Abstract
When lipid synthesis is limited in HepG2 cells, apoprotein B100 (apoB100) is not secreted but rapidly degraded by the ubiquitin-proteasome pathway. To investigate apoB100 biosynthesis and secretion further, the physical and functional states of apoB100 destined for either degradation or lipoprotein assembly were studied under conditions in which lipid synthesis, proteasomal activity, and microsomal triglyceride transfer protein (MTP) lipid-transfer activity were varied. Cells were pretreated with a proteasomal inhibitor (which remained with the cells throughout the experiment) and radiolabeled for 15 min. During the chase period, labeled apoB100 remained associated with the microsomes. Furthermore, by crosslinking sec61β to apoB100, we showed that apoB100 remained close to the translocon at the same time apoB100–ubiquitin conjugates could be detected. When lipid synthesis and lipoprotein assembly/secretion were stimulated by adding oleic acid (OA) to the chase medium, apoB100 was deubiquitinated, and its interaction with sec61β was disrupted, signifying completion of translocation concomitant with the formation of lipoprotein particles. MTP participates in apoB100 translocation and lipoprotein assembly. In the presence of OA, when MTP lipid-transfer activity was inhibited at the end of pulse labeling, apoB100 secretion was abolished. In contrast, when the labeled apoB100 was allowed to accumulate in the cell for 60 min before adding OA and the inhibitor, apoB100 lipidation and secretion were no longer impaired. Overall, the data imply that during most of its association with the endoplasmic reticulum, apoB100 is close to or within the translocon and is accessible to both the ubiquitin-proteasome and lipoprotein-assembly pathways. Furthermore, MTP lipid-transfer activity seems to be necessary only for early translocation and lipidation events.
Apolipoprotein B100 (apoB100) is a 4,536-aa polypeptide and is the major structural protein of the liver-derived very low density and low density lipoproteins. Hepatic lipoprotein assembly begins when apoB100 is cotranslationally translocated across the endoplasmic reticulum (ER) membrane (1) and interacts with the luminal microsomal triglyceride transfer protein (MTP; refs. 2–4). MTP catalyzes the initial transfer of lipid to nascent apoB100, with subsequent lipoprotein maturation steps occurring in the ER and possibly the Golgi apparatus (5, 6). Studies of cultured primary hepatocytes and transformed liver cells of human and nonhuman origin have established that significant control over apoB100 secretion can be exerted at the posttranslational level by the targeting of the nascent protein to degradation. This presecretory degradation is increased when the availability of lipid ligands of apoB100 is limited by inadequate levels of either lipid synthesis or MTP-mediated lipid-transfer activity (7, 8). In recent studies with the human hepatocarcinoma cell line HepG2, a standard model of lipoprotein metabolism, we and others have shown that most, if not all, of the degradation of apoB100 that occurs when lipid availability is limited is mediated by the ubiquitin-proteasome pathway (9–11).
Although the components of this pathway are cytosolic, a number of recent reports have shown that a variety of membrane-associated and secretory proteins can be targeted to the proteasome for degradation in eukaryotic cells (reviewed in refs. 12 and 13). The precise mechanism by which these proteins become substrates for a cytosolic protease has remained elusive. Based on results for major histocompatibility complex class I molecules, it has been hypothesized that a protein translocated into the ER can be fully “dislocated” back into the cytosol and subsequently attacked by the proteasome (14). The need for complete dislocation, which would require some form of reverse translocation, may not be general, given the finding that proteasomes can be found in association with the cytosolic face of the ER (15, 16). Thus, the initial attack of the proteasome could be directed against a protein domain that is or becomes exposed to the cytosol while other domains remain segregated by the ER membrane. Consistent with this possibility is the finding that two yeast ubiquitin-conjugating enzymes essential for ER-associated proteasomal degradation, Ubc6p and Ubc7p, can be localized to the ER. Ubc6p does so as an integral membrane protein (17), and Ubc7p does so by docking onto the membrane protein Cue1p (18).
The extremely large size of apoB100, its potential to pause or arrest during translocation (19, 20), as well as its multiple hydrophobic β-sheet domains (21), which would favor membrane interactions, led us to consider whether full translocation followed by complete dislocation was a likely path from the ER to the proteasome. Key elements of the degradation pathway can be on the cytosolic side of the ER membrane; this finding suggested that apoB100 that was incompletely translocated or partially dislocated could be the proximal substrate of the ubiquitin-proteasome machinery.
We hypothesized, therefore, that even after translation, the apoB100 polypeptide could remain in close proximity to the translocon. While in close proximity, it could be either targeted for degradation or assembled as a lipoprotein, depending on the availability of lipid ligands. This hypothesis was tested primarily by examining the cellular localization and functional state of apoB100 protected from degradation by inhibition of the proteasome. As described below, the results not only support the hypothesis but further suggest that apoB100 destined to become part of a lipoprotein has a prolonged interaction with the ER membrane; during this time, apoB100 becomes associated with its lipid ligands in a process that depends on MTP only at an early stage of the assembly process.
EXPERIMENTAL PROCEDURES
Materials.
Rabbit anti-human apoB100 and anti-human apoprotein AI (apoAI) antisera were from Calbiochem; mouse anti-human apoB100 antiserum was from Caltag (South San Francisco, CA). Rabbit anti-sec61β and anti-sec61α antisera were gifts from Tom A. Rapoport (Harvard University, Cambridge, MA; ref. 22) and Arthur E. Johnson (Texas A & M, College Station, TX). Rabbit anti-ubiquitin antiserum was from StressGen Biotechnologies (Victoria, Canada). Antisera and associated reagents for immunofluorescence studies are described below.
[35S]methionine/cysteine was from DuPont/NEN. Lactacystin was from the laboratory of E. J. Corey (Harvard University; ref. 23) and MG132 was from Calbiochem. Minimum essential medium (MEM), methionine-free medium, glutamine, and penicillin/streptomycin were from Life Technologies (Gaithersburg, MD). Fetal bovine serum and oleic acid (OA) were from Sigma.
Growth of HepG2 Cells.
HepG2 cells were grown as described (24). Briefly, cells were grown on collagen-coated plates in MEM containing 10% fetal bovine serum, 2 mM glutamine, and 100 μg/ml penicillin/streptomycin and were used at 70–90% confluence for all experiments.
Treatment and Metabolic Labeling of Cells.
For metabolic radiolabeling of proteins, cells were washed twice with PBS and preincubated in labeling medium (methionine-free MEM with 1% fetal bovine serum) for 60 min. Cells were then pulsed for 15 min with 100 μCi/ml [35S]methionine/cysteine in labeling medium, washed twice with chase medium (MEM with 1% fetal bovine serum, 2 mM methionine, and 0.6 mM cysteine), and then incubated in chase medium for indicated time periods. Lactacystin (10 μM) was added to cells 60 min before labeling; the equivalent volume of dimethyl sulfoxide was added to control cells. OA (0.8 mM) was added as a 5:1 molar complex with BSA (25).
Immunoprecipitation.
Immunoprecipitation of apoB100 or apoAI was performed as described (10). For conditioned medium, aliquots were removed from the culture wells, and protease inhibitors were added. For lysates, cells were washed twice with ice-cold PBS and solubilized overnight at 4°C in lysis buffer containing 130.5 mM NaCl, 4.35 mM EDTA, 43.5 mM Tris (pH 7.6), 62.5 mM sucrose, 1.37% Triton X-100, 0.5% deoxycholate, 0.087% SDS, and protease inhibitors. Cell lysates were clarified by centrifugation for 5 min at 3500 × g. Aliquots of lysate or conditioned medium were diluted 2- to 3-fold into NET buffer (150 mM NaCl/5 mM EDTA/50 mM Tris, pH 7.6/1% Triton X-100/0.1% SDS) and incubated with antiserum to human apoB100 or apoAI for 6–8 h at 4°C. Protein A-Sepharose CL-4B (Pharmacia) was added to a final concentration of 0.5% and incubation continued overnight at 4°C. Immunoprecipitated samples were washed three times with NET buffer and eluted by boiling for 4 min in sample buffer (125 mM Tris, pH 6.8/4% SDS/20% glycerol/10% 2-mercaptoethanol). Samples were analyzed by gel electrophoresis on a 3–17% gradient gel and quantified by a PhosphorImager (Molecular Dynamics).
ApoB100–ubiquitin conjugates were detected, and their identity was confirmed by sequential immunoprecipitation analysis with rabbit anti-ubiquitin and anti-apoB100 antisera, as described (10).
Cell Fractionation.
HepG2 cells were pretreated with 10 μM lactacystin for 1 h and labeled with [35S]methionine/cysteine for 15 min. After medium was removed, the cells were washed twice in chase medium and incubated in lactacystin-containing chase medium for 0, 20, 40, or 60 min. Cells were then disrupted by sonication. After low-speed centrifugation (10,000 × g, 10 min, 4°C) to remove nuclei and unbroken cells, the supernatant was fractionated by centrifugation at 100,000 × g for 1 h at 4°C. ApoB100 was then immunoprecipitated from the microsomal (pellet) and cytosolic (supernatant) fractions.
Crosslinking of ApoB100 and sec61β.
HepG2 cells were pretreated with a proteasome inhibitor (either MG132 or lactacystin at a final concentration of 10 μM) for 1 h and then labeled for 15 min by adding 100 μCi/ml of [35S]methionine/cysteine. The medium was removed, and the cells were washed twice with chase medium and then incubated in chase medium containing the proteasome inhibitor for 0, 20, 40, or 60 min in the presence or absence of OA (0.8 mM). At the end of each chase period, cells were washed with ice-cold PBS and harvested in crosslinking buffer (PBS/0.4% digitonin/0.5 mM EDTA). The permeable cell suspension was incubated on ice for 1 h with the homobifunctional chemical crosslinking reagent dithiobis(succinimidyl propionate) (final concentration of 0.25 mM added as a concentrated solution in dimethyl sulfoxide). The reaction was quenched by incubation in 20 mM Tris (pH 7.5) for 15 min. ApoB100–sec61β crosslinks were recovered by sequential immunoprecipitation; an aliquot of cell suspension was first immunoprecipitated with anti-sec61β antiserum. The immunoprecipitate was dissolved by boiling in sample buffer, diluted in NET, and immunoprecipitated with anti-apoB100 antiserum. The immunoprecipitate was analyzed by SDS/PAGE and fluorography. In preliminary experiments, verification that the resulting bands were apoB100 was obtained by western-blot analysis of selected samples with apoB100 antiserum and the Renaissance Western blot chemiluminescent reagent (DuPont/NEN).
Immunofluorescence of ApoB100.
Cells were fixed with 3% paraformaldehyde for 15 min on ice, made permeable with 0.1% Triton X-100 for 15 min on ice, and blocked with 1% BSA for 15 min at room temperature. The permeable cells were then incubated with the primary (specific) antibodies in 0.1% BSA for 1 h at room temperature. After washing, cells were incubated with secondary fluorescent-labeled anti-IgG in 0.1% BSA for 1 h at room temperature. Texas Red-conjugated goat anti-rabbit IgG and fluorescein isothiocyanate-conjugated goat anti-mouse IgG secondary antibodies were obtained from Jackson ImmunoResearch. For colocalization analysis, the primary antisera used were against apoB100, the ER resident protein SSRα (a gift from Chris Nicchitta, Duke University, Durham, NC), the 58-kDa Golgi-specific protein (Molecular Probes), and the intermediate compartment marker ERGIC53 (a gift from Hans-Peter Hauri, Biocentre of the University of Basel, Basel, Switzerland). Fluorescent images were collected by a Nikon Optiphot-2 microscope connected to a Sony Digital Photo camera.
MTP Inhibition Effects on ApoB100.
HepG2 cells were pretreated, radiolabeled, and chased under conditions described above. An inhibitor of the lipid-transfer activity of MTP (Pfizer CP-10447; ref. 26) was added to the medium at 100 μM either 60 min before, at the start of, or 60 min after labeling and maintained in the medium until cells were harvested. Aliquots of cell lysates containing equal amounts of trichloroacetic acid-precipitable radioactivity were used for immunoprecipitation. Immunoprecipitates were separated by SDS/PAGE, and the apoB100 bands were quantified by fluorography and densitometry.
RESULTS
ApoB100 Protected from Degradation by the Proteasome Is Associated Stably with Microsomes.
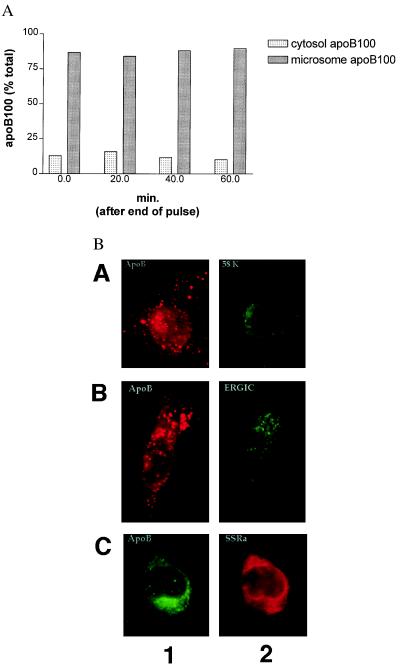
We reported (10) that in steady state, the majority of cellular apoB100 was associated with the microsomal fraction in fatty acid-deprived HepG2 cells treated with lactacystin. To determine whether this partitioning represents a stable association or a precursor state for a cytosolic pool that is the proximal substrate for degradation by the proteasome, a pulse–chase protocol was used. HepG2 cells were pretreated with 10 μM lactacystin for 1 h, labeled with [35S]methionine/cysteine for 15 min, and chased in lactacystin-containing medium for 0, 20, 40, or 60 min. Cell lysates were then separated by ultracentrifugation into cytosolic (supernatant) and microsomal (pellet) fractions, and apoB100 was immunoprecipitated from each. As shown in Fig. 1A, the percentage of labeled apoB100 associated with the microsomes was essentially unchanged throughout the chase period. From two separate experiments, this percentage was estimated to be ≈85%. This result was similar to those for two control secretory proteins, fibronectin and apoAI (data not shown), suggesting that the apoB100 found in the supernatant fraction was not a bona fide cytosolic pool but was protein either released by damaged microsomes or associated with small microsomes that failed to pellet. Overall, the results imply that when lipid synthesis is not stimulated and proteasomal degradation is blocked, metabolically labeled apoB100 stays associated with the microsome and is not transferred to the cytosol as a soluble protein.
Figure 1.
ApoB100 protected from degradation by the proteasome is associated stably with microsomes. (A) HepG2 cells were treated with lactacystin starting 30 min before pulse labeling with [35S]methionine/cysteine. At the end of the 15-min labeling period, the medium was changed to one that was radioisotope-free. At the indicated times in the chase period, cell lysates were collected and separated into supernatant (cytosol) and pellet (microsomes) fractions by ultracentrifugation. Labeled apoB100 in each fraction was measured by immunoprecipitation-SDS/PAGE analysis, and the percentage of distribution was calculated. The results shown are the averages of two separate experiments. (B) HepG2 cells were treated with lactacystin for 2 h and then subjected to double-label immunofluorescence analysis by using antisera to apoB100 and markers for the Golgi apparatus (58-kDa protein; row A), the intermediate compartment (ERGIC; row B), or the ER (SSRα, indicated by SSRa; row C).
ApoB100 that Accumulates After Proteasomes Are Inhibited Is Localized to the ER.
We employed immunofluorescence analysis to examine the cellular localization of proteasome-protected apoB100 further. Cells were treated with lactacystin for 2 h as above, and by double-labeling techniques, they were immunostained for apoB100 and SSRα, an ER-membrane protein (27), ERGIC53, an intermediate compartment protein (28), or the 58-kDa Golgi-specific protein (29). There was no crossreaction of the secondary fluorescent antibodies with the primary antibody to apoB100 and vice versa (data not shown). The apoB100 fluorescent signal overlapped with the ER-specific marker SSRα (Fig. 1B, row C), suggesting that, under these conditions, apoB100 remained associated with the ER. There are, however, punctate regions of apoB100 immunofluorescence that may represent apoB100 concentrated in extensions of the ER membrane (see Discussion). The considerably smaller fraction of punctate regions that overlaps with either the intermediate compartment or Golgi marker (Fig. 1B, rows A and B) may represent the limited post-ER transport of apoB100 that can occur at low levels of lipid synthesis (30). Overall, taken with the cell-fractionation experiments, the bulk of the microsomal pool most likely represents ER-associated apoB100 stabilized by proteasomal inhibition.
ER-Associated ApoB100 Is in Close Proximity to sec61β Before and During Lipoprotein Assembly.
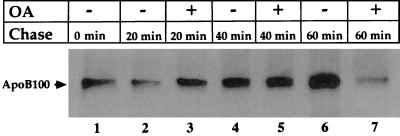
To define better the molecular nature of the interaction between the ER membrane and the apoB100 that accumulates on lactacystin treatment, we investigated whether this pool is associated with the translocon, a major protein of which is sec61β. HepG2 cells were pretreated with lactacystin, pulse-labeled as described in Fig. 1A, and chased in lactacystin-containing medium in the presence or absence of OA. Cell suspensions were placed on ice, treated with the crosslinking reagent dithiobis(succinimidyl propionate) or dimethyl sulfoxide (control), and sequentially immunoprecipitated with antisera against sec61β and anti-apoB100. SDS/PAGE analysis showed that when apoB100 was protected from proteasomal degradation, it assumed a stable (i.e., for at least 60 min) and intimate orientation relative to sec61β (Fig. 2, −OA lanes). When lipid synthesis and apoB100 secretion were stimulated by OA, the interaction with sec61β was not disrupted until after 40 min of chase (compare the 40 and 60 min −OA and +OA lanes), implying that most, if not all, of lipoprotein assembly occurred while apoB100 remained associated with the ER membrane in close proximity to the translocon. Similar results were obtained for the interaction between another translocon protein, sec61α, and apoB100 (data not shown).
Figure 2.
ER-associated apoB100 is in close proximity to the translocon protein sec61β before and during lipoprotein assembly. HepG2 cells were treated with lactacystin or MG132 starting 30 min before pulse labeling with [35S]methionine/cysteine. At the end of the 15-min labeling period, the medium was changed to one that was isotope-free and contained either OA complexed to BSA (+) or BSA alone (−). At the indicated times in the chase period, cells were subjected to dithiobis(succinimidyl propionate)-crosslinking and sequential immunoprecipitation analysis with sec61β antiserum followed by apoB100 antiserum to isolate the labeled apoB100 that was in close proximity to the translocon. The immunoprecipitates were resolved by SDS/PAGE, and the labeled apoB100 bands were visualized by fluorography.
The specificity of the interaction between sec61β and apoB100 was indicated by the characteristics of the final band. The appearance of the final band depended on the presence of the dithiobis(succinimidyl propionate) crosslinker. The final band was absent when anti-sec61β antibody was omitted from the first immunoprecipitation reaction (data not shown).
Proteasome-Protected ApoB100 Can Be Recruited to Lipoprotein Assembly and the Secretory Pathway.
To probe the functional state of the ER-associated apoB100 pool that accumulates after proteasomal inhibition, we investigated whether apoB100 was still able to be secreted. Lactacystin-treated HepG2 cells were pulse-labeled for 15 min as described in Fig. 1A and chased for 1 h in lactacystin-containing medium. Without lactacystin, after 1 h of chase, >80% of the labeled apoB100 degraded (24), implying that a domain (or domains) had to have been accessible to the cytosol.
After 1 h of chase, aliquots of cells were immediately lysed, and samples of conditioned medium were taken. OA complexed to BSA or BSA alone was added to the wells of remaining cells. Aliquots of conditioned medium were removed 30, 90, and 150 min after the addition of OA. ApoB100 was immunoprecipitated from lysate and medium samples. As shown in Fig. 3, the stimulation of lipid synthesis by OA treatment led to a significant increase in the secretion of apoB100; in 3 separate experiments, from 70% to 90% of the cellular apoB100 present at the time of OA addition was secreted within 2.5 h. Because peak incorporation of radiolabel into apoB100 occurs 20 min after the beginning of the chase period (25, 31, 32), this secreted apoB100 had been associated with the ER for at least 40 min after its translation seemed to be complete (as judged by the electrophoretic mobility of cellular and secreted apoB100 on SDS/PAGE). Control studies were performed in which the accumulation of apoAI or trichloroacetic acid-precipitable 35S-labeled protein in conditioned medium was unaffected by OA addition (data not shown). These control studies indicated that this stimulation of apoB100 secretion was specific. Taken with the cell-fractionation, immunofluorescence, and crosslinking data, these results imply that the pool of apoB100 that accumulates when the proteasome is inhibited remains associated with the ER membrane in proximity to the translocon and can be assembled into a lipoprotein and secreted. Furthermore, whatever the translocation status of this apoB100 pool, both the proteasomal and lipoprotein-assembly pathways must have access to it, because, as noted above, without lactacystin, >80% of nascent apoB100 would have been degraded by the proteasome in the absence of OA (24).
Figure 3.
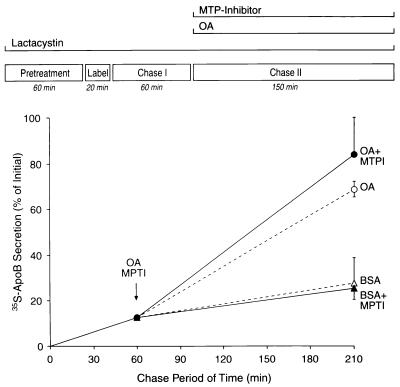
Recruitment of proteasome-protected apoB100 to lipoprotein assembly and the secretory pathway and the effects of MTP inhibition. HepG2 cells were treated as summarized at the top of the figure. At the end of the labeling period, samples from three wells were measured to determine the initial cellular pool of labeled apoB100 by immunoprecipitation-SDS/PAGE analysis of lysates. At the ends of the chase I and II periods, conditioned media samples from three wells were taken to measure the cumulative secretion of labeled apoB100. Some wells of HepG2 cells were similarly treated and analyzed, except that an inhibitor of MTP lipid-transfer activity was added at the end of chase I (+MTPI). The results are expressed as the mean percentage (± standard error) of the initial cellular pool.
MTP Lipid-Transfer Activity Is Required Only for the Initial Lipidation of ApoB100.
MTP lipid-transfer activity has been shown to be required for cotranslational lipidation of apoB100 and its subsequent secretion as a lipoprotein. Recent reports suggest that MTP is not required for the conversion of a “primordial” apoB100–lipid complex to a fully lipidated lipoprotein, as shown, for example, in studies of rat hepatoma McArdle-RH7777 cells (33). There are, however, some data to the contrary (34). To investigate the role of MTP lipid-transfer activity in the assembly and secretion of the ER-associated pool of apoB100 protected from the proteasome, HepG2 cells were treated with lactacystin, pulse-labeled, and chased in medium containing lactacystin (control medium), lactacystin plus an inhibitor of MTP lipid-transfer activity (26), lactacystin plus OA, or lactacystin plus OA and the MTP inhibitor. The data are summarized in Fig. 3.
After a 20-min pulse-labeling, only ≈13% of initially labeled apoB100 was recovered 60 min later from the control medium (chase I). When the chase was extended for an additional 150 min (chase II), ≈28% of initially labeled apoB100 was secreted. Secretion during chase II increased to ≈70% of initially labeled apoB100 when OA was present. Treatment of cells with the MTP-inhibitor at the start of chase II did not affect the secretion of labeled apoB100 into either the control (≈25%) or the OA-containing (≈84%) media. By contrast, secretion of apoB100 independent of the OA content of the medium was abolished when the MTP-inhibitor was added either 60 min before or during the pulse-labeling (data not shown). Of note, sucrose-gradient ultracentrifugation showed that the majority of apoB100 lipoproteins secreted during chase II in the absence of OA had a density of >1.020 g/ml. In contrast, in the presence of OA, there was a significant fraction of less dense apoB100 lipoproteins (density was <1.020 g/ml, indicating an increase in lipid content) that was independent of the inhibition of MTP lipid-transfer activity (data not shown). Thus, although inhibition of MTP at the beginning of lipoprotein assembly decreased apoB100 secretion, MTP inhibition after apoB100 had been allowed to enter and remain in the ER-associated pool did not affect lipoprotein assembly and secretion, suggesting that the lipid-transfer activity of MTP is no longer needed after early lipidation is accomplished.
Recruitment into the Secretory Pathway Involves Deubiquitination.
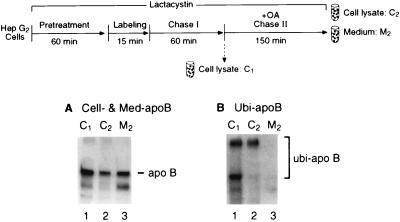
HepG2 cells were pretreated with lactacystin and pulse-labeled for 15 min as described in Fig. 1A. After a 1-h chase (chase I), aliquots of cells were lysed, and OA was added to the medium of the remaining cells as in Fig. 3. The conditioned medium was collected 2.5 h after OA addition (chase II). Lysate and medium samples were immunoprecipitated with anti-ubiquitin antibody, resuspended, and reimmunoprecipitated with anti-apoB100 antibody. As shown in Fig. 4A, by the end of chase II, labeled apoB100 in the cell decreased, whereas that in the conditioned medium increased correspondingly, with the total (cell plus medium) remaining relatively constant (data not shown). In Fig. 4B, note that high molecular-weight material corresponding to lightly and more heavily ubiquitinated apoB100 (bottom and top of the bracket, respectively) was in cell lysates taken at the time OA was added at the end of chase I. However, no ubiquitination of the secreted apoB100 could be detected at the end of chase II, implying that the ER-associated apoB100 was deubiquitinated before secretion. The material remaining in the cell at the end of chase II (Fig. 4B, lane 2) probably represents apoB100 that is so heavily ubiquitinated that it may be an exceptionally difficult substrate for deubiquitinating enzymes, thereby impeding its movement into the ER. Taken with the finding that the intensity of the lightly ubiquitinated apoB100 in the cell lysate at the end of chase II was only ≈10% of what it was at the start (C2 vs. C1; Fig. 4B), these data suggest that factors that control the extent of ubiquitination may, in part, regulate the net amount of apoB100 ultimately secreted as a lipoprotein.
Figure 4.
Recruitment into the secretory pathway of apoB100 protected from degradation by the proteasome involves deubiquitination. HepG2 cells were treated as summarized at the top of the figure. At the ends of chase I (C1) and chase II (C2), cells were harvested to measure labeled apoB100 (A) or to identify cellular apoB100–ubiquitin conjugates (B) by immunoprecipitation-SDS/PAGE analysis of lysates. In addition, at the end of chase II, conditioned medium (M2) was taken for a similar analysis of the content of labeled apoB100 (A) or of ubiquitin–apoB100 conjugates (B). The lower bands in A represent a reported (24) crossreacting nonspecific protein.
These data and the following further support the suggestion that the ER-associated apoB100 pool is in a bitopic state with domains in the cytosol and ER lumen: (i) apoB100 can be ubiquitinated cotranslationally (35); (ii) it is unlikely that a ubiquitinated substrate fully translocates into the ER lumen; and (iii) all known deubiquitinating enzymes are located outside of the ER (36).
DISCUSSION
A number of proteins known to undergo endoplasmic reticulum-associated degradation have been shown to be substrates of the cytosolic ubiquitin-proteasome pathway. These include both proteins with mutations leading to misfolding (e.g., CFTR, refs. 37 and 38; carboxypeptidase yscY, ref. 39; and α1-anti-trypsin Z, ref. 40) and wild-type, though unassembled, oligomeric proteins (e.g., major histocompatibility complex class I molecules, refs. 14 and 41, and T cell receptor α, ref. 42). ApoB100 is an unusual member of this family in that it is a wild-type single-chain polypeptide whose secretory rate can be regulated by altering the fraction targeted for proteasomal degradation. This targeting depends on the availability of lipid ligands (9, 10). Nevertheless, all these proteins pose the same fundamental topological question: how is an ER-associated protein ultimately degraded by a cytosolic protease?
To address this question for apoB100, we set out to trap (by limiting lipid synthesis and inhibiting the proteasome) and characterize an intermediate pool in the degradation pathway. As shown in Fig. 3, the majority of this pool could be recruited to lipoprotein assembly when OA was used to stimulate lipid synthesis well after apoB100 translation was expected to be complete, at a time when ordinarily (i.e., in the absence of lactacystin) >80% of nascent apoB100 would have been degraded by the proteasome (24). A possibility that cannot be excluded at present, given the limits of gel electrophoresis to resolve minor differences in molecular weights, is that apoB100 translation may not have been terminated completely, so that the ribosome could remain attached to the ER membrane and tether nearly complete apoB100 to, or in the vicinity of, the translocon. Nonetheless, this pool represents a physiologically relevant state of apoB100 and not an artifact of proteasome inhibition, as suggested by its ability to be assembled into lipoproteins and by its secretion being blocked by the ER to Golgi transport inhibitor brefeldin A (data not shown), which also disrupts apoB100-lipoprotein secretion in HepG2 cells not treated with proteasomal inhibitors (24). Interestingly, the intermediate pool contained ubiquitinated apoB100 that apparently was deubiquitinated before secretion (Fig. 4).
The location of the intermediate pool seems to be the ER, because metabolically labeled apoB100 protected from the proteasome stayed associated with microsomal membranes in close proximity to the translocon and did not accumulate in the cytosol (Figs. 1A and 2). This location agrees with studies of HepG2 cells by Ingram and Shelness (43), who found that the microsomal topology of newly synthesized apoB100, assessed by its sensitivity to exogenous protease treatment, did not change for at least 60 min when degradation was inhibited by ALLN, which is also a proteasomal inhibitor. The location of the intermediate pool in the ER is also supported by the fact that proteasome-protected apoB100 colocalized predominately with an immunofluorescent marker of the ER (Fig. 1B). Based on recent results for a viral glycoprotein (44), the punctate appearance of the immunofluorescence may represent apoB100 concentrated in specialized extensions of the ER membrane. It is interesting to note the report of Raposo et al. (45) who found that misfolded major histocompatibility complex class I molecules targeted for proteasomal degradation in TAP1-deficient mice were shown to accumulate in a structure believed to be an expansion of the ER-Golgi intermediate compartment. A similar phenomenon, though on a reduced scale, may also occur in HepG2 cells under our experimental conditions, given the overlap of a small fraction of the punctate apoB100-containing regions with the ER-Golgi intermediate compartment (Fig. 1B, row B). Nevertheless, the overall findings show that the vast majority of apoB100 in the intermediate pool is ER-associated and accessible to both the ubiquitin-proteasome and lipoprotein-assembly pathways.
A simple explanation for our data is that there is direct attack of the proteasome on a domain(s) exposed to the cytosol, as has been suggested for some membrane-bound proteins, such as the CD3-δ subunit of the T cell antigen receptor (46). Presumably, in such a model there is an activity, such as the intrinsic ATPase activities of cytosolic hsp70 or the proteasome, that can serve as a molecular motor to extract the remaining domains from or through the ER membrane (47). Note that an extraction process may also involve ubiquitination, which, in addition to “tagging” proteins to be processed by the proteasome, may also contribute to the unidirectional transport of proteins from the ER lumen (18).
The fact that apoB100 domains are exposed to the cytosol is supported by a large body of evidence gathered from widely different approaches. This evidence includes the ubiquitination of apoB100 during its translation (35), the protease susceptibility of microsomal apoB100 in HepG2 cells and rat hepatocytes (30, 48, 49), the detection of apoB100 epitopes by immunofluorescence in semipermeable HepG2 cells (50), the interaction between cytosolic hsp70 and apoB100 in HepG2 cells (10, 51), and the transient cytosolic exposure of translating apoB100 in a cell-free system (19). Thus, it is certainly possible that apoB100 is a proteasome substrate that is attacked “directly” while still associated with the ER membrane.
An alternate paradigm for endoplasmic reticulum-associated degradation has been suggested by studies of other substrates, such as the major histocompatibility complex class I heavy chain (14, 41), in which there is complete translocation followed by retrotranslocation or dislocation through a sec61-containing structure. In that case, accumulation of a cytosolic pool of the substrate protein is detected on inhibition of the ubiquitin-proteasome pathway by pharmacologic or genetic methods (14, 39). There are a number of arguments against this model of the trafficking of apoB100 to the ubiquitin-proteasome pathway. (i) The targeting of apoB100 for degradation can occur during its translation (11, 35). (ii) As noted above, no cytosolic accumulation of apoB100 was detected after the proteasome was inhibited. (iii) The ability of apoB100 to be recruited quantitatively into the secretory pathway 40–60 min after translation was completed (or nearly completed, see above) in the presence of lactacystin makes it unlikely that it had been dislocated into the cytosol and then recruited back into the ER lumen.
Rather than competing with each other, the dislocation and direct-attack models potentially represent two ends of a continuum in which the transport processes (forward or retrograde movement of a protein across the ER membrane) and the proteolytic pathway (ubiquitination and proteasomal degradation) act in concert. In the case where a cytosolic intermediate is found, the retrograde movement of the protein may be faster than the multistep ubiquitin-proteasome pathway, so that the protein is dislocated fully before it is degraded. For apoB100, in which no cytosolic intermediate was found (Fig. 1A), the pool targeted to the proteasome may not have been translocated completely into the ER. ApoB100 may be unique in this regard, because translocational pausing (19) or inhibition of lipoprotein assembly (35) may expose the nascent chain to the cytosol, permitting cotranslational ubiquitination and maintaining interactions with the translocon proteins sec61α (this report and ref. 52), sec61β (Fig. 2 and ref. 52), and TRAM (53).
A reasonable scenario that summarizes our data is that, at some point during its translation, apoB100 assumes a bitopic orientation relative to the ER membrane with the potential to be committed to either lipoprotein assembly or proteasomal degradation. If lipid synthesis or MTP lipid-transfer activity is sufficient, lipoprotein assembly initiates. Much of the subsequent lipidation, however, apparently can be accomplished without the lipid transfer mediated by MTP (Fig. 3). Furthermore, based on the analysis of metabolically labeled apoB100, particularly in the cell-fractionation (Fig. 1A) and crosslinking studies (Fig. 2), much of the lipoprotein-assembly process seems to be accomplished while the protein is associated with the ER membrane in close proximity to, perhaps even still within, a translocon.
In addition to the data in this report and the material reviewed earlier in this discussion, support for such a model comes from studies showing that apoB100 in rat primary hepatocytes has a prolonged residence in the ER (31) and that most apoB100 in rat hepatocytes and hepatoma cells is associated with the ER membrane with relatively few lipoprotein particles found in the ER lumen (54, 55). Furthermore, the apoB100-containing lipoproteins in the lumen are lipidated fully, as can be observed in their diameter or flotation behavior (54), suggesting that lipoprotein assembly occurred while the apoB100 was associated with the ER membrane.
Recently, Olofsson and coworkers (33) provided direct evidence that supports this suggestion. They extracted from microsomal membranes of rat hepatoma cells apoB100 lipoproteins that are not lipidated fully and presumably represent intermediates in the assembly process (33). In the same study, it was also noted, as we found in HepG2 cells, that MTP-mediated lipidation is important only during the initial lipoprotein-assembly process. Overall, the fact that many results among different model systems (rat hepatocytes, rat hepatoma cells, HepG2, and cell-free) agree to a significant extent makes it likely that the proposed model provides a framework for the general understanding of the mechanisms by which apoB100 lipoproteins are assembled and secreted.
Acknowledgments
We thank Jill F. Fisher for editorial assistance and the reviewers for their insightful comments. These studies were supported by a grant from the American Heart Association/New York City Affiliate (to E.A.F.), National Institutes of Health Grants HL58541 (to E.A.F.) and HL55638 (to H.N.G.), a grant from the Medical Research Council of Canada (to J.D.A.), and a grant from the Alberta Heritage Foundation for Medical Research (to J.D.A.).
ABBREVIATIONS
- apoAI
apoprotein AI
- apoB100
apoprotein B100
- ER
endoplasmic reticulum
- MTP
microsomal triglyceride transfer protein
- OA
oleic acid
References
- 1.Borén J, Graham L, Wettesten M, Scott J, White A, Olofsson S. J Biol Chem. 1992;267:9858–9867. [PubMed] [Google Scholar]
- 2.Wu X, Zhou M, Huang L-S, Wetterau J, Ginsberg H N. J Biol Chem. 1996;271:10277–10281. doi: 10.1074/jbc.271.17.10277. [DOI] [PubMed] [Google Scholar]
- 3.Patel S B, Grundy S M. J Biol Chem. 1996;271:18686–18694. doi: 10.1074/jbc.271.31.18686. [DOI] [PubMed] [Google Scholar]
- 4.Hussain M M, Bakillah A, Jamil H. Biochemistry. 1997;21:13060–13067. doi: 10.1021/bi971395a. [DOI] [PubMed] [Google Scholar]
- 5.Vance J E, Vance D E. Annu Rev Nutr. 1990;10:337–356. doi: 10.1146/annurev.nu.10.070190.002005. [DOI] [PubMed] [Google Scholar]
- 6.Yao Z, McLeod R S. Biochim Biophys Acta. 1994;1212:152–166. doi: 10.1016/0005-2760(94)90249-6. [DOI] [PubMed] [Google Scholar]
- 7.Dixon J L, Ginsberg H N. J Lipid Res. 1993;34:167–177. [PubMed] [Google Scholar]
- 8.Yao Z, Tran K, McLeod R S. J Lipid Res. 1997;38:1937–1953. [PubMed] [Google Scholar]
- 9.Yeung S J, Chen S H, Chan L. Biochemistry. 1996;35:13843–13848. doi: 10.1021/bi9618777. [DOI] [PubMed] [Google Scholar]
- 10.Fisher E A, Zhou M, Mitchell D M, Wu X, Omura S, Wang H, Goldberg A L, Ginsberg H N. J Biol Chem. 1997;272:20427–20434. doi: 10.1074/jbc.272.33.20427. [DOI] [PubMed] [Google Scholar]
- 11.Benoist F, Grand-Perret T. J Biol Chem. 1997;272:20435–20442. doi: 10.1074/jbc.272.33.20435. [DOI] [PubMed] [Google Scholar]
- 12.Bonifacino J S. Nature (London) 1996;384:405–406. doi: 10.1038/384405a0. [DOI] [PubMed] [Google Scholar]
- 13.Kopito R R. Cell. 1997;88:427–430. doi: 10.1016/s0092-8674(00)81881-4. [DOI] [PubMed] [Google Scholar]
- 14.Wiertz E J, Tortorella D, Bogyo M, Yu J, Mothes W, Jones T R, Rapoport T A, Ploegh H L. Nature (London) 1996;384:432–438. doi: 10.1038/384432a0. [DOI] [PubMed] [Google Scholar]
- 15.McGee T P, Cheng H H, Kumagi H, Omura S, Simoni R D. J Biol Chem. 1996;271:25630–25638. doi: 10.1074/jbc.271.41.25630. [DOI] [PubMed] [Google Scholar]
- 16.Yang Y, Fruh K, Ahn K, Peterson P A. J Biol Chem. 1995;270:27687–27694. doi: 10.1074/jbc.270.46.27687. [DOI] [PubMed] [Google Scholar]
- 17.Sommer T, Jentsch S. Nature (London) 1993;365:176–179. doi: 10.1038/365176a0. [DOI] [PubMed] [Google Scholar]
- 18.Biederer T, Volkwein C, Sommer T. Science. 1997;278:1806–1809. doi: 10.1126/science.278.5344.1806. [DOI] [PubMed] [Google Scholar]
- 19.Chuck S L, Lingappa V R. Cell. 1992;68:9–21. doi: 10.1016/0092-8674(92)90202-n. [DOI] [PubMed] [Google Scholar]
- 20.Bonnardel J A, Davis R A. J Biol Chem. 1995;270:28892–28896. doi: 10.1074/jbc.270.48.28892. [DOI] [PubMed] [Google Scholar]
- 21.Segrest J P, Jones M K, Mishra V K, Anantharamaiah G M, Garber D W. Arterioscler Thromb. 1994;14:1674–1685. doi: 10.1161/01.atv.14.10.1674. [DOI] [PubMed] [Google Scholar]
- 22.Gorlich D, Rapoport T A. Cell. 1993;75:615–630. doi: 10.1016/0092-8674(93)90483-7. [DOI] [PubMed] [Google Scholar]
- 23.Fenteany G, Standaert R F, Lane W S, Choi S, Corey E J, Schreiber S L. Science. 1995;268:726–731. doi: 10.1126/science.7732382. [DOI] [PubMed] [Google Scholar]
- 24.Dixon J L, Furukawa S, Ginsberg H N. J Biol Chem. 1991;266:5080–5086. [PubMed] [Google Scholar]
- 25.Wang H, Chen X, Fisher E A. J Clin Invest. 1993;91:1380–1389. doi: 10.1172/JCI116340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haghpassand M, Wilder D, Moberly J B. J Lipid Res. 1996;37:1468–1480. [PubMed] [Google Scholar]
- 27.Gilmore R. Curr Opin Cell Biol. 1991;3:580–584. doi: 10.1016/0955-0674(91)90026-u. [DOI] [PubMed] [Google Scholar]
- 28.Kappeler F, Itin C, Schindler R, Hauri H P. J Biol Chem. 1994;269:6279–6281. [PubMed] [Google Scholar]
- 29.Saraste J, Palade G E, Farquhar M G. J Cell Biol. 1987;105:2021–2029. doi: 10.1083/jcb.105.5.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Furukawa S, Sakata N, Ginsberg H N, Dixon J L. J Biol Chem. 1992;267:22630–22638. [PubMed] [Google Scholar]
- 31.Borchardt R A, Davis R A. J Biol Chem. 1987;262:16394–16402. [PubMed] [Google Scholar]
- 32.Sparks J D, Sparks C E. J Biol Chem. 1990;265:8854–8862. [PubMed] [Google Scholar]
- 33.Rustaeus S, Stillemark P, Lindberg K, Gordon D, Olofsson S. J Biol Chem. 1998;273:5196–5203. doi: 10.1074/jbc.273.9.5196. [DOI] [PubMed] [Google Scholar]
- 34.Wang Y, McLeod R S, Yao Z. J Biol Chem. 1997;272:12272–12278. doi: 10.1074/jbc.272.19.12272. [DOI] [PubMed] [Google Scholar]
- 35.Zhou M, Fisher E A, Ginsberg H N. J Biol Chem. 1998;273:24649–24653. doi: 10.1074/jbc.273.38.24649. [DOI] [PubMed] [Google Scholar]
- 36.Hochstrasser M. Annu Rev Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- 37.Jensen T J, Loo M A, Pind S, Williams D B, Goldberg A L, Riordan J R. Cell. 1995;83:129–135. doi: 10.1016/0092-8674(95)90241-4. [DOI] [PubMed] [Google Scholar]
- 38.Ward C L, Omura S, Kopito R R. Cell. 1995;83:121–127. doi: 10.1016/0092-8674(95)90240-6. [DOI] [PubMed] [Google Scholar]
- 39.Hiller M M, Finger A, Schweiger M, Wolf D H. Science. 1996;273:1725–1728. doi: 10.1126/science.273.5282.1725. [DOI] [PubMed] [Google Scholar]
- 40.Qu D, Teckman J H, Omura S, Perlmutter D H. J Biol Chem. 1996;271:22791–22795. doi: 10.1074/jbc.271.37.22791. [DOI] [PubMed] [Google Scholar]
- 41.Hughes E A, Hammond C, Cresswell P. Proc Nat Acad Sci USA. 1997;94:1896–1901. doi: 10.1073/pnas.94.5.1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu H, Kaung G, Kobayashi S, Kopito R R. J Biol Chem. 1997;272:20800–20804. doi: 10.1074/jbc.272.33.20800. [DOI] [PubMed] [Google Scholar]
- 43.Ingram M F, Shelness G S. J Lipid Res. 1996;37:2202–2214. [PubMed] [Google Scholar]
- 44.Presley J F, Cole N B, Schroer T A, Hirschberg K, Zaal K J, Lippincott-Schwartz J. Nature (London) 1997;389:81–85. doi: 10.1038/38001. [DOI] [PubMed] [Google Scholar]
- 45.Raposo G, van Santen H M, Leijendekker R, Geuze H J, Ploegh H L. J Cell Biol. 1995;131:1403–1419. doi: 10.1083/jcb.131.6.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang M, Omura S, Bonifacino J S, Weissman A M. J Exp Med. 1998;187:835–846. doi: 10.1084/jem.187.6.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mayer T U, Braun T, Jentsch S. EMBO J. 1998;17:3251–3257. doi: 10.1093/emboj/17.12.3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Davis R A, Thrift R N, Wu C C, Howell K E. J Biol Chem. 1990;265:10005–10011. [PubMed] [Google Scholar]
- 49.Du E Z, Kurth J, Wang S-L, Humiston P, Davis R A. J Biol Chem. 1994;269:24169–24176. [PubMed] [Google Scholar]
- 50.Du X, Stoops J D, Mertz J R, Stanley C M, Dixon J L. J Cell Biol. 1998;141:585–599. doi: 10.1083/jcb.141.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou M, Wu X, Huang L, Ginsberg H N. J Biol Chem. 1995;270:25220–25224. doi: 10.1074/jbc.270.42.25220. [DOI] [PubMed] [Google Scholar]
- 52.Chen Y, Le-Caherec F, Chuck S L. J Biol Chem. 1998;273:11887–11894. doi: 10.1074/jbc.273.19.11887. [DOI] [PubMed] [Google Scholar]
- 53.Hegde R S, Voigt S, Rapoport T A, Lingappa V R. Cell. 1998;92:621–631. doi: 10.1016/s0092-8674(00)81130-7. [DOI] [PubMed] [Google Scholar]
- 54.Rusiñol A, Verkade H, Vance J E. J Biol Chem. 1993;268:3555–3562. [PubMed] [Google Scholar]
- 55.Borén J, Rustaeus S, Olofsson S-O. J Biol Chem. 1994;41:25879–25888. [PubMed] [Google Scholar]