Abstract
The human type VII collagen gene (COL7A1) recently has been identified as an immediate-early response gene for transforming growth factor β (TGF-β)/SMAD signaling pathway. In this study, by using MDA-MB-468 SMAD4−/− breast carcinoma cells, we demonstrate that expression of SMAD4 is an absolute requirement for SMAD-mediated promoter activity. We also demonstrate that the SMAD binding sequence (SBS) representing the TGF-β response element in the region −496/−444 of the COL7A1 promoter functions as an enhancer in the context of a heterologous promoter. Electrophoretic mobility-shift assays with nuclear extracts from COS-1 cells transfected with expression vectors for SMADs 1–5 indicate that SMAD3 forms a complex with a migration similar to that of the endogenous TGF-β-specific complex observed in fibroblast extracts. Electrophoretic mobility-shift assays using recombinant glutathione S-transferase-SMAD fusion proteins indicate that both SMAD4 and C-terminally truncated SMAD3, but not SMAD2, can bind the COL7A1 SBS. Coexpression of SMAD3 and SMAD4 in COS-1 cells leads to the formation of two complexes: a DNA/protein complex containing SMAD3 alone and another slower-migrating complex containing both SMAD3 and SMAD4, the latter complex not being detected in fibroblasts. Maximal transactivation of COL7A1 SBS-driven promoters in either MDA-MB-468 carcinoma cells or fibroblasts requires concomitant overexpression of SMAD3 and SMAD4. These data may represent the first identification of a functional homomeric SMAD3 complex regulating a human gene.
Keywords: gene expression/signal transduction/transcription factors
Members of the transforming growth factor β (TGF-β) family of growth factors signal through serine/threonine kinase transmembrane receptors. On activation by TGF-β or related ligands, signaling from the receptors to the nucleus is mediated by phosphorylation of cytoplasmic mediators called SMADs (reviewed in refs. 1 and 2). The receptor-associated SMADs, such as SMAD1, SMAD2, SMAD3, and SMAD5, interact directly with, and are phosphorylated by, activated TGF-β receptor type I (TBRI or BMPRI) (3–6). They are ligand-specific and form, on phosphorylation, heteromeric complexes with SMAD4. The latter functions as a common mediator for all SMAD pathways (7–9). These complexes then are translocated into the nucleus, where they function as transcription factors, possibly in association with other proteins, such as the recently described Fast1 and Fast2 (7, 10, 11). The third group of SMAD proteins, the inhibitory SMADs such as SMAD6 or SMAD7, prevent phosphorylation and/or nuclear translocation of receptor-associated SMADs (12–15).
Most of the knowledge of the biological endpoints of SMAD pathways has been gained from studies in Xenopus laevis, Caenorhabditis elegans, and Drosophila melanogaster by using genetic screens aimed at understanding the downstream events of serine-threonine kinase receptors of the TGF-β family (reviewed in refs. 1 and 2). Although SMADs play a major role in TGF-β signaling in mammals, very few of the human targets for SMADs have been identified thus far. They include plasminogen activator inhibitor-1, (16, 17), p21 (18), Jun-B (19), and type VII collagen (COL7A1) (20). In the latter case, we have demonstrated that TGF-β up-regulation of COL7A1 gene expression in dermal fibroblasts is mediated by rapid and transient binding of a SMAD-containing complex to the region −496/−444 of the COL7A1 promoter. This SMAD binding sequence (SBS) was shown to be composed of two distinct binding sites, located at both ends of the −496/−444 region, whose simultaneous presence is required for SMAD/DNA complex formation. Sequence analysis of the SBS revealed the presence of several repeats, notably two adjacent CAGA repeats in the 5′ end and a GCCGGCG stretch in the 3′ end (20), matching the consensus Drosophila Mad binding sequence (21). Of interest, similar CAGA boxes have been identified in the human plasminogen activator inhibitor-1 promoter (17) and through a screen of SMAD-binding DNA sequences using a random pool of oligonucleotides (22). In both studies, it was shown that multimers of these CAGA boxes can bind SMAD3 and SMAD4 and can confer TGF-β responsiveness when cloned upstream of a minimal promoter, otherwise unresponsive to the growth factor.
In this study, we undertook a mechanistic analysis of SMAD-mediated transcription of COL7A1 by TGF-β. We demonstrate that, although SMAD4 is required for transcriptional activation, only SMAD3 was detected in the protein/DNA complexes formed between the COL7A1 promoter SBS and nuclear extracts from TGF-β-treated fibroblasts.
MATERIALS AND METHODS
Cell Cultures.
Human dermal fibroblast cultures, established by explanting tissue specimens obtained from neonatal foreskins, were used in passages 3–6. COS-1 cells were purchased from the American Type Culture Collection. MDA-MB-468, a cell line derived from a patient with breast carcinoma, has a homozygous deletion of the complete SMAD4 coding region (23). All cells were grown in DMEM supplemented with 10% heat-inactivated fetal calf serum, 2 mM glutamine, and antibiotics [100 units/ml penicillin, 50 μg/ml streptomycin-G, and 0.25 μg/ml Fungizone (GIBCO/BRL)]. Human recombinant TGF-β1 was a kind gift from R & D Systems. It will be referred to as TGF-β throughout the text.
Plasmid Constructs.
The various COL7A1 promoter 5′ deletion/chloramphenicol acetyltransferase (CAT) constructs cloned into promoterless pBS0CAT vector (24) and N-terminally Flag-tagged and C-terminally Myc-tagged SMAD expression vectors have been described (25, 26). Cloning of wild-type and mutant SBS fragments into pBLCAT5 was performed according to standard protocols (24). Bacterial glutathione S-transferase (GST)-SMAD fusion protein expression vectors were described previously (25, 26) or were constructed by using appropriate restriction enzymes.
Transient Cell Transfections and CAT Assays.
Transient cell transfections of human dermal fibroblasts were performed with the calcium phosphate/DNA coprecipitation procedure (27). COS-1 and MDA-MB-468 (SMAD4−/−) breast carcinoma cells were transfected by using lipofectamine as described (26). After appropriate incubation periods (see Figure legends), the cells were rinsed once with PBS, were harvested by scraping, and were lysed in 200 μl of reporter lysis buffer (Promega). pRSV-β-galactosidase was cotransfected in every experiment, and the β-galactosidase activities, measured according to a standard protocol (28), were used to monitor transfection efficiency. Unless stated otherwise, aliquots corresponding to identical β-galactosidase activity were used for each CAT assay with [14C]chloramphenicol as substrate (29) by using thin layer chromatography. After autoradiography, the plates were cut and counted by liquid scintillation to quantify the acetylated [14C]chloramphenicol.
Electrophoretic Mobility-Shift Assays (EMSAs).
A fragment spanning the region −496/−444 of the COL7A1 promoter, corresponding to the TGF-β response element binding a SMAD complex, was used as a probe as described (20). Nuclear extracts were isolated by using a small scale preparation (30), were aliquoted in small fractions to avoid repetitive freeze-thawing, and were stored at −80°C until use. The protein concentration in the extracts was determined by using a commercial assay kit (Bio-Rad). For supershift experiments, nuclear extracts (5–7 μg) were incubated overnight with antisera before the binding reaction. Mouse monoclonal anti-Flag (M2) and anti-Myc antibodies were from Kodak and Zymed, respectively.
Recombinant Proteins.
Full length or truncated GST-fusion SMAD proteins were expressed in Escherichia coli and were purified as directed (Pharmacia). In brief, proteins were overexpressed in DH5α cells by induction with 0.5 mM isopropyl β-d-thiogalactoside for 1 h at 37°C. Cells then were lysed by ultrasonication in PBS containing 0.5% Triton X-100 and 0.25 mM 4-(2-aminoethyl)benzenesulfonyl fluoride (Sigma). Proteins were collected on glutathione-Sepharose beads (Pharmacia). Beads were washed extensively, and GST fusion proteins were eluted overnight at 4°C in 10 mM reduced glutathione and 50 mM Tris⋅HCl (pH 8.0) containing 0.1% Triton X-100. A GST–bullous pemphigoid antigen 2 protein (31) did not bind any of the DNA fragments shown to interact with the SMAD proteins (not shown), indicating that GST itself does not bind the DNA sequences used in our study.
Western Blotting.
Proteins from lysates of transfected COS-1 cells were resolved by SDS-10% PAGE and were electrotransferred to nitrocellulose membrane (Hybond, Amersham). The blots were blocked in 0.1% Tween-PBS containing 5% nonfat dry milk. The anti-Flag M2 antibody was added to the blocking solution at 1:1,000 for 1 h at room temperature. The blots were washed three times with Tween-PBS, and the second antibody (horseradish peroxidase-conjugated sheep anti-mouse IgG, Santa Cruz Biotechnology) was added at 1:5,000 for 1 h at room temperature. After three washes in Tween-PBS, the blots were developed with ECL (Amersham) and were exposed on Kodak XAR5 film.
RESULTS
SMAD4 Is Required for COL7A1 Promoter Activity and Inducibility by TGF-β.
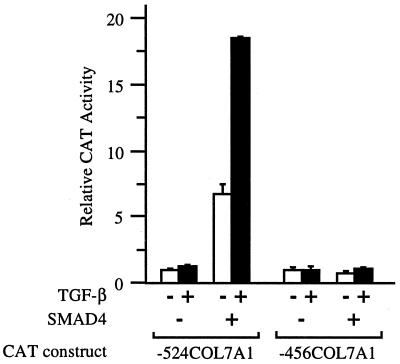
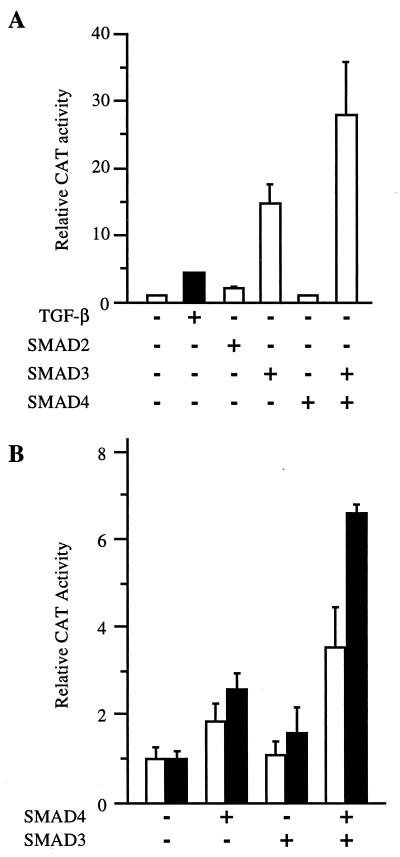
To investigate the role of SMAD4 in the inducibility of the COL7A1 response by TGF-β, SMAD4−/− MDA-MB-468 human breast carcinoma cells were transfected with either −524COL7A1/CAT or −456COL7A1/CAT constructs, and promoter response to TGF-β was measured. As shown in Fig. 1, little basal promoter activity was detected with −524COL7A1/CAT in these SMAD4−/− cells, and no activation by TGF-β was seen. Overexpression of SMAD4 increased both basal and TGF-β-induced promoter activity. The rescue of COL7A1 promoter activity by SMAD4 was not observed with the construct −456COL7A1/CAT, which lacks the 5′ element of SMAD-binding region (20), indicating that the bipartite SMAD binding element within the −496/−444 region of promoter is responsible for the effects of SMAD4 on −524COL7A1/CAT activity. These data indicate that the SMAD signaling pathway is involved in TGF-β up-regulation of COL7A1 gene expression. It cannot be excluded that the increased basal activity in response to SMAD4 overexpression may reflect promoter activation by endogenously produced TGF-β (32) and that SMAD members other than SMAD4 may be involved.
Figure 1.
SMAD4 is required for efficient transcription of the COL7A1 promoter and its responsiveness to TGF-β. COS-1 cells were transfected with either the −524COL7A1/CAT or the −456COL7A1/CAT construct of the human COL7A1 promoter, in the absence (−) or in the presence (+) of TGF-β, without (−) or with (+) a Smad4 expression vector. TGF-β (10 ng/ml) was added to the medium 4 h later, and the incubation continued for 40 h. In this and the subsequent figures, cell extracts were assayed for CAT activity with [14C]chloramphenicol as a substrate by using identical amounts of β-galactosidase activity. The relative CAT activity (mean ± SD) of at least two experiments performed in duplicate is shown in the form of a bar graph.
The COL7A1 SBS Confers TGF-β Responsiveness to a Heterologous Promoter.
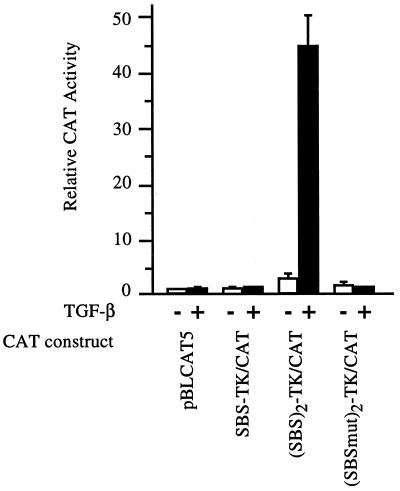
To determine whether the SBS could function as an enhancer in the context of a heterologous promoter, one or two copies of the −496/−444 SBS were cloned upstream of the thymidine kinase (TK) promoter in the pBLCAT5 vector and were used in transient cell transfection experiments. As shown in Fig. 2, a tandem repeat, but not a single copy, of the COL7A1 SBS could confer TGF-β responsiveness to the otherwise unresponsive TK promoter in pBLCAT5. These data indicate that the previously identified SMAD binding sequence can function as an enhancer element driving the transcriptional activation of a heterologous promoter when present as a dimer. The lack of responsiveness of SBS-TK/CAT, containing a single copy of the SBS, is not surprising because it is a common phenomenon observed with other well characterized enhancers, such as NF-κB or AP-1 binding sites, which require multiple tandem copies to be functional in the context of a heterologous promoter. Introduction of a triple point mutation (CCAGACAGA to AACGACAGA) that disrupts the first CAGA repeat within the 5′ portion of the SBS totally abolished TGF-β responsiveness. It has been shown that the integrity of this CAGA box is essential for SMAD binding (20). Indeed, EMSAs using a similarly mutated SBS probe confirmed the lack of SMAD binding (not shown). Together, these data indicate that SMAD binding to the COL7A1 SBS (20) is directly responsible for TGF-β activation of promoter activity (Fig. 2).
Figure 2.
A COL7A1 SBS tandem repeat confers TGF-β responsiveness to a heterologous promoter. Confluent fibroblast cultures were transfected with several constructs derived from pBLCAT5 by the calcium phosphate/DNA coprecipitation procedure. These constructs contained one copy of the COL7A1 SBS (SBS-TK/CAT), two copies of the SBS [(SBS)2-TK/CAT], or two copies of a mutated SBS [(SBSmut)2-TK/CAT]. After glycerol shock, the cultures were incubated in fresh medium containing 1% fetal calf serum, without (−) or with (+) TGF-β (10 ng/ml) added to the medium 4 h later. After 40 h of incubation, cell extracts were assayed for CAT activity. Results are the mean ± SD of three independent experiments.
Flag-SMAD3 Binds the COL7A1 SBS in COS-1 Cell Extracts.
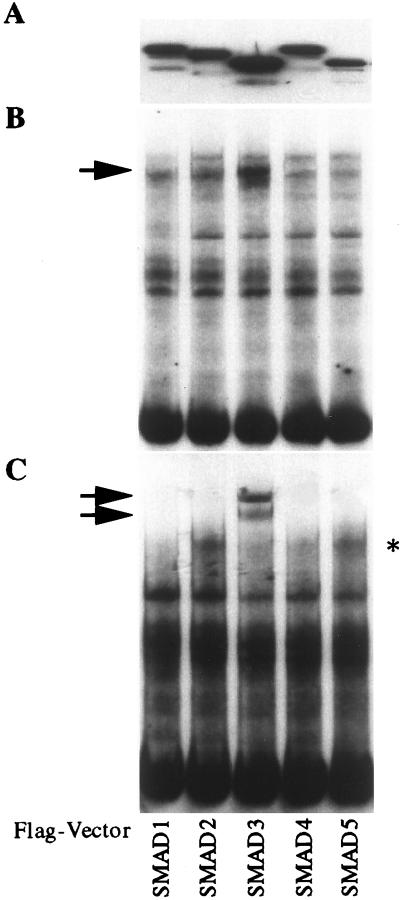
To identify the member(s) of the SMAD family present in the TGF-β-induced complex that binds the COL7A1 SBS, COS-1 cells were transfected with Flag-tagged SMAD1, SMAD2, SMAD3, SMAD4, or SMAD5 expression vectors. The expression of each Flag-tagged SMAD protein was assessed by Western blotting with an anti-Flag antibody in each nuclear extract preparation. As shown in Fig. 3A, each Flag-SMAD protein was expressed at a high level. EMSAs using the COL7A1 fragment −496/−444 as a probe revealed that a strong protein/DNA complex formed in SMAD3-transfected COS-1 cell extracts (Fig. 3B). Supershift assays with the M2 anti-Flag antibody identified Flag-SMAD3 as part of this strong complex whereas the antibody did not supershift the faint complex observed in the extracts from cells transfected with either SMAD1, SMAD2, SMAD4, or SMAD5 expression vectors (Fig. 3C). The latter faint band (Fig. 3C, *) is therefore likely to represent endogenous SMAD3-containing binding activity because it is also present in untransfected cells (not shown).
Figure 3.
Flag-SMAD3 binds the COL7A1 SBS. COS-1 cells were transfected with expression vectors encoding various SMADs tagged with the Flag epitope as indicated, and nuclear extracts were prepared 40 h later. (A) Western blot of each of the nuclear extract preparations, using a Flag antibody. Note that each Flag-SMAD protein is present at high levels in their respective extracts. (B) EMSA using the COL7A1 SBS as a probe. Note the strong binding present in nuclear extracts containing Flag-SMAD3 (arrow). (C) Supershift assay using 3 μg/lane of anti-Flag antibody. Note that the faint band (∗) is likely to represent endogenous Smad3-containing binding activity.
The supershift created by the mAb (anti-Flag) appeared as a doublet, suggesting the presence of at least two SMAD3 molecules within the complex. It should be noted that protein/DNA complex formation was not enhanced in SMAD4-Flag transfected cells, although SMAD4 was present in the nuclear extracts (see Western blot in Fig. 3A) and is required for optimal promoter activity (Fig. 1).
The COL7A1 SBS Binds Recombinant SMAD3 and SMAD4 but not SMAD2.
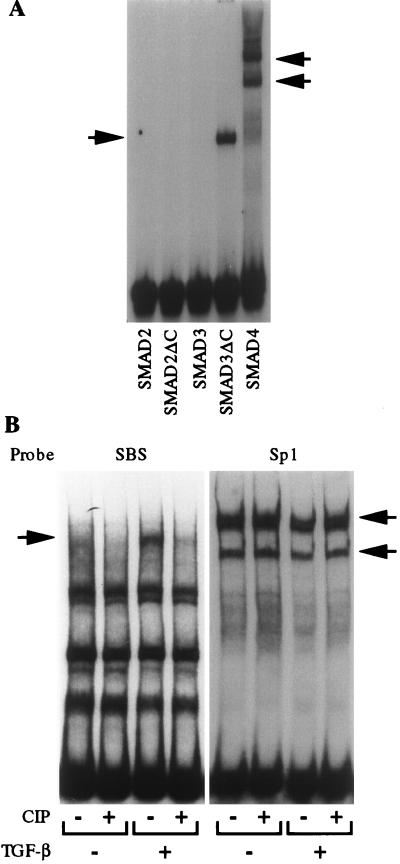
Both SMAD2 and SMAD3 have been shown to be specific mediators of the TGF-β/activin pathways whereas SMAD4 is common to all TGF-β family members (reviewed in refs. 1 and 2). To determine the binding specificity of the COL7A1 SBS, EMSA experiments were performed by using recombinant GST-SMAD fusion proteins. To circumvent the problem of phosphorylation requirement of SMAD2 or SMAD3 for DNA binding (see reviews in refs. 1 and 2), C-terminally truncated fusion proteins SMAD2ΔC and SMAD3ΔC also were generated. As shown in Fig. 4A, SMAD2, SMAD2ΔC, and SMAD3 did not bind the SBS whereas both SMAD3ΔC and SMAD4 efficiently bound the COL7A1 TGF-β response element.
Figure 4.
(A) Recombinant GST-SMAD3ΔC and GST-SMAD4 bind the COL7A1 SBS. EMSAs were performed by using 100 ng of E. coli-derived GST-fusion proteins of SMAD2, SMAD2ΔC, SMAD3, SMAD3ΔC, and SMAD4, using the COL7A1 SBS as a probe and 10 μg/lane of BSA. Note that only SMAD3ΔC and SMAD4 could bind the probe. (B) Binding of TGF-β-induced SMAD-containing complex to the SBS requires phosphorylation. EMSAs were performed by using either the COL7A1 SBS promoter fragment (left) or a consensus Sp1 oligonucleotide (right) as probes. Nuclear extracts from either control (−) or TGF-β-treated (+) fibroblast cultures were digested (+) or not (−) with calf intestine phosphatase (CIP, 0.1 unit/ml, 10 min), and their ability to bind either probe was tested.
The ability of SMAD3ΔC but not the full length SMAD3 to bind the SBS suggests that SBS/protein complex formation in vivo may require SMAD phosphorylation. To address this possibility, nuclear extracts from TGF-β-treated fibroblasts were digested with calf intestine phosphatase before EMSA, using either the SBS or a consensus Sp1 oligonucleotide as probes. As shown in Fig. 4B, calf intestine phosphatase digestion of the nuclear extracts completely abolished binding of the TGF-β specific complex to the SBS (Fig. 4B, left). In contrast, Sp1 binding activity, unchanged by TGF-β, was not altered by calf intestine phosphatase treatment (Fig. 4B, right), in accordance with previous observations (33). These data indicate that phosphorylation is required for SBS/SMAD complex formation.
Coexpression of SMAD3 and SMAD4 Results in Optimal COL7A1 Promoter Transactivation.
The capacity of SMAD3 to bind the COL7A1 SBS led us to investigate its potential role as a transcriptional activator of COL7A1. In a first set of experiments, fibroblasts were cotransfected with expression vectors for SMAD2, SMAD3, and SMAD4, together with (SBS)2-TK/CAT. As shown in Fig. 5A, significant induction of promoter activity was detected when SMAD3, but not SMAD2 or SMAD4, was coexpressed. Maximal transactivation was observed when both SMAD3 and SMAD4 were coexpressed, indicating that endogenous SMAD4 levels may have been limiting when over-expressing SMAD3 alone.
Figure 5.
SMAD3 and SMAD4 synergize to activate −524COL7A1/CAT and (SBS)2-TK/CAT. (A) Human dermal fibroblasts were cotransfected with (SBS)2-TK/CAT and either SMAD2, SMAD3, and/or SMAD4 expression vectors. Cell extracts were prepared and assayed for CAT activity 40 h later. (B) SMAD4−/− MDA-MB-468 cells were transfected with −524COL7A1/CAT (open bars) or (SBS)2-TK/CAT (closed bars) constructs, together with SMAD3 and/or SMAD4 expression vectors. After 40 h, cell extracts were prepared and assayed for CAT activity. Results are the mean ± SD of three independent experiments.
Next, MDA-MB-468 cells were transfected with either the −524COL7A1/CAT or (SBS)2-TK/CAT constructs, together with expression vectors for either SMAD3 or SMAD4, either alone or in combination. As shown in Fig. 5B, both constructs were equally responsive to SMAD4 over-expression, as expected from our earlier observations (Fig. 1), but the highest promoter activity again was observed when both SMAD3 and SMAD4 were coexpressed.
SMAD4 May Not Participate in the Protein/DNA Complex Induced by TGF-β.
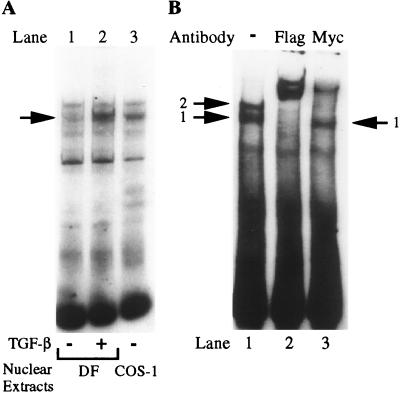
The next set of experiments was designed to characterize the components of the SMAD/DNA complex formed with the COL7A1 TGF-β response element. First, the migratory pattern of the TGF-β-induced fibroblast SBS-binding complex was compared with that of Flag-SMAD3-transfected COS-1 cell extracts. As shown in Fig. 6A, the two complexes could not be differentiated on the basis of their electrophoretic mobility, suggesting that they may be identical in composition.
Figure 6.
SMAD3 but not SMAD4 may participate in SBS/protein complex formation. (A) Nuclear extracts were prepared from dermal fibroblasts (DF) treated (+) or not (−) with TGF-β for 30 min or from Flag-SMAD3 transfected cells. Their ability to bind the COL7A1 SBS was compared by EMSA. Note the identical migratory pattern of the TGF-β-specific band with the Flag-SMAD3/DNA complex. (B) Nuclear extracts from Flag-SMAD3 and SMAD4-Myc-transfected COS-1 cells were used in EMSA and were supershifted with 3 μg of either anti-Flag or anti-Myc antibodies. Note that the anti-Myc antibody does not supershift complex 1.
Next, we attempted to understand the discrepancy between the absolute requirement for SMAD4 in COL7A1/SBS-mediated transactivation (Fig. 1) and the absence of SMAD4 binding activity in SMAD4-Flag transfected COS-1 cells (Fig. 3B). For this purpose, COS-1 cells were cotransfected with both SMAD3-Flag and SMAD4-Myc expression vectors. EMSAs using the COL7A1 SBS as a probe revealed that the corresponding nuclear extracts had the ability to generate two distinct complexes (Fig. 6B, lane 1). The fastest complex (1) has an electrophoretic mobility comparable to that of the TGF-β-specific complex induced in fibroblasts and to that present in SMAD3-transfected COS-1 cells (Fig. 6A) whereas complex 2 has a slower mobility. An anti-Flag antibody supershifted both complexes, indicating the presence of SMAD3 in each of them (Fig. 6A, lane 2). In contrast, an anti-Myc antibody efficiently supershifted complex 2 but not complex 1 (Fig. 6A, lane 3), indicating that SMAD4 is present only in the upper complex, which does not exist in nuclear extracts from TGF-β-treated fibroblasts. Collectively, these results suggest that, although SMAD4 is required for transcriptional activation of the COL7A1 promoter by TGF-β, it may not be present in the complex forming with the SBS, in which we could only detect SMAD3.
DISCUSSION
Although much has been learned recently about the cytoplasmic events involved in TGF-β signal transduction, little is known about the nuclear events leading to activation of specific genes. Several lines of evidence suggest that SMAD proteins activate their targets by both direct and indirect binding to DNA and that the mechanisms may be isoform specific. Drosophila Mad, SMAD3, and SMAD4 all have been shown to bind DNA directly whereas SMAD2 likely requires a DNA-binding intermediate (1, 2). Once bound to DNA, the mechanism of transcriptional activation by SMAD proteins remains elusive, although a complex interaction with multiple elements of the transcriptional machinery, and in particular CBP/p300 (34–36) or MSG1 (37), is likely.
Our data suggest that the interaction of SMAD proteins with cis elements depends on the context in which these elements are presented. A SMAD binding sequence has been identified (20) that contains two CAGA motifs similar to sequences identified both in the plasminogen activator inhibitor-1 and Jun-B promoters and through a screen of SMAD-binding DNA sequences (17, 19, 22). Recently, it was demonstrated that SMAD proteins exist as hetero- and homooligomers in vivo (38). Specifically, it was shown that SMAD3 homomers, as well as SMAD3/SMAD4 heteromeric complexes, can bind DNA. The in vivo functions of SMAD3 oligomers, capable of forming DNA-binding complexes, have not been determined. SMAD3 and SMAD4 also were identified as part of the complexes induced by TGF-β and binding the CAGA box identified in the plasminogen activator inhibitor-1 promoter (17). In the latter case, TGF-β was shown to induce the formation of two distinct complexes binding the CAGA sequence and containing SMAD3, but the authors failed to identify the specific components of each of the complexes. Support for the hypothesis that SMAD4 may not be part of the DNA-binding complex also is provided by a recent study of Drosophila development (39). Specifically, MEDEA, the Drosophila ortholog for SMAD4, was shown to be dispensable for omb gene activation by Mad during wing development.
In fibroblasts, TGF-β induces only one SMAD-containing complex to bind the COL7A1 SBS (20). However, when SMAD3 and SMAD4 were simultaneously over-expressed in COS-1 cells, two complexes were detected in EMSA, in a pattern similar to that observed by using other SMAD binding sequences (17, 38). We have determined that the upper complex, not found in fibroblasts, is composed of both SMAD3 and SMAD4. Analysis of the lower complex, which comigrates with the fibroblast TGF-β-specific complex, indicates that it contains SMAD3. No SMAD4 could be detected. At this time, we cannot rule out that technical limitations, such as the lack of accessibility of the SMAD4 epitopes to antibodies used in supershift assays, may be responsible for the lack of SMAD4 detection in the lower complex in SMAD3/4-transfected COS-1 cells and in the TGF-β-specific fibroblast complex binding the SBS. However, to our knowledge, there is to date no clear evidence demonstrating an absolute requirement for SMAD4 in an endogenous complex. Therefore, it must be considered that, although SMAD4 is clearly necessary for COL7A1 promoter transactivation and has the ability to bind the COL7A1 SBS, the TGF-β-induced complex may be composed of SMAD3 only.
The binding of SMAD3 and SMAD4 to the COL7A1 promoter, as observed with recombinant GST-SMAD proteins, appears to be specific and physiologically relevant. SMAD1 and SMAD5 did not bind the identified element, although they were clearly expressed in the nucleus of transfected COS-1 cells. SMAD2, another intermediate in the TGF-β signaling pathway and, like Smad3, a substrate for the TGF-β type I receptor, did not bind the COL7A1 element either when transiently expressed in COS-1 cells or as a bacterially expressed fusion protein. Consistent with previously published data (16), full length, bacterially produced SMAD4 bound quite well, but truncation of the MH2 domain of SMAD3 was required to relieve the autoinhibition of the MH1 binding domain. In vivo, this inhibition is likely to be relieved by phosphorylation of SMAD3 in the extreme C terminus. This model is supported by our finding that treatment of fibroblast nuclear extracts with phosphatase eliminated detectable DNA binding. The specificity for SMAD3 binding was demonstrated further by the complete lack of binding of a truncated version of SMAD2.
The process by which SMAD proteins activate transcription remains elusive, although our data suggest that several mechanisms may be at work in the COL7A1 promoter. There is no transcriptional activation, consistent with results obtained with 3TP-Lux and other reporter constructs, in the absence of SMAD4, (16, 17, 36). Overexpression of SMAD3 failed to rescue reporter gene activity in the absence of SMAD4, demonstrating convincingly that expression of SMAD4 is an absolute requirement for transcriptional activation in this context. Nevertheless, coexpression of both SMAD3 and SMAD4 is required for maximal activation of both −524COL7A1/CAT and (SBS)2-TK/CAT constructs.
Activation of the (SBS)2-TK/CAT after transient overexpression of both SMAD3 and SMAD4 in the absence of ligand is consistent with results obtained in other laboratories with different promoters (9, 16, 17). These data suggest a dual mechanism of activation of the COL7A1 promoter and perhaps multiple functions for SMAD proteins in transcriptional activation. SMADs may provide the initial impetus for transcription by direct DNA binding and transcriptional activation and also may induce other genes that are necessary for maintenance of transcriptional activity. Interactions of SMAD proteins in the later stages of transcription may occur in the absence of direct DNA binding, perhaps through interactions with other transcription factors (34–37).
These results implicate TGF-β and SMAD proteins in the pathogenesis of diseases involving COL7A1 and other genes that contain similar elements. Our identification of a specific sequence that is required for SMAD binding and activation suggests that mutations in this sequence could lead to disregulation of COL7A1 by TGF-β. Although complete loss of SMAD3 or SMAD4 is likely to be lethal, persons heterozygous at these loci might be prone to epidermal defects characterized by loss of COL7A1 expression. This study, therefore, identifies a potential link between SMAD signal transduction and loss of a specific gene in human disease.
Acknowledgments
The expert technical assistance of Ying-Jee Song is acknowledged gratefully. The authors thank Dr. Peter ten Dijke (Ludwig Institute for Cancer Research, Uppsala, Sweden) for the generous gift of the Smad2 construct. This work was supported by National Institutes of Health Grants R29-AR43751 (to A.M.) and RO1-AR41439 and PO1-AR38923 (to J.U.) and a Dermatology Foundation research fellowship (to L.V.).
ABBREVIATIONS
- CAT
chloramphenicol acetyltransferase
- EMSA
electrophoretic mobility-shift assay
- SBS
SMAD binding sequence
- TGF-β
transforming growth factor-β
- TK
thymidine kinase
- GST
glutathione S-transferase
References
- 1.Massagué J, Hata A, Liu F. Trends Cell Biol. 1997;7:187–192. doi: 10.1016/S0962-8924(97)01036-2. [DOI] [PubMed] [Google Scholar]
- 2.Heldin C H, Miyazono K, ten Dijke P. Nature (London) 1997;390:465–471. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- 3.Macias-Silva M, Abdollah S, Hoodless P A, Pirone R, Attisano L, Wrana J L. Cell. 1996;87:1215–1224. doi: 10.1016/s0092-8674(00)81817-6. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Y, Feng X-H, Wu R-Y, Derynck R. Nature (London) 1996;383:168–172. doi: 10.1038/383168a0. [DOI] [PubMed] [Google Scholar]
- 5.Liu F, Pouponnot C, Massagué J. Genes Dev. 1997;11:3157–3167. doi: 10.1101/gad.11.23.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakao A, Imamura T, Souchelnytskyi S, Kawabata M, Ishisaki A, Oeda E, Tamaki K, Hanai J, Heldin C H, Miyazono K, et al. EMBO J. 1997;16:5353–5362. doi: 10.1093/emboj/16.17.5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lagna G, Hata A, Hemmati-Brivanlou A, Massagué J. Nature (London) 1996;383:832–836. doi: 10.1038/383832a0. [DOI] [PubMed] [Google Scholar]
- 8.Candia A F, Watabe T, Hawley S H B, Onichtchouk D, Zhang Y, Derynck R, Niehrs C, Cho K W Y. Development (Cambridge, UK) 1997;24:4467–4480. doi: 10.1242/dev.124.22.4467. [DOI] [PubMed] [Google Scholar]
- 9.Wu H, Wade M, Krall L, Grisham J, Xiong Y, Van Dyke T. Genes Dev. 1996;10:245–260. doi: 10.1101/gad.10.3.245. [DOI] [PubMed] [Google Scholar]
- 10.Chen X, Weisberg E, Fridmacher V, Watanabe M, Naco G, Whitman M. Nature (London) 1997;389:85–89. doi: 10.1038/38008. [DOI] [PubMed] [Google Scholar]
- 11.Labbé E, Silvestri C, Hoodless P A, Wrana J L. Mol Cell. 1998;2:109–120. doi: 10.1016/s1097-2765(00)80119-7. [DOI] [PubMed] [Google Scholar]
- 12.Hayashi H, Abdollah S, Qiu Y, Cai J, Xu Y Y, Grinnell B W, Richardson M A, Topper J N, Gimbrone M A, Jr, Wrana J L, et al. Cell. 1997;89:1165–1173. doi: 10.1016/s0092-8674(00)80303-7. [DOI] [PubMed] [Google Scholar]
- 13.Nakao A, Afrakhte M, Moren A, Nakayama T, Christian J L, Heuchel R, Itoh S, Kawabata M, Heldin N E, Heldin C H, et al. Nature (London) 1997;389:631–635. doi: 10.1038/39369. [DOI] [PubMed] [Google Scholar]
- 14.Imamura T, Takase M, Nishihara A, Oeda E, Hanai L L, Kawabata M, Miyazono K. Nature (London) 1997;389:622–626. doi: 10.1038/39355. [DOI] [PubMed] [Google Scholar]
- 15.Hata A, Lagna G, Massagué J, Hemmati-Brivanlou A. Genes Dev. 1998;12:186–197. doi: 10.1101/gad.12.2.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yingling J M, Datto M B, Wong C, Frederick J P, Liberati N T, Wang X-F. Mol Cell Biol. 1997;17:7019–7028. doi: 10.1128/mcb.17.12.7019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dennler S, Itoh S, Vivien D, ten Dijke P, Huet S, Gauthier J-M. EMBO J. 1998;17:3091–3100. doi: 10.1093/emboj/17.11.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moustakas A, Kardassis D. Proc Natl Acad Sci USA. 1998;95:6733–6738. doi: 10.1073/pnas.95.12.6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jonk L J, Itoh S, Heldin C H, ten Dijke P, Kruijer W. J Biol Chem. 1998;273:21145–21152. doi: 10.1074/jbc.273.33.21145. [DOI] [PubMed] [Google Scholar]
- 20.Vindevoghel L, Kon A, Lechleider R J, Uitto J, Roberts A B, Mauviel A. J Biol Chem. 1998;273:13053–13057. doi: 10.1074/jbc.273.21.13053. [DOI] [PubMed] [Google Scholar]
- 21.Kim J, Johnson K, Chen H J, Carroll S, Laughton A. Nature (London) 1997;388:304–308. doi: 10.1038/40906. [DOI] [PubMed] [Google Scholar]
- 22.Zawel L, Le Dai J, Buckhaults P, Zhou S, Kinzler K W, Vogelstein B, Kern S E. Mol Cell. 1998;1:611–617. doi: 10.1016/s1097-2765(00)80061-1. [DOI] [PubMed] [Google Scholar]
- 23.Schutte M, Hruban R H, Hedrick L, Cho K R, Nadasdy G M, Weinstein C L, Bova G S, Isaacs W B, Cairns P, Nawroz H, et al. Cancer Res. 1996;56:2527–2530. [PubMed] [Google Scholar]
- 24.Vindevoghel L, Chung K-Y, Davis A, Kouba D, Kivirikko S, Alder H, Uitto J, Mauviel A. J Biol Chem. 1997;272:10196–10204. doi: 10.1074/jbc.272.15.10196. [DOI] [PubMed] [Google Scholar]
- 25.Lechleider R J, de Caestecker M P, Dehejta A, Polymeropoulos M H, Roberts A B. J Biol Chem. 1997;271:17617–17620. doi: 10.1074/jbc.271.30.17617. [DOI] [PubMed] [Google Scholar]
- 26.de Caestecker M P, Parks W T, Frank C J, Castagnino P, Botaro P, Roberts A B, Lechleider R J. Genes Dev. 1998;12:1587–1592. doi: 10.1101/gad.12.11.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gorman C, Moffat L F, Howard B H. Mol Cell Biol. 1982;2:1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 29.Graham F L, van der Eb A J. Virology. 1973;52:456–463. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 30.Andrews N C, Faller D V. Nucleic Acids Res. 1991;19:2499. doi: 10.1093/nar/19.9.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li K, Tamai K, Tan E M, Uitto J. J Biol Chem. 1993;268:8825–8834. [PubMed] [Google Scholar]
- 32.de Caestecker M P, Hemmati P, Larisch-Bloch S, Ajmera R, Roberts A B, Lechleider R J. J Biol Chem. 1997;272:13690–13696. doi: 10.1074/jbc.272.21.13690. [DOI] [PubMed] [Google Scholar]
- 33.Jackson S P, MacDonald J J, Lees-Miller S P, Tjian R. Cell. 1990;63:155–165. doi: 10.1016/0092-8674(90)90296-q. [DOI] [PubMed] [Google Scholar]
- 34.Janknecht R, Wells N J, Hunter T. Genes Dev. 1998;12:2114–2119. doi: 10.1101/gad.12.14.2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feng X-H, Zhang Y, Wu R-Y, Derynck R. Genes Dev. 1998;12:2153–2163. doi: 10.1101/gad.12.14.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Topper J N, DiChiara M R, Brown J D, Williams A J, Falb D, Collins T, Gimbrone M A., Jr Proc Natl Acad Sci USA. 1998;95:9506–9511. doi: 10.1073/pnas.95.16.9506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shioda T, Lechleider R J, Dunwoodie S L, Li H, Yahata T, de Caestecker M P, Fenner M H, Roberts A B, Isselbacher K J. Proc Natl Acad Sci USA. 1998;95:9785–9790. doi: 10.1073/pnas.95.17.9785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kawabata M, Inoue H, Hanyu A, Imamura T, Miyazono K. EMBO J. 1998;17:4056–4065. doi: 10.1093/emboj/17.14.4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wisotzkey R G, Mehra A, Sutherland D J, Dobens L L, Liu X, Dohrmann C, Attisano L, Raftery L A. Development (Cambridge, UK) 1998;125:1433–1445. doi: 10.1242/dev.125.8.1433. [DOI] [PubMed] [Google Scholar]