Abstract
The Saccharomyces cerevisiae myosin-V, Myo2p, has been implicated in the polarized movement of several organelles and is essential for yeast viability. We have shown previously that Myo2p is required for the movement of a portion of the lysosome (vacuole) into the bud and consequently for proper inheritance of this organelle during cell division. Class V myosins have a globular carboxyl terminal tail domain that is proposed to mediate localization of the myosin, possibly through interaction with organelle-specific receptors. Here we describe a myo2 allele whose phenotypes support this hypothesis. vac15–1/myo2–2 has a single mutation in this globular tail domain, causing defects in vacuole movement and inheritance. Although a portion of wild-type Myo2p fractionates with the vacuole, the myo2–2 gene product does not. In addition, the mutant protein does not concentrate at sites of active growth, the predominant location of wild-type Myo2p. Although deletion of the tail domain is lethal, the myo2–2 gene product retains the essential functions of Myo2p. Moreover, myo2–2 does not cause the growth defects and lethal genetic interactions seen in myo2–66, a mutant defective in the actin-binding domain. These observations suggest that the myo2–2 mutation specifically disrupts interactions with selected myosin receptors, namely those on the vacuole membrane and those at sites of polarized growth.
The polarized movement of membranous organelles is critical for many diverse processes in eukaryotic cells. For instance, lysosomes redistribute between perinuclear locations and the peripheral cytoplasm during receptor-mediated endocytosis, macrophage activation, and antigen-presenting dendritic cell maturation (1, 2, 3). In addition, mitochondria concentrate at sites of high energy demand and redistribute in response to various stimuli (4). Also, polarized secretion requires the directed movement of secretory vesicles to apical cell surfaces. Roles for microtubules and microfilaments in organelle transport processes have been defined, and the involvement of several motor proteins, including kinesin, dynein, and myosin, has been demonstrated. However, little is known about how these motors interact with various organelles.
In the budding yeast Saccharomyces cerevisiae, cell growth and thus secretion is asymmetrically directed toward buds and also toward the tips of mating pheromone-induced projections. In addition, subcellular organelles are partitioned between mother and bud via directed movement of a portion of the organelle into the growing bud. We recently have begun to elucidate the molecules required for partitioning of the yeast lysosome (vacuole). Vacuole partitioning requires actin, profilin, an armadillo repeat protein (Vac8p), and the class V unconventional myosin Myo2p (5, 6). This process is dynamic and coordinated with the cell cycle (7, 8). Before bud emergence, a small portion of the vacuole is polarized along actin cables to the presumptive bud site (5, 6). The vacuole then extends into the emerging bud via a tubular-vesicular segregation structure. In certain mutant alleles of actin and myo2, this directed movement of the vacuole does not occur, suggesting that the vacuole segregation structure is moved along an actin track by a myosin motor. The locations of actin cables and Myo2p relative to the vacuole membrane are consistent with this model (5).
In addition to its role in directed vacuole transport, Myo2p is implicated in post-Golgi vesicle transport and polarized growth. At the nonpermissive temperature, myo2–66 fails to form new buds and accumulates 80- to 100-nm vesicles in the mother cell (9, 10). Accordingly, myo2–66 is lethal in combination with a subset of late-acting secretory pathway (sec) mutants (10). In addition, Myo2p has been implicated in transport of the chitosome to the plasma membrane (11). myo2–66 displays a disrupted actin cytoskeleton at the nonpermissive temperature (12), leading to the suggestion that Myo2p may act in these processes by organizing the actin cytoskeleton.
The class V unconventional myosins chicken brain myosin-V, mouse dilute, and yeast MYO2 are involved in polarized movement of membranous organelles (9, 10, 13). Mouse dilute mutants have a lightened coat color and die of seizures a few weeks after birth. This lightened coat color is caused by defects in melanosome transport to dendritic processes in melanocytes. The seizures also likely are caused by defects in organelle movement. dilute mice and rats lack the usual extensions of smooth endoplasmic reticulum into dendritic spines of brain Purkinje cells (14, 15), yet the morphology and cytoskeletal organization of growth cones appears normal (16). Additionally, myosin-V has been shown to associate with synaptic vesicles and may be required for Ca2+-dependent exocytosis (17).
The class V myosins are distinguished by a unique tail that contains a coiled-coil region thought to mediate dimerization and a globular domain postulated to mediate myosin–cargo interactions through specific receptors (18, 19). In this report, we describe vac15–1/myo2–2, a point mutation in the globular tail domain of Myo2p that causes a pronounced vacuole inheritance defect and mislocalization of Myo2p without severe deleterious effects on cell growth. Fractionation studies demonstrate that wild-type Myo2p but much less of the mutant myo2–2 gene product associates with the vacuole. These data strongly support the hypothesis that Myo2p is the motor for vacuole movement into the bud. Moreover, the results provide strong genetic evidence that the globular tail domain mediates specific interactions of Myo2p with membranes.
METHODS
Yeast Strains and Media.
Yeast strains are related to LWY7235 (MATa, ura3–52, leu2–3, 112, his3-Δ200, trp1-Δ901, lys2–801, suc2-Δ9) (20) unless otherwise specified. LWY2947 (myo2Δ∷TRP1, pMYO2) was generated by using pΔM∷TRP1 as described (12). Other strains are NY57 (MATa, ura3–52, sec9–4), NY64 (sec15–1), NY130, NY405 (sec4–8), and NY878 (sec7–1) (10) and SLY55 (MATa, ura3, leu2, his4, smy1Δ∷LEU2) (21). RH2580 (MATa, cmd1–1, his3, his4, ura3, leu2, trp1, bar1) was obtained from H. Riezman (Univ. of Basel). Unless specified, yeast were grown at 24°C in 1% yeast extract, 2% peptone, and 2% dextrose medium. Standard yeast genetic techniques were used (22).
Plasmids.
For pRS413-MYO2, a 5-kilobase ClaI fragment containing MYO2 was subcloned from pMYO2 (9) into pRS413. The 1.6-kilobase EcoRI fragment containing the carboxyl terminus of MYO2 was removed to make pMYE, and the 90-bp AflII fragment was removed to make pMYA.
Immunofluorescence Labeling.
Indirect immunofluorescence was performed as described (5). Cells were fixed for 2–3 h in 3.7% formaldehyde in the culture medium. The cell wall then was removed by digestion with oxalyticase (Enzogenetics, Corvallis, OR) in SP buffer (1.2 M sorbitol/0.1 M potassium phosphate, pH 6.5/1% 2-mercaptoethanol). Cells were applied to polyethyleneimine coated 8-well slides (ICN) and were treated for 6 min in −20°C methanol and then for 30 s in −20°C acetone. For Myo2p staining, affinity-purified rabbit polyclonal Myo2p antiserum (12) was used at a 1:10 dilution and was incubated for 4 h. The Myo2p antibody was detected with goat anti-rabbit IgG (1:200) followed by CY2-conjugated donkey anti-goat IgG (1:400) (Jackson ImmunoResearch). For actin localization, goat anti-yeast actin serum was affinity-purified against yeast actin bound to Affigel-15 resin (Bio-Rad). This antibody was used at a 1:200 dilution and detected with CY2-conjugated donkey anti-goat IgG (1:400). Vacuole membranes were visualized with mouse mAb against the 60-kDa subunit of the vacuolar ATPase (Molecular Probes) (1:50) detected with Lissamine Rhodamine conjugated donkey anti-mouse IgG (Jackson ImmunoResearch).
In Vivo Labeling of Vacuoles.
Vacuoles were labeled in vivo with N-(3-triethylammoniumpropyl)-4-(6–(4-(diethylamino)phenyl)hexatrienyl) pyridinium dibromide (FM4–64) (Molecular Probes) as described (5). FM4–64 was added to log phase cultures to a final concentration of 80 μM. Cultures labeled in synthetic medium were buffered with 20 mM Pipes (pH 6.8). After 1 h of labeling, the label was chased in fresh liquid medium. For the cell sorting experiment, 1-ml samples were removed at indicated times and were kept on ice in the dark. Analysis was performed with an EPICS 753 fluorescence-activated cell sorter (Coulter) with an excitation 488 nm and an emission of 670/14.
Confocal and Conventional Microscopy.
Confocal images for vacuole polarization studies were obtained with an MRC-1024 laser scanning confocal (Bio-Rad) mounted on a Nikon Optiphot microscope. For each field, a z-series of 5–10 1-μm slices was scanned and projected to a single view by using the MRC-1024 lasersharp software (Bio-Rad). Adobe photoshop (Adobe Systems, Mountain View, CA) was used for further processing of images. All other microscopy used a Zeiss Axioskop with an MC-100 35-mm camera. Actin and Myo2p images were captured on TMAX-100 film, and FM4–64 images were captured on TMAX-400 film. Equal exposure times were used for any images to be compared.
Yeast Fractionation and Vacuole Purification.
Subcellular fractionation was performed as described (6). Cells were grown to mid-log phase (OD600 0.5–1.0) and were harvested, washed, and resuspended in ice-cold lysis buffer (0.3 M sorbitol/10 mM Tris/0.1 M NaCl/1 mM MgCl2/1 mM EDTA, pH 7.5) plus 1 mM phenylmethylsulfonyl fluoride and 1× Complete protease inhibitor mixture (Boehringer Mannheim). Cells were vortexed with glass beads. The resultant extract was cleared of cell debris by centrifugation for 5 min at 500 × g. Subsequent centrifugation (10 min at 13,000 × g) generated a low-speed pellet (P13). The supernatant then was centrifuged at 100,000 × g for 1 h to generate a high-speed pellet (P100) and supernatant (S100).
Vacuoles were purified by flotation on a Ficoll (Sigma) step-gradient as described (23) except a modified buffer (10 mM Na-Pipes, pH 6.8/0.2 M sorbitol/25 mM KCl/1 mM MgCl2/1 mM EGTA) was used. Phenylmethylsulfonyl fluoride (1 mM) and 1× Complete protease inhibitor mixture were added to the cell suspension and the 0% Ficoll layers of the gradient. After removal from the gradient, vacuoles were pelleted by centrifugation for 10 min at 13,000 × g and were washed twice in the gradient buffer.
Western Blot Analysis.
SDS/PAGE sample buffer (24) was added to equal OD600 units of cell lysate (fractionation) or protein amounts as judged by Bradford assay (purified vacuoles). Samples were heated at 80°C for 3 min and then were loaded on a 5% polyacrylamide SDS gel. After electrophoresis, proteins were transferred to Hybond ECL nitrocellulose (Amersham) at 30 V for 22 h.
For detection of Myo2p, the membrane was incubated overnight with affinity-purified Myo2p antibody (1:100) then with horseradish peroxidase-conjugated goat anti-rabbit IgG (1:2500) (Bio-Rad). Bands were visualized by using ECL (Amersham). For detection of alkaline phosphatase (ALP), the membrane was incubated with rabbit polyclonal ALP antibody (1:40,000) followed by horseradish peroxidase-conjugated goat anti-rabbit IgG (1:50,000).
RESULTS
vac15–1 Is Suppressed by MYO2.
We isolated vac15–1 through methanesulfonic acid ethyl ester mutagenesis followed by a fluorescence-activated cell sorting enrichment for cells defective in vacuole inheritance as described (25). vac15–1 cells display normal vacuole morphology and a severe vacuole inheritance defect (Fig. 1A). vac15–1 cells are viable at a wide range of growth temperatures (18°C to 37°C; data not shown).
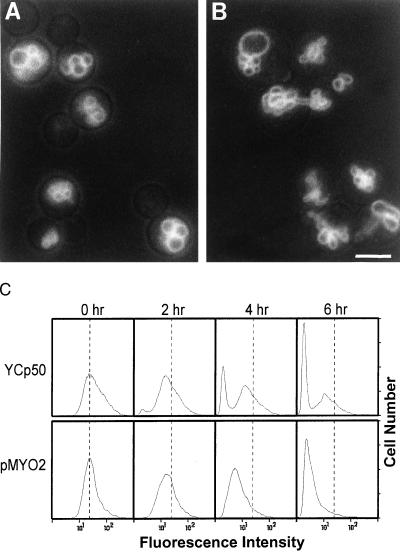
Figure 1.
The vac15–1/myo2–2 segregation defect is corrected by MYO2 on a low copy plasmid. Cells of strain LWY1655 (vac15–1/myo2–2) carrying YCp50 (vector) (A) or pMYO2 (B) were labeled with the vacuole specific dye FM4–64. After 1 h of labeling, the cells were washed, and the label was chased for 3 h in fresh synthetic media lacking uracil. (Bar = 5 μm.) (C) Flow cytometric analysis. Cells were labeled as above, and the label was chased for the indicated timepoints. For each panel, 10,000 cells were analyzed.
As an initial determination of whether vac15–1 represented a mutation in a gene already known to affect vacuole inheritance, vac15–1 cells were transformed with a low copy plasmid containing MYO2 or with the vector alone. Cells were labeled with FM4–64, a dye stably maintained in the vacuole membrane (26), and the label was chased for approximately one cell doubling. As expected, vac15–1 cells containing the vector plasmid displayed the vacuole inheritance defect. Most buds lacked fluorescent label, indicating that they had not received vacuolar material from the mother cells, and no segregation structures projecting from the mother vacuoles were seen (Fig. 1A). In contrast, vac15–1 cells containing the MYO2 plasmid displayed wild-type vacuoles. All buds contained a labeled, inherited vacuole, and many segregation structures connecting the mother and bud vacuoles were present (Fig. 1B).
Complementation of the vac15–1 inheritance defect by MYO2 in a larger population of cells was examined by fluorescence-activated cell sorting. Cells were labeled and chased as described above. Samples were removed at specific times and were analyzed (Fig. 1C). The results corroborate those obtained by fluorescence microscopy. After 2 h of chase, vac15–1 carrying the vector plasmid displayed the bimodal distribution characteristic of vac mutants (25). Both a brightly labeled peak of mother cells and a low fluorescence-intensity peak of daughter cells were seen. The brightly labeled peak remained at a near-constant intensity throughout the chase, indicating that the labeled mother cells retained most of their label. Conversely, vac15–1 carrying the MYO2 plasmid displayed a wild-type distribution of labeled cells. A single peak of cells decreased in fluorescence intensity as the cells doubled, indicating that fluorescent material was divided between mother and daughter cells.
Linkage analysis was performed to further investigate the relationship of vac15–1 to MYO2. vac15–1 was mated with myo2–66, and 15 of the resultant tetrads were examined. No wild-type progeny were recovered, demonstrating that vac15–1 is closely linked to the MYO2 gene locus.
vac15–1/myo2–2 Specifies a Mutation in the Tail Domain of Myo2p.
The vac15–1 allele was recovered from the genome of LWY1655 by gap repair of pRS413-MYO2 with the 3.3-kilobase BstEII-AflII fragment of MYO2 removed. The resultant plasmid, pNLC1, did not complement the myo2–2 vacuole inheritance defect but was able to support growth in a myo2Δ strain. Sequencing from BstEII to ClaI (beyond the carboxyl-terminus of MYO2) identified a single point mutation that alters the ggt codon for Gly1248 to gat, which encodes aspartic acid. This mutation is in the globular region of the MYO2 tail domain, which has been suggested to function in vesicle binding (18).
To confirm that the vac15–1 defect is caused by a single mutation in the MYO2 tail and not an additional mutation outside of the sequenced fragment, the mutant gene was regenerated by subcloning an EcoRI fragment of pNLC1 containing the tail region into pMYE. This construct behaved identically to pNLC1. These data confirm that vac15–1 is an allele of MYO2; accordingly, we refer to vac15–1 as myo2–2.
The MYO2 Tail Domain Is Essential.
Defects caused by the myo2–2 mutation support the idea that the tail domain mediates interaction with cargo. To further investigate the role of this domain, we generated two deletion plasmids and transformed them into myo2Δ containing pMYO2. pMYE is missing the globular tail domain. pMYA has a small, in-frame 30-aa deletion in the tail domain. Loss of the URA3-selected wild-type MYO2 plasmid pMYO2 on media supplemented with 5-fluoroorotic acid resulted in inviability for strains transformed with pMYA or pMYE (Fig. 2). Thus, neither deletion is capable of supporting growth as the sole copy of MYO2, indicating that an intact tail is required for Myo2p function. Moreover, Western blot analysis of cell extracts using antibody against the Myo2p motor domain suggests that both Myo2p truncations are expressed. The requirement for an intact tail domain implies that the myo2–2 mutation does not grossly disrupt the structure or function of this domain.
Figure 2.
The MYO2 tail is essential for viability. LWY2947 (myo2Δ∷TRP1, pMYO2) cells were transformed with pRS413-MYO2, pMYE, or pMYA. Transformants were streaked onto synthetic media lacking histidine and uracil (sc-his-ura) and synthetic media containing 5-fluoroorotic acid (5 FOA). Cells containing pMYE or pMYA as their sole copy of MYO2 are not viable.
The myo2–2 Gene Product Is Localized Improperly.
In wild-type cells, Myo2p concentrates in caps at the presumptive bud site and tips of small buds, at tips of mating pheromone-induced projections, and at the bud neck before cytokinesis (12, 27). These areas correspond to sites of active growth. Additionally, Myo2p antibody stains cytoplasmic spots (12) that are often coincident with the vacuolar membrane (5).
We examined Myo2p localization in myo2–2 by immunofluorescence. Surprisingly, the myo2–2 gene product does not concentrate at sites of active growth (Fig. 3F). Instead, only the punctate cytoplasmic staining is seen. Myo2p localization is similar in a myo2–2/myo2–2 homozygous diploid (data not shown). Western blot analysis confirmed that the antibody against Myo2p recognizes the myo2–2 gene product and that the mutant protein is expressed at a normal level (data not shown).
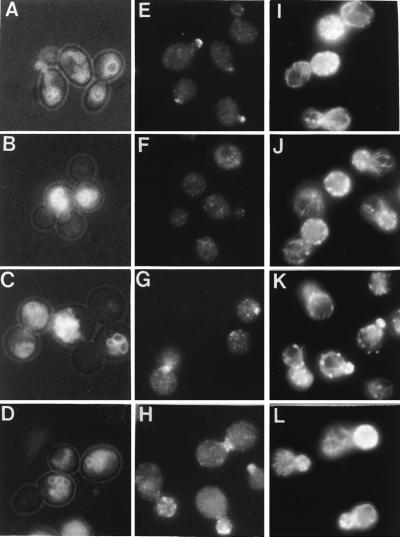
Figure 3.
The myo2–2 gene product does not concentrate at sites of active growth. Shown are LWY2947 cells carrying a plasmid with the indicated myo2 allele: MYO2 (A, E, and I), myo2–2 (B, F, and J), myo2–4 (C, G, and K), and myo2–5 (D, H, and L). (A–D) FM4–64 labeling. The vacuole inheritance defect of myo2–4 and myo2–5 cells is similar to that of myo2–2. (E–L) Indirect immunofluorescence. Cells were fixed and stained with antibody against Myo2p (E–H) or antibody against yeast actin (I–L). Although the myo2–2 gene product localizes abnormally, myo2–2 cells have a normal actin cytoskeleton.
To address the question of whether MYO2 tail mutations could cause vacuole inheritance defects independent of mislocalization, we investigated Myo2p localization in myo2 mutants generated by random mutagenesis of the MYO2 tail region (N. Hutchings and L.S.W., unpublished data). Four strains with vacuole inheritance defects similar to myo2–2 (Fig. 3 C and D, and data not shown) were examined. Two strains (myo2–4 and myo2–5) displayed wild-type localization of Myo2p (Fig. 3 G and H) and two (myo2–6 and myo2–7) displayed defects in Myo2p localization similar to myo2–2 (data not shown). This observation suggests that Myo2p tail determinants for vacuole inheritance are distinct from determinants of localization to sites of polarized growth.
One proposed role for Myo2p is organization of the actin cytoskeleton (12). Thus, we investigated actin organization in these mutants. If localization of Myo2p at sites of active growth is necessary for organization or maintenance of a normal actin cytoskeleton, myo2–2 cells should display an abnormal actin cytoskeleton. However, the actin organization in myo2–2 and the other myo2 mutants was indistinguishable from wild-type (Fig. 3 I, J, K, and L; for myo2–6 and myo2–7, data not shown). Additionally, we investigated the localization of the myo2–2 gene product in mating projections (Fig. 4). Cells were treated with mating pheromone. The myo2–2 gene product does not localize to the tips of mating projections, although these projections are readily formed.
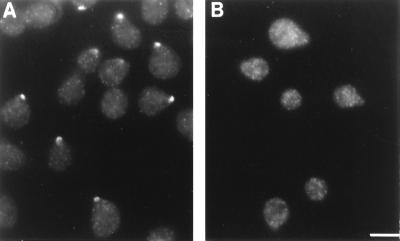
Figure 4.
Localization of Myo2p in cells responding to mating pheromone. LWY7235 (wild-type) (A) and LWY1655 (myo2–2) (B) cells were treated with 10 μg/ml α factor for 2 h and then were fixed and stained with antibody against Myo2p. Although myo2–2 cells readily form projections, the myo2–2 gene product does not localize to the tip. (Bar = 5 μm.)
myo2–2 Does Not Display the Same Synthetic Lethal Interactions as myo2–66.
Synthetic lethal interactions between myo2–66 and a variety of late secretory (sec) and other mutants (10, 21, 27, 28, 29) suggests the involvement of Myo2p in secretion, polarized growth, and organization of the actin cytoskeleton. These synthetic lethal interactions raise the possibility that the vacuole inheritance defect in myo2–66 may be indirect. Therefore, we crossed myo2–2 with sec2–41, sec4–8, sec9–4, sec7–1, sec15–1, cmd1–1, and Δsmy1 strains and sporulated the resultant diploids. All crosses produced viable double mutants with the same defects seen in the original single mutants. To confirm that the viability differences between myo2–2 and myo2–66 double mutants was not caused by strain background differences, we crossed LWY2752 (myo2–66) with sec2–41, sec9–4, sec7–1, sec15–1, and Δsmy1 and examined at least 10 tetrads from each. Patterns of viability were consistent with earlier reports. These results suggest that the myo2–2 mutation in the Myo2p tail causes specific defects in vacuole inheritance and localization of Myo2p, in contrast to the actin-binding domain mutation myo2–66, which appears to disrupt several processes.
myo2–2 Cells Are Defective in Vacuole Migration to the Presumptive Bud Site.
Before bud emergence, the vacuole aligns along actin cables and a portion juxtaposes next to the ring of cortical actin that marks the new bud site (5, 6). This is the earliest identified landmark in vacuole inheritance. To determine whether myo2–2 cells are defective in this event, we performed double-label immunofluorescence of the vacuole membrane and actin. Unbudded cells in which the actin cytoskeleton was clearly polarized were examined. Cells were scored as containing polarized vacuoles if the vacuole was positioned adjacent to the presumptive bud site (Fig. 5 A, B, E, and F). Cells displaying a random distribution of the vacuole within the cell with respect to the presumptive bud site were scored as unpolarized (Fig. 5 C, D, G, and H). Cells not clearly categorizable were scored as ambiguous. A population of 69 myo2–2 cells were scored (Table 1) and compared with a previously scored population of wild-type cells (6). As an internal control, wild-type cells were labeled in parallel, and a small population was scored. Cells from wild-type and myo2–2 populations were randomized and scored “blind.” A majority of myo2–2 cells had an unpolarized vacuole. Moreover, the extent of the defect is similar to that seen for vac8Δ cells (6), suggesting that Myo2p and Vac8p function at a similar early step in vacuole inheritance.
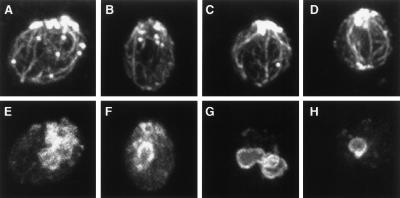
Figure 5.
myo2–2 cells are defective in polarization of the vacuole to the presumptive bud site. Cells were processed for immunofluorescence and were double-labeled with actin antibody (A–D) and antibody against the 60-kDa vacuolar ATPase (E–H). (A, B, E, and F) LWY7235 (wild-type). (C, D, G, and H) LWY2599 (myo2–2).
Table 1.
Polarization of the vacuole in unbudded cells
| Polarized | Unpolarized | Ambiguous | |
|---|---|---|---|
| Wild-type, n = 112 | 73% | 26% | 1% |
| myo2–2, n = 69 | 22% | 57% | 22% |
Cultures of wild-type and myo2–2 cells were processed for immunofluorescence, and were stained for actin and the vacuole membrane. Confocal images were obtained, and individual unbudded cells were evaluated for polarization of the vacuole to the presumptive bud site.
Some Myo2p Is Vacuole-Associated.
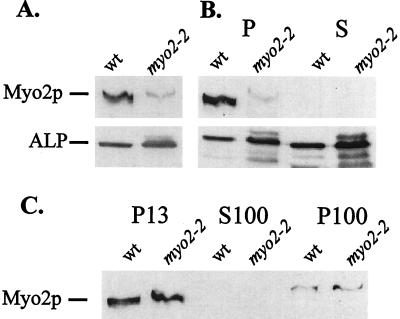
Immunofluorescence experiments demonstrate that some punctate cytoplasmic Myo2p colocalizes with the vacuole membrane and with the leading edge of the segregation structure, usually in the tip of the bud (5). To investigate Myo2p localization further, we analyzed vacuoles prepared on a Ficoll flotation gradient. Wild-type and myo2–2 vacuoles were separated by SDS/PAGE and were immunoblotted for Myo2p and the vacuolar integral membrane protein ALP (Fig. 6A). Myo2p was found in wild-type vacuole preparations. This Myo2p could not be washed off the vacuoles with the isolation buffer but could be removed with the addition of 0.5 M NaCl (data not shown); possibly, Myo2p associates with the vacuole as a peripheral membrane protein. Intensity of the Myo2p signal was compared by densitometry over a range of protein amounts loaded (5–20 μg) to assess relative amounts of wild-type and mutant Myo2p. Results averaged from three experiments suggest that at least four times as much wild-type as myo2–2 protein associates with vacuoles. Both wild-type and mutant Myo2p were found in the Triton X-100 insoluble pellet whereas most of the ALP was extractable (Fig. 6B).
Figure 6.
The myo2–2 gene product is defective in binding to the vacuole. Vacuoles were isolated from LWY7235 (wild-type) and LWY2599 (myo2–2) on a Ficoll gradient. (A) Vacuole protein (10 μg) was separated on a 5% acrylamide SDS gel and was blotted for Myo2p and the vacuolar integral membrane protein ALP. More wild-type Myo2p than myo2–2 gene product cofractionates with the vacuole. (B) Vacuoles were treated with 1% Triton X-100 on ice for 10 min and then were pelleted for 10 min at 12,500 × g. The resulting pellets and supernatants were immunoblotted as described above. Triton X-100 extracts most of the membrane protein ALP into the supernatant, yet Myo2p is insoluble. (C) Cell extracts from LWY7235 and LWY2599 were fractionated by differential centrifugation. Equal OD600 units of each fraction were immunoblotted as described above. Both wild-type Myo2p and the myo2–2 gene product were found primarily in the low speed pellet. The blots shown are representative of at least three experiments.
To assess association of the myo2–2 gene product with other membranes, extracts from wild-type and myo2–2 cells were fractionated by differential centrifugation and were analyzed by immunoblot for Myo2p (Fig. 6C). For both wild-type and myo2–2 cells, most of the Myo2p was found in the low-speed pellet (P13). The P13 contains membranous organelles, including the vacuole, endoplasmic reticulum, and the plasma membrane. These results suggest that both wild-type Myo2p and the myo2–2 gene product associate with membranes, yet the mutant protein is specifically defective in association with vacuoles, which represent only a fraction of the membranes found in the P13. A small amount of Myo2p was found in the high-speed pellet (P100), which contains smaller vesicles. No detectable Myo2p was found in the soluble (S100) fractions.
DISCUSSION
Genetic Evidence of Myo2p Tail Domain Function.
The globular carboxyl-terminal tail domain of class V unconventional myosins has been proposed to mediate myosin-V localization within the cell and/or binding to vesicular cargo, perhaps through cargo-specific receptors (18). In the case of yeast Myo2p, this region is required for essential functions. Characterization of vac15–1/myo2–2 demonstrates that selected defects in myosin-V function can result from a single amino acid substitution in the globular tail domain. In contrast to myo2–66, a mutation in the actin-binding domain causing temperature sensitivity and multiple other defects, myo2–2 has defects in vacuole movement and localization of Myo2p to regions of polarized growth. These specific phenotypes, together with detection of the mutant Myo2p by both immunofluorescence and immunoblotting, indicate that the mutation does not grossly affect protein structure or stability.
In a simple model, full function of the myosin-V actin-binding head domain would be required generally for movement whereas the tail domain may interact differently with various cargoes. Thus, a head-domain mutation, like myo2–66, might cause defects in all Myo2p-dependent processes whereas a tail-domain mutation like myo2–2 could cause a relatively narrow range of defects. Other point mutations in the Myo2p tail might affect other functions of Myo2p.
Studies in other systems also support a role for the tail domain in myosin-V interaction with cargo. Biochemical evidence suggests that the tail domain mediates myosin-V association with synaptic vesicles (17), and analysis of mouse dilute tail-domain mutants supports the idea that specific regions of the tail domain are important for selected cargo interactions (30).
A Direct Role for Myo2p in Vacuole Movement.
Identification and characterization of myo2–2 provides critical support to the hypothesis that Myo2p serves as the vacuolar motor. myo2–2 cells grow relatively normally and do not display many of the defects seen in myo2–66, suggesting that vacuole partitioning is not a secondary defect. Moreover, we now demonstrate that Myo2p fractionates with vacuoles and that the myo2–2 gene product is defective in this association. Finally, we have generated additional MYO2 tail mutants that display vacuole inheritance defects. Together, these observations strongly suggest that Myo2p has a direct role in vacuole inheritance.
It seems likely that Myo2p acts as a conventional motor transporting the vacuole and other organelles and not solely as a tether for cargo that has been transported by other means. The gene products of myo2–66 and two of our additional myo2 tail mutant alleles localize normally at the permissive temperature (ref. 12 and Fig. 3), yet vacuole inheritance is impaired severely in these cells (5). Thus, proper localization of Myo2p to the bud tip is not sufficient for vacuole movement. Moreover, overexpression of the myo2–66 suppressor SMY1 (21) in myo2–2 cells appears to restore localization of the mutant gene product to sites of polarized growth (K. Beningo, S. Lillie, and S. Brown, personal communication) yet does not affect the vacuole inheritance defect (unpublished data). Myo2p association with the vacuole also suggests that Myo2p does not merely act in the bud tip as a tether for the vacuole. Moreover, a similar localization of the mammalian class V myosin, dilute, to lysosomes and melanosomes has been demonstrated (31, 32), suggesting that Myo2p and dilute function in an analogous fashion.
Localization of Myo2p Is Not Required for its Essential Function.
Myo2p localizes most prominently at areas of polarized growth, and this localization was presumed to be important for Myo2p function (12). Thus, it is surprising to find that, in myo2–2 cells, the mutant Myo2p does not concentrate at these sites. The relatively normal growth of myo2–2 cells despite the mislocalization of Myo2p suggests that concentration of Myo2p at sites of polarized growth is not required for its essential functions. Furthermore, Myo2p does not act at these sites to organize actin or promote polarized growth. Moreover, calcofluor staining of chitin reveals normal axial distribution of bud scars in myo2–2 haploids and a random budding pattern in diploids (unpublished data) in contrast to the delocalized chitin distribution seen in myo2–66 cells at the nonpermissive temperature.
The mislocalization of myo2–2 protein suggests that the MYO2 tail is important for both interactions with cargo like the vacuole and concentration of Myo2p at sites of active growth. This second role is consistent with reports that some Myo2p localizes to sites of active growth even in the absence of an intact actin cytoskeleton (33). Thus, Myo2p localization is not entirely a consequence of transport along actin but may reflect interactions with proteins at sites of polarized growth.
How Does Myo2p Associate with the Vacuole?
Consistent with Myo2p colocalization with vacuoles via immunofluorescence (5), we find that Myo2p fractionates with isolated vacuoles. Moreover, much less mutant Myo2p from myo2–2 cells is present on the vacuole. These data suggest that the glycine to aspartic acid change in the tail disrupts Myo2p interaction with the vacuole. Myo2p can be extracted from vacuoles by high salt (data not shown) and thus may associate with the vacuole as a peripheral membrane protein, perhaps via a protein receptor. Of interest, Myo2p cannot be extracted from the vacuole membrane with Triton X-100. Similarly, organelle-associated vertebrate myosin-V is resistant to extraction by Triton X-100 (16, 34). Triton X-100 insolubility may indicate that Myo2p associates with the vacuole through vacuole-associated actin. Indeed, we have demonstrated actin association with isolated vacuoles by both fluorescence microscopy and immunoblotting (E. Kauffman and L.S.W., unpublished results). However, a simple association of Myo2p with actin cannot account for its fractionation in the P13. Under the conditions used for differential fractionation, the majority of actin fractionates in the S100, where no Myo2p is detected. A second model is that the Triton X-100 insolubility may result from Myo2p receptor association with a specialized membrane domain. Triton X-100-insoluble patches on the plasma membrane have been described (35). This model may explain how a specific portion of the vacuole membrane is recruited to move and become the segregation structure.
The identity of vacuolar or other receptors for myosins are unknown. However, many vac mutants with defects similar to myo2–2 have been isolated (ref. 26 and unpublished data), some of which may represent defects in the vacuolar Myo2p receptor or other components of the transport machinery. Furthermore, genetic screens based on the use of myo2–2 may provide a valuable approach toward identification of the vacuolar Myo2p receptor.
Acknowledgments
We thank Fusheng Tang for construction of pMYE and pMYA, Nathan Hutchings for random mutagenesis of the MYO2 tail region, Cecilia Bonangelino, Yong-Xu Wang, and Dr. Kent Hill for helpful discussions, and Emily Kauffman and Grant Gerdes for excellent technical assistance. We are grateful to Justin Fishbaugh and the University of Iowa Flow Cytometry Facility for help with cell sorting and analysis, Tom Monninger and the University of Iowa Central Microscopy Research Facility for use of the confocal microscope, Drs. Sue Lillie, Susan Brown, and Karen Beningo for strains, pΔM∷TRP1, and affinity-purified Myo2p antibody and for sharing unpublished results, Drs. Howard Riezman and Peter Novick for strains, Samara Reck-Peterson and Dr. Mark Mooseker for affinity-purified Myo2p motor domain antibody, and Drs. John Cooper and Scott Emr for actin and ALP antisera, respectively. This work was supported by the following awards to L.S.W.: National Science Foundation CAREER Award MCB 96-00867, National Institutes of Health Grant GM50403, and a gift from the Carver Charitable Trust. N.L.C. was supported in part by National Institutes of Health/National Institute on Aging Grant T32 AG 00214 to the Interdisciplinary Research Training Program on Aging, University of Iowa.
ABBREVIATIONS
- ALP
alkaline phosphatase
- FM4–64
N-(3-triethylammoniumpropyl)-4-(6–(4-(diethylamino)phenyl)hexatrienyl) pyridinium dibromide
References
- 1.Herman B, Albertini D F. Nature (London) 1983;304:738–740. doi: 10.1038/304738a0. [DOI] [PubMed] [Google Scholar]
- 2.Phaire-Washington L, Silverstein S C, Wang E. J Cell Biol. 1980;86:641–685. doi: 10.1083/jcb.86.2.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pierre P, Turley S J, Gatti E, Hull M, Meltzer J, Mirza A, Inaba K, Steinman R M, Mellman I. Nature (London) 1997;388:787–792. doi: 10.1038/42039. [DOI] [PubMed] [Google Scholar]
- 4.Hollenbeck P J. Front Biosci. 1996;1:D91–D102. doi: 10.2741/a118. [DOI] [PubMed] [Google Scholar]
- 5.Hill K L, Catlett N L, Weisman L S. J Cell Biol. 1996;135:1535–1549. doi: 10.1083/jcb.135.6.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y-X, Catlett N L, Weisman L S. J Cell Biol. 1998;140:1063–1074. doi: 10.1083/jcb.140.5.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weisman L S, Bacallao R, Wickner W. J Cell Biol. 1987;105:1539–1547. doi: 10.1083/jcb.105.4.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weisman L S, Wickner W. Science. 1988;241:589–591. doi: 10.1126/science.3041591. [DOI] [PubMed] [Google Scholar]
- 9.Johnston G C, Prendergast J A, Singer R A. J Cell Biol. 1991;113:539–551. doi: 10.1083/jcb.113.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Govindan B, Bowser R, Novick P. J Cell Biol. 1995;128:1055–1068. doi: 10.1083/jcb.128.6.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Santos B, Snyder M. J Cell Biol. 1997;136:95–110. doi: 10.1083/jcb.136.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lillie S H, Brown S S. J Cell Biol. 1994;125:825–842. doi: 10.1083/jcb.125.4.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Provance D W, Jr, Wei M, Ipe V, Mercer J A. Proc Natl Acad Sci USA. 1996;93:14554–14558. doi: 10.1073/pnas.93.25.14554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dekker-Ohno K, Hayasaka S, Takagishi Y, Oda S, Wakasugi N, Mikoshiba K, Inouye M, Yamamura H. Brain Res. 1996;714:226–230. doi: 10.1016/0006-8993(95)01560-4. [DOI] [PubMed] [Google Scholar]
- 15.Takagishi Y, Oda S, Hayasaka S, Dekker-Ohno K, Shikata T, Inouye M, Yamamura H. Neurosci Lett. 1996;215:169–172. doi: 10.1016/0304-3940(96)12967-0. [DOI] [PubMed] [Google Scholar]
- 16.Evans L L, Hammer J, Bridgman P C. J Cell Sci. 1997;110:439–449. doi: 10.1242/jcs.110.4.439. [DOI] [PubMed] [Google Scholar]
- 17.Prekeris R, Terrian D M. J Cell Biol. 1997;137:589–1601. doi: 10.1083/jcb.137.7.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mooseker M, Cheney R. Annu Rev Cell Dev Biol. 1995;11:633–675. doi: 10.1146/annurev.cb.11.110195.003221. [DOI] [PubMed] [Google Scholar]
- 19.Cheney R E, O’Shea M K, Heuser J E, Coelho M V, Wolenski J S, Espreafico E M, Forscher P, Larson R E, Mooseker M S. Cell. 1993;75:13–23. doi: 10.1016/S0092-8674(05)80080-7. [DOI] [PubMed] [Google Scholar]
- 20.Bonangelino C J, Catlett N L, Weisman L S. Mol Cell Biol. 1997;17:6847–6858. doi: 10.1128/mcb.17.12.6847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lillie S H, Brown S S. Nature (London) 1992;356:358–361. doi: 10.1038/356358a0. [DOI] [PubMed] [Google Scholar]
- 22.Kaiser C, Michaelis S, Mitchell A. Methods in Yeast Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1994. [Google Scholar]
- 23.Condradt B, Shaw J, Vida T, Emr S, Wickner W. J Cell Biol. 1992;119:1469–1479. doi: 10.1083/jcb.119.6.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y-X, Zhao H, Harding T M, Gomes de Mesquita D S, Woldringh C L, Klionsky D J, Munn A L, Weisman L S. Mol Biol Cell. 1996;7:1375–1389. doi: 10.1091/mbc.7.9.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vida T A, Emr S D. J Cell Biol. 1995;128:779–792. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brockerhoff S E, Stevens R C, Davis T N. J Cell Biol. 1994;124:315–323. doi: 10.1083/jcb.124.3.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hermann G J, King E J, Shaw J M. J Cell Biol. 1997;137:141–153. doi: 10.1083/jcb.137.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu H, Bretscher A. J Cell Biol. 1992;118:285–299. doi: 10.1083/jcb.118.2.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang J-D, Mermall V, Strobel M C, Russell L B, Mooseker M S, Copeland N G, Jenkins N A. Genetics. 1998;148:1963–1972. doi: 10.1093/genetics/148.4.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nascimento A A C, Amaral R G, Bizario J C S, Larson R E, Espreafico E M. Mol Biol Cell. 1997;8:1971–1988. doi: 10.1091/mbc.8.10.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu X, Bowers B, Wei Q, Kocher B, Hammer J A., III J Cell Sci. 1997;110:847–859. doi: 10.1242/jcs.110.7.847. [DOI] [PubMed] [Google Scholar]
- 33.Ayscough K R, Stryker J, Pokala N, Sanders M, Crews P, Drubin D G. J Cell Biol. 1997;137:399–416. doi: 10.1083/jcb.137.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Evans L L, Lee A J, Bridgman P C, Mooseker M S. J Cell Sci. 1998;111:2055–2066. doi: 10.1242/jcs.111.14.2055. [DOI] [PubMed] [Google Scholar]
- 35.Simons K, Ikonen E. Nature (London) 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]