heparin binding glycoproteins that associate with the cell surface and extracellular matrix.
WISP proteins exhibit the modular architecture of the CCN family, characterized by four conserved cysteine-rich domains (Fig. 3B) (21). The N-terminal domain, which includes the first 12 cysteine residues, contains a consensus sequence (GCGCCXXC) conserved in most insulin-like growth factor (IGF)-binding proteins (BP). This sequence is conserved in WISP-2 and WISP-3, whereas WISP-1 has a glutamine in the third position instead of a glycine. CTGF recently has been shown to specifically bind IGF (22) and a truncated nov protein lacking the IGF-BP domain is oncogenic (23). The von Willebrand factor type C module (VWC), also found in certain collagens and mucins, covers the next 10 cysteine residues, and is thought to participate in protein complex formation and oligomerization (24). The VWC domain of WISP-3 differs from all CCN family members described previously, in that it contains only six of the 10 cysteine residues (Fig. 3 A and B). A short variable region follows the VWC domain. The third module, the thrombospondin (TSP) domain is involved in binding to sulfated glycoconjugates and contains six cysteine residues and a conserved WSxCSxxCG motif first identified in thrombospondin (25). The C-terminal (CT) module containing the remaining 10 cysteines is thought to be involved in dimerization and receptor binding (26). The CT domain is present in all CCN family members described to date but is absent in WISP-2 (Fig. 3 A and B). The existence of a putative signal sequence and the absence of a transmembrane domain suggest that WISPs are secreted proteins, an observation supported by an analysis of their expression and secretion from mammalian cell and baculovirus cultures (data not shown).
Expression of WISP mRNA in Human Tissues.
Tissue-specific expression of human WISPs was characterized by PCR analysis on adult and fetal multiple tissue cDNA panels. WISP-1 expression was seen in the adult heart, kidney, lung, pancreas, placenta, ovary, small intestine, and spleen (Fig. 3C). Little or no expression was detected in the brain, liver, skeletal muscle, colon, peripheral blood leukocytes, prostate, testis, or thymus. WISP-2 had a more restricted tissue expression and was detected in adult skeletal muscle, colon, ovary, and fetal lung. Predominant expression of WISP-3 was seen in adult kidney and testis and fetal kidney. Lower levels of WISP-3 expression were detected in placenta, ovary, prostate, and small intestine.
In Situ Localization of WISP-1 and WISP-2.
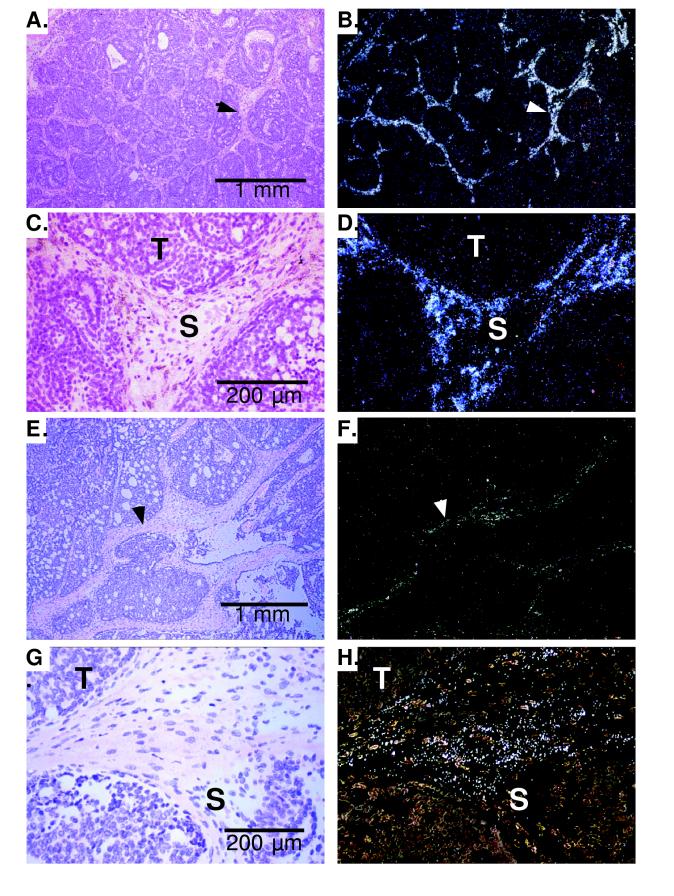
Expression of WISP-1 and WISP-2 was assessed by in situ hybridization in mammary tumors from Wnt-1 transgenic mice. Strong expression of WISP-1 was observed in stromal fibroblasts lying within the fibrovascular tumor stroma (Fig. 4 A–D). However, low-level WISP-1 expression also was observed focally within tumor cells (data not shown). No expression was observed in normal breast. Like WISP-1, WISP-2 expression also was seen in the tumor stroma in breast tumors from Wnt-1 transgenic animals (Fig. 4 E–H). However, WISP-2 expression in the stroma was in spindle-shaped cells adjacent to capillary vessels, whereas the predominant cell type expressing WISP-1 was the stromal fibroblasts.
Figure 4.
(A, C, E, and G) Representative hematoxylin/eosin-stained images from breast tumors in Wnt-1 transgenic mice. The corresponding dark-field images showing WISP-1 expression are shown in B and D. The tumor is a moderately well-differentiated adenocarcinoma showing evidence of adenoid cystic change. At low power (A and B), expression of WISP-1 is seen in the delicate branching fibrovascular tumor stroma (arrowhead). At higher magnification, expression is seen in the stromal(s) fibroblasts (C and D), and tumor cells are negative. Focal expression of WISP-1, however, was observed in tumor cells in some areas. Images of WISP-2 expression are shown in E–H. At low power (E and F), expression of WISP-2 is seen in cells lying within the fibrovascular tumor stroma. At higher magnification, these cells appeared to be adjacent to capillary vessels whereas tumor cells are negative (G and H).
Chromosome Localization of the WISP Genes.
The chromosomal location of the human WISP genes was determined by radiation hybrid mapping panels. WISP-1 is approximately 3.48 cR from the meiotic marker AFM259xc5 [logarithm of odds (lod) score 16.31] on chromosome 8q24.1 to 8q24.3, in the same region as the human locus of the novH family member (27) and roughly 4 Mbs distal to c-myc (28). Preliminary fine mapping indicates that WISP-1 is located near D8S1712 STS. WISP-2 is linked to the marker SHGC-33922 (lod = 1,000) on chromosome 20q12–20q13.1. Human WISP-3 mapped to chromosome 6q22–6q23 and is linked to the marker AFM211ze5 (lod = 1,000). WISP-3 is approximately 18 Mbs proximal to CTGF and 23 Mbs proximal to the human cellular oncogene MYB (27, 29).
Amplification and Aberrant Expression of WISPs in Human Colon Tumors.
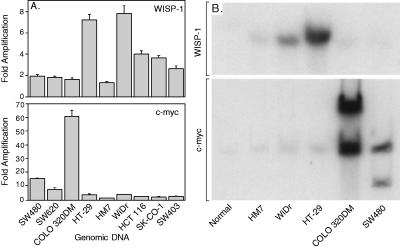
Amplification of protooncogenes is seen in many human tumors and has etiological and prognostic significance. For example, in a variety of tumor types, c-myc amplification has been associated with malignant progression and poor prognosis (30). Because WISP-1 resides in the same general chromosomal location (8q24) as c-myc, we asked whether it was a target of gene amplification, and, if so, whether this amplification was independent of the c-myc locus. Genomic DNA from human colon cancer cell lines was assessed by quantitative PCR and Southern blot analysis. (Fig. 5 A and B). Both methods detected similar degrees of WISP-1 amplification. Most cell lines showed significant (2- to 4-fold) amplification, with the HT-29 and WiDr cell lines demonstrating an 8-fold increase. Significantly, the pattern of amplification observed did not correlate with that observed for c-myc, indicating that the c-myc gene is not part of the amplicon that involves the WISP-1 locus.
Figure 5.
Amplification of WISP-1 genomic DNA in colon cancer cell lines. (A) Amplification in cell line DNA was determined by quantitative PCR. (B) Southern blots containing genomic DNA (10 μg) digested with EcoRI (WISP-1) or XbaI (c-myc) were hybridized with a 100-bp human WISP-1 probe (amino acids 186–219) or a human c-myc probe (located at bp 1901–2000). The WISP and myc genes are detected in normal human genomic DNA after a longer film exposure.
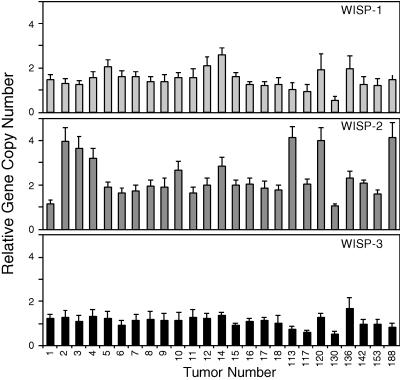
We next examined whether the WISP genes were amplified in a panel of 25 primary human colon adenocarcinomas. The relative WISP gene copy number in each colon tumor DNA was compared with pooled normal DNA from 10 donors by quantitative PCR (Fig. 6). The copy number of WISP-1 and WISP-2 was significantly greater than one, approximately 2-fold for WISP-1 in about 60% of the tumors and 2- to 4-fold for WISP-2 in 92% of the tumors (P < 0.001 for each). The copy number for WISP-3 was indistinguishable from one (P = 0.166). In addition, the copy number of WISP-2 was significantly higher than that of WISP-1 (P < 0.001).
Figure 6.
Genomic amplification of WISP genes in human colon tumors. The relative gene copy number of the WISP genes in 25 adenocarcinomas was assayed by quantitative PCR, by comparing DNA from primary human tumors with pooled DNA from 10 healthy donors. The data are means ± SEM from one experiment done in triplicate. The experiment was repeated at least three times.
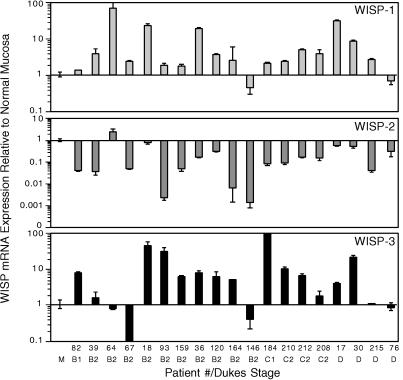
The levels of WISP transcripts in RNA isolated from 19 adenocarcinomas and their matched normal mucosa were assessed by quantitative PCR (Fig. 7). The level of WISP-1 RNA present in tumor tissue varied but was significantly increased (2- to >25-fold) in 84% (16/19) of the human colon tumors examined compared with normal adjacent mucosa. Four of 19 tumors showed greater than 10-fold overexpression. In contrast, in 79% (15/19) of the tumors examined, WISP-2 RNA expression was significantly lower in the tumor than the mucosa. Similar to WISP-1, WISP-3 RNA was overexpressed in 63% (12/19) of the colon tumors compared with the normal mucosa. The amount of overexpression of WISP-3 ranged from 4- to >40-fold.
Figure 7.
WISP RNA expression in primary human colon tumors relative to expression in normal mucosa from the same patient. Expression of WISP mRNA in 19 adenocarcinomas was assayed by quantitative PCR. The Dukes stage of the tumor is listed under the sample number. The data are means ± SEM from one experiment done in triplicate. The experiment was repeated at least twice.
DISCUSSION
One approach to understanding the molecular basis of cancer is to identify differences in gene expression between cancer cells and normal cells. Strategies based on assumptions that steady-state mRNA levels will differ between normal and malignant cells have been used to clone differentially expressed genes (31). We have used a PCR-based selection strategy, SSH, to identify genes selectively expressed in C57MG mouse mammary epithelial cells transformed by Wnt-1.
Three of the genes isolated, WISP-1, WISP-2, and WISP-3, are members of the CCN family of growth factors, which includes CTGF, Cyr61, and nov, a family not previously linked to Wnt signaling.
Two independent experimental systems demonstrated that WISP induction was associated with the expression of Wnt-1. The first was C57MG cells infected with a Wnt-1 retroviral vector or C57MG cells expressing Wnt-1 under the control of a tetracyline-repressible promoter, and the second was in Wnt-1 transgenic mice, where breast tissue expresses Wnt-1, whereas normal breast tissue does not. No WISP RNA expression was detected in mammary tumors induced by polyoma virus middle T antigen (data not shown). These data suggest a link between Wnt-1 and WISPs in that in these two situations, WISP induction was correlated with Wnt-1 expression.
It is not clear whether the WISPs are directly or indirectly induced by the downstream components of the Wnt-1 signaling pathway (i.e., β-catenin-TCF-1/Lef1). The increased levels of WISP RNA were measured in Wnt-1-transformed cells, hours or days after Wnt-1 transformation. Thus, WISP expression could result from Wnt-1 signaling directly through β-catenin transcription factor regulation or alternatively through Wnt-1 signaling turning on a transcription factor, which in turn regulates WISPs.
The WISPs define an additional subfamily of the CCN family of growth factors. One striking difference observed in the protein sequence of WISP-2 is the absence of a CT domain, which is present in CTGF, Cyr61, nov, WISP-1, and WISP-3. This domain is thought to be involved in receptor binding and dimerization. Growth factors, such as TGF-β, platelet-derived growth factor, and nerve growth factor, which contain a cystine knot motif exist as dimers (32). It is tempting to speculate that WISP-1 and WISP-3 may exist as dimers, whereas WISP-2 exists as a monomer. If the CT domain is also important for receptor binding, WISP-2 may bind its receptor through a different region of the molecule than the other CCN family members. No specific receptors have been identified for CTGF or nov. A recent report has shown that integrin αvβ3 serves as an adhesion receptor for Cyr61 (33).
The strong expression of WISP-1 and WISP-2 in cells lying within the fibrovascular tumor stroma in breast tumors from Wnt-1 transgenic animals is consistent with previous observations that transcripts for the related CTGF gene are primarily expressed in the fibrous stroma of mammary tumors (34). Epithelial cells are thought to control the proliferation of connective tissue stroma in mammary tumors by a cascade of growth factor signals similar to that controlling connective tissue formation during wound repair. It has been proposed that mammary tumor cells or inflammatory cells at the tumor interstitial interface secrete TGF-β1, which is the stimulus for stromal proliferation (34). TGF-β1 is secreted by a large percentage of malignant breast tumors and may be one of the growth factors that stimulates the production of CTGF and WISPs in the stroma.
It was of interest that WISP-1 and WISP-2 expression was observed in the stromal cells that surrounded the tumor cells (epithelial cells) in the Wnt-1 transgenic mouse sections of breast tissue. This finding suggests that paracrine signaling could occur in which the stromal cells could supply WISP-1 and WISP-2 to regulate tumor cell growth on the WISP extracellular matrix. Stromal cell-derived factors in the extracellular matrix have been postulated to play a role in tumor cell migration and proliferation (35). The localization of WISP-1 and WISP-2 in the stromal cells of breast tumors supports this paracrine model.
An analysis of WISP-1 gene amplification and expression in human colon tumors showed a correlation between DNA amplification and overexpression, whereas overexpression of WISP-3 RNA was seen in the absence of DNA amplification. In contrast, WISP-2 DNA was amplified in the colon tumors, but its mRNA expression was significantly reduced in the majority of tumors compared with the expression in normal colonic mucosa from the same patient. The gene for human WISP-2 was localized to chromosome 20q12–20q13, at a region frequently amplified and associated with poor prognosis in node negative breast cancer and many colon cancers, suggesting the existence of one or more oncogenes at this locus (36–38). Because the center of the 20q13 amplicon has not yet been identified, it is possible that the apparent amplification observed for WISP-2 may be caused by another gene in this amplicon.
A recent manuscript on rCop-1, the rat orthologue of WISP-2, describes the loss of expression of this gene after cell transformation, suggesting it may be a negative regulator of growth in cell lines (16). Although the mechanism by which WISP-2 RNA expression is down-regulated during malignant transformation is unknown, the reduced expression of WISP-2 in colon tumors and cell lines suggests that it may function as a tumor suppressor. These results show that the WISP genes are aberrantly expressed in colon cancer and suggest that their altered expression may confer selective growth advantage to the tumor.
Members of the Wnt signaling pathway have been implicated in the pathogenesis of colon cancer, breast cancer, and melanoma, including the tumor suppressor gene adenomatous polyposis coli and β-catenin (39). Mutations in specific regions of either gene can cause the stabilization and accumulation of cytoplasmic β-catenin, which presumably contributes to human carcinogenesis through the activation of target genes such as the WISPs. Although the mechanism by which Wnt-1 transforms cells and induces tumorigenesis is unknown, the identification of WISPs as genes that may be regulated downstream of Wnt-1 in C57MG cells suggests they could be important mediators of Wnt-1 transformation. The amplification and altered expression patterns of the WISPs in human colon tumors may indicate an important role for these genes in tumor development.
Acknowledgments
We thank the DNA synthesis group for oligonucleotide synthesis, T. Baker for technical assistance, P. Dowd for radiation hybrid mapping, K. Willert and R. Nusse for the tet-repressible C57MG/Wnt-1 cells, V. Dixit for discussions, and D. Wood and A. Bruce for artwork.
ABBREVIATIONS
- TGF
transforming growth factor
- CTGF
connective tissue growth factor
- SSH
suppression subtractive hybridization
- VWC
von Willebrand factor type C module
Footnotes
References
- 1.Cadigan K M, Nusse R. Genes Dev. 1997;11:3286–3305. doi: 10.1101/gad.11.24.3286. [DOI] [PubMed] [Google Scholar]
- 2.Dale T C. Biochem J. 1998;329:209–223. doi: 10.1042/bj3290209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nusse R, Varmus H E. Cell. 1982;31:99–109. doi: 10.1016/0092-8674(82)90409-3. [DOI] [PubMed] [Google Scholar]
- 4.van Ooyen A, Nusse R. Cell. 1984;39:233–240. doi: 10.1016/0092-8674(84)90209-5. [DOI] [PubMed] [Google Scholar]
- 5.Tsukamoto A S, Grosschedl R, Guzman R C, Parslow T, Varmus H E. Cell. 1988;55:619–625. doi: 10.1016/0092-8674(88)90220-6. [DOI] [PubMed] [Google Scholar]
- 6.Brown J D, Moon R T. Curr Opin Cell Biol. 1998;10:182–187. doi: 10.1016/s0955-0674(98)80140-3. [DOI] [PubMed] [Google Scholar]
- 7.Molenaar M, van de Wetering M, Oosterwegel M, Peterson-Maduro J, Godsave S, Korinek V, Roose J, Destree O, Clevers H. Cell. 1996;86:391–399. doi: 10.1016/s0092-8674(00)80112-9. [DOI] [PubMed] [Google Scholar]
- 8.Korinek V, Barker N, Willert K, Molenaar M, Roose J, Wagenaar G, Markman M, Lamers W, Destree O, Clevers H. Mol Cell Biol. 1998;18:1248–1256. doi: 10.1128/mcb.18.3.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Munemitsu S, Albert I, Souza B, Rubinfeld B, Polakis P. Proc Natl Acad Sci USA. 1995;92:3046–3050. doi: 10.1073/pnas.92.7.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He T C, Sparks A B, Rago C, Hermeking H, Zawel L, da Costa L T, Morin P J, Vogelstein B, Kinzler K W. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 11.Diatchenko L, Lau Y F, Campbell A P, Chenchik A, Moqadam F, Huang B, Lukyanov S, Lukyanov K, Gurskaya N, Sverdlov E D, Siebert P D. Proc Natl Acad Sci USA. 1996;93:6025–6030. doi: 10.1073/pnas.93.12.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown A M, Wildin R S, Prendergast T J, Varmus H E. Cell. 1986;46:1001–1009. doi: 10.1016/0092-8674(86)90699-9. [DOI] [PubMed] [Google Scholar]
- 13.Wong G T, Gavin B J, McMahon A P. Mol Cell Biol. 1994;14:6278–6286. doi: 10.1128/mcb.14.9.6278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shimizu H, Julius M A, Giarre M, Zheng Z, Brown A M, Kitajewski J. Cell Growth Differ. 1997;8:1349–1358. [PubMed] [Google Scholar]
- 15.Hashimoto Y, Shindo-Okada N, Tani M, Nagamachi Y, Takeuchi K, Shiroishi T, Toma H, Yokota J. J Exp Med. 1998;187:289–296. doi: 10.1084/jem.187.3.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang R, Averboukh L, Zhu W, Zhang H, Jo H, Dempsey P J, Coffey R J, Pardee A B, Liang P. Mol Cell Biol. 1998;18:6131–6141. doi: 10.1128/mcb.18.10.6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grotendorst G R. Cytokine Growth Factor Rev. 1997;8:171–179. doi: 10.1016/s1359-6101(97)00010-5. [DOI] [PubMed] [Google Scholar]
- 18.Kireeva M L, Mo F E, Yang G P, Lau L F. Mol Cell Biol. 1996;16:1326–1334. doi: 10.1128/mcb.16.4.1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Babic A M, Kireeva M L, Kolesnikova T V, Lau L F. Proc Natl Acad Sci USA. 1998;95:6355–6360. doi: 10.1073/pnas.95.11.6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martinerie C, Huff V, Joubert I, Badzioch M, Saunders G, Strong L, Perbal B. Oncogene. 1994;9:2729–2732. [PubMed] [Google Scholar]
- 21.Bork P. FEBS Lett. 1993;327:125–130. doi: 10.1016/0014-5793(93)80155-n. [DOI] [PubMed] [Google Scholar]
- 22.Kim H S, Nagalla S R, Oh Y, Wilson E, Roberts C T, Jr, Rosenfeld R G. Proc Natl Acad Sci USA. 1997;94:12981–12986. doi: 10.1073/pnas.94.24.12981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Joliot V, Martinerie C, Dambrine G, Plassiart G, Brisac M, Crochet J, Perbal B. Mol Cell Biol. 1992;12:10–21. doi: 10.1128/mcb.12.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mancuso D J, Tuley E A, Westfield L A, Worrall N K, Shelton-Inloes B B, Sorace J M, Alevy Y G, Sadler J E. J Biol Chem. 1989;264:19514–19527. [PubMed] [Google Scholar]
- 25.Holt G D, Pangburn M K, Ginsburg V. J Biol Chem. 1990;265:2852–2855. [PubMed] [Google Scholar]
- 26.Voorberg J, Fontijn R, Calafat J, Janssen H, van Mourik J A, Pannekoek H. J Cell Biol. 1991;113:195–205. doi: 10.1083/jcb.113.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martinerie C, Viegas-Pequignot E, Guenard I, Dutrillaux B, Nguyen V C, Bernheim A, Perbal B. Oncogene. 1992;7:2529–2534. [PubMed] [Google Scholar]
- 28.Takahashi E, Hori T, O’Connell P, Leppert M, White R. Cytogenet Cell Genet. 1991;57:109–111. doi: 10.1159/000133124. [DOI] [PubMed] [Google Scholar]
- 29.Meese E, Meltzer P S, Witkowski C M, Trent J M. Genes Chromosomes Cancer. 1989;1:88–94. doi: 10.1002/gcc.2870010114. [DOI] [PubMed] [Google Scholar]
- 30.Garte S J. Crit Rev Oncog. 1993;4:435–449. [PubMed] [Google Scholar]
- 31.Zhang L, Zhou W, Velculescu V E, Kern S E, Hruban R H, Hamilton S R, Vogelstein B, Kinzler K W. Science. 1997;276:1268–1272. doi: 10.1126/science.276.5316.1268. [DOI] [PubMed] [Google Scholar]
- 32.Sun P D, Davies D R. Annu Rev Biophys Biomol Struct. 1995;24:269–291. doi: 10.1146/annurev.bb.24.060195.001413. [DOI] [PubMed] [Google Scholar]
- 33.Kireeva M L, Lam S C T, Lau L F. J Biol Chem. 1998;273:3090–3096. doi: 10.1074/jbc.273.5.3090. [DOI] [PubMed] [Google Scholar]
- 34.Frazier K S, Grotendorst G R. Int J Biochem Cell Biol. 1997;29:153–161. doi: 10.1016/s1357-2725(96)00127-6. [DOI] [PubMed] [Google Scholar]
- 35.Wernert N. Virchows Arch. 1997;430:433–443. doi: 10.1007/s004280050053. [DOI] [PubMed] [Google Scholar]
- 36.Tanner M M, Tirkkonen M, Kallioniemi A, Collins C, Stokke T, Karhu R, Kowbel D, Shadravan F, Hintz M, Kuo W L, et al. Cancer Res. 1994;54:4257–4260. [PubMed] [Google Scholar]
- 37.Brinkmann U, Gallo M, Polymeropoulos M H, Pastan I. Genome Res. 1996;6:187–194. doi: 10.1101/gr.6.3.187. [DOI] [PubMed] [Google Scholar]
- 38.Bischoff J R, Anderson L, Zhu Y, Mossie K, Ng L, Souza B, Schryver B, Flanagan P, Clairvoyant F, Ginther C, et al. EMBO J. 1998;17:3052–3065. doi: 10.1093/emboj/17.11.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morin P J, Sparks A B, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler K W. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 40.Lu L H, Gillett N. Cell Vision. 1994;1:169–176. [Google Scholar]