Abstract
Stabilization of biologically active peptides is a major goal in peptide-based drug design. Cyclization is an often-used strategy to enhance resistance of peptides towards protease degradation and simultaneously improve their affinity for targets by restricting their conformational flexibility. Amongst the various cyclization strategies, the use of thioether crosslinks has been successful for various peptides including enkephalin. The synthesis of these thioethers can be arduous, especially for longer peptides. Described herein is an enzymatic strategy taking advantage of the lantibiotic synthetase LctM that dehydrates Ser and Thr residues to the corresponding dehydroalanine and dehydrobutyrine residues and catalyzes the Michael-type addition of Cys residues to form thioether crosslinks. The use of LctM to prepare thioether containing analogs of enkephalin, contryphan, and inhibitors of human tripeptidyl peptidase II and spider venom epimerase is demonstrated.
Interest in peptide-based materials for use in human therapeutics has greatly increased in recent years, and fully synthetic peptide drugs have increasingly reached the clinic.1 The proteolytic instability of peptides still presents a limitation, however, for widespread utilization of peptide therapeutics. An often employed strategy for the design of peptide-based drugs with improved selectivity and decreased proteolytic susceptibility involves cyclization to constrain their conformational flexibility.2 A notable example features the stabilization of enkephalin by the introduction of a thioether crosslink between two alanines (Figure 1), which increased the bioactivity of the compound by several orders of magnitude due to increased biostability.3 Other studies have also shown the increased stability of peptides and proteins by thioether crosslinks.4,5 Thioether crosslinks between two alanine residues are called lanthionines and their synthesis has received much attention.6 Despite important recent advances,7–12 at present it is still difficult to introduce these structures efficiently into synthetic peptides, especially for large peptides. One promising route to these structures is through the biosynthetic machinery for lantibiotics. These compounds are ribosomally synthesized and post-translationally modified antimicrobial peptides.13,14 The first step in the modification process of class II lantibiotics involves phosphorylation of Ser and Thr residues and subsequent elimination of the phosphate group to generate dehydroalanines and Z-dehydrobutyrines, respectively (Figure 2).15 In a succeeding step, intramolecular Michael-type addition by Cys residues to the dehydroamino acids forms the lanthionine (from Ser) and methyllanthionine (from Thr) crosslinks. Lacticin 481 synthetase (LctM) carries out both steps in a regio- and stereoselective fashion as illustrated in Figure 3.16 The enzyme only carries out posttranslational modification of the structural region of its peptide substrate LctA. Although not absolutely required for activity and not modified itself, the leader peptide greatly enhances the efficiency of LctM.17 In previous studies we have shown that LctM tolerates variants of the LctA substrate peptide, including substitution of Ser, Thr, and Cys residues with nonproteinogenic analogs.18–20 Furthermore, we17,18 and Moll and coworkers21–24 have shown that non-lantibiotic peptides attached to the leader sequence of lantibiotics are substrates for the biosynthetic enzymes, and that the leader and structural peptide can be linked by non-peptidic structures. We here report the use of LctM for the preparation of a series of peptides with diverse biological properties including the previously mentioned lanthionine-containing enkephalin derivative, a thioether analog of the conotoxin contryphan (2), and inhibitors of spider venom epimerase (3) and human tripeptidyl exopeptidase II (4) (Figure 1).
Figure 1.
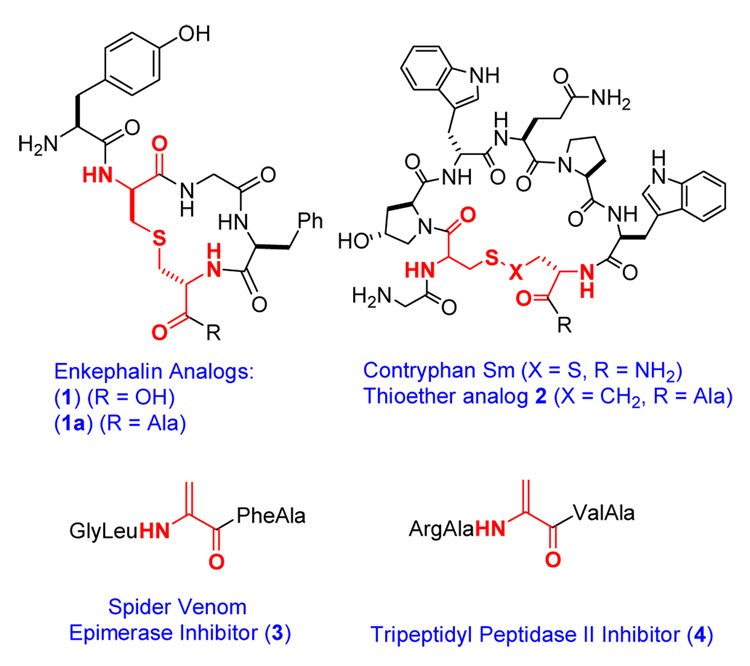
Compounds prepared using lacticin 481 synthetase.
Figure 2.
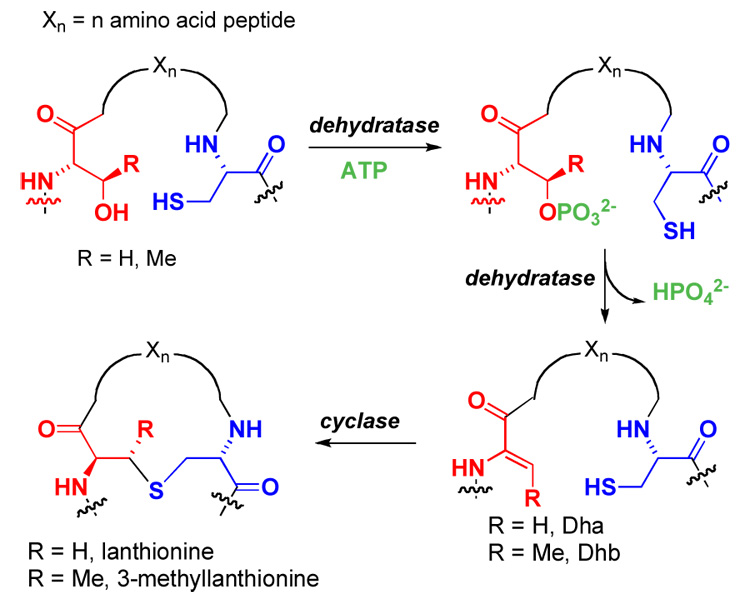
Post-translational modifications in lantibiotic biosynthesis.
Figure 3.

Biosynthetic pathway for the formation of lacticin 481 by the bifunctional enzyme LctM.
Enkephalins are thought to constitute the physiological ligand for the human δ-opioid receptor. The disadvantages of enkephalins as therapeutic agents have been well documented and several approaches have been reported to overcome the rapid proteolytic degradation, poor receptor subtype selectivity, and relatively poor bioavailability. Goodman and coworkers prepared and tested a series of lanthionine containing enkephalin analogs and determined their relative in vivo activity. Structure 1 (Figure 1) displayed an ED50 of 0.0018 nM compared to a value of 15 nM for morphine as an agonist for the μ- and δ-opioid receptors in mouse models. Degradation studies attributed the subnanomolar analgesic potencies to increased biostability.3 In this work we pursued an in vitro enzymatic approach in which the enkephalin peptide was attached to the C-terminus of the lacticin 481 leader peptide with a Lys inserted right before the enkephalin peptide (Figure 4). This Lys allows proteolytic removal of the leader peptide and isolation of the enkephalin analog.
Figure 4.
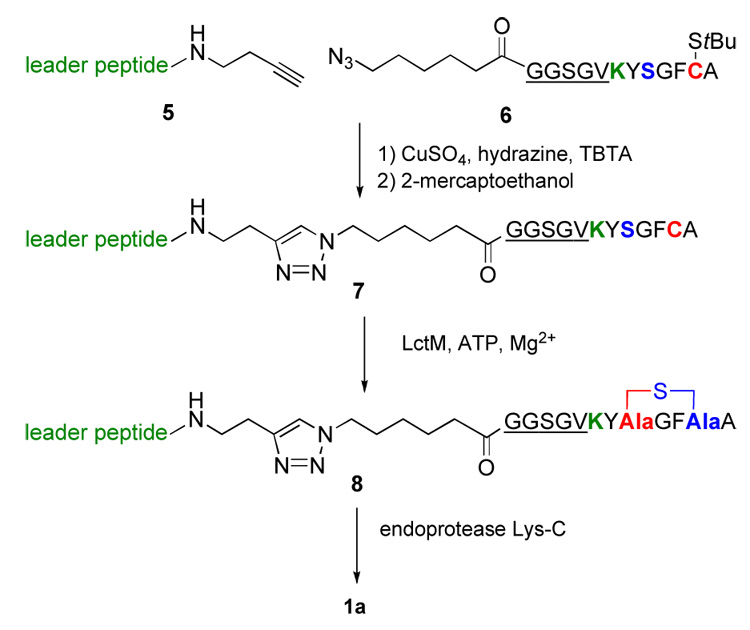
Enzymatic route towards enkephalin analog 1a.
The preparation of the substrate for modification by LctM was achieved using a ligation strategy that relied on the Cu(I)-catalyzed [2+3] cycloaddition25 of an alkyne moiety appended to the C-terminus of the leader peptide17 (5) and an azide attached to the N-terminus of a peptide comprising a linker peptide (underlined), the aforementioned Lys (green), and the cyclic enkephalin precursor (Figure 4, 6). All peptides were prepared by standard Fmoc-based solid phase peptide synthesis. Upon generation of the ligation product, it was incubated with lacticin 481 synthetase in the presence of ATP and MgCl2. Subsequent analysis by MALDI mass spectrometry demonstrated that the peptide had been dehydrated (Supporting Information). Furthermore, treatment with the thiol derivatizing agent p-hydroxymercuribenzoic acid (PMBA)26,27 revealed the absence of a thiol group in the product indicating cyclization had taken place. This conclusion was further supported by the absence of an adduct upon treatment of the enzymatic product with 2- mercaptoethanol, demonstrating that a reactive Michael acceptor was not present. Based on our previous studies as well as unsuccessful in vivo attempts to cyclize this peptide using the nisin biosynthetic machinery,21 it is very likely that the cyclization step is performed by the enzyme but we cannot rule out that some of the cyclic product arises from non-enzymatic background reaction. If so, it would not affect the stereochemistry of cyclization because both the non-enzymatic and enzymatic cyclization exhibit high stereoselectivity for the D-isomer for peptides in which the Dha and Cys are separated by two amino acids,10–12,28 including previous studies on the enkephalin peptide.29 After having established that peptide 8 was cyclized, it was incubated with endoproteinase Lys-C, and the product was purified by HPLC providing a peptide with the expected molecular weight for 1a as determined by ESI MS (Supporting Information).
Conotoxins are constrained, cyclic, post-translationally modified peptides found in the venom of carnivorous cone snails. They usually contain one or more disulfides and have high affinities for various cell surface receptors. As such they have received much attention as potential therapeutics.30 Several approaches have been reported to stabilize the reduction-sensitive disulfide bridges, including the use of diselenide analogs (9)31 or thioethers (10)32 (Figure 5). We decided to explore the use of LctM for the preparation of analogs of conotoxins in which the disulfide bond would be replaced by a thioether linkage of the same length (Figure 5, 11). We envisioned this could be achieved by an enzyme catalyzed Michael-type addition of a homocysteine to a dehydroalanine. The contryphans are a family of peptides containing a single disulfide as well as other posttranslational modifications such as d-Trp and 4-trans-hydroxyPro (O).33–37 The precursor peptide for contryphan-Sm was prepared by standard Fmoc-based SPPS. At the site of the C-terminal Cys of contryphan-Sm, a homocysteine was incorporated protected as an ethyldisulfide. Upon linking peptide 12 to the leader peptide using the Cu(I)-catalyzed [2+3] cycloaddition, the homocysteine residue was deprotected with 2- mercaptoethanol and the resulting peptide 13 was incubated with LctM and ATP/MgCl2. Analysis of the assay product by MALDI-mass spectrometry demonstrated that the peptide had been dehydrated although significantly more enzyme was required for complete conversion. Treatment with PMBA showed that the thiol of homocysteine was not present in the final product, consistent with the formation of a homolanthionine ring structure 14 (Supporting Information). Proteolysis with trypsin resulted in cleavage behind an Arg that had been inserted intentionally.
Figure 5.
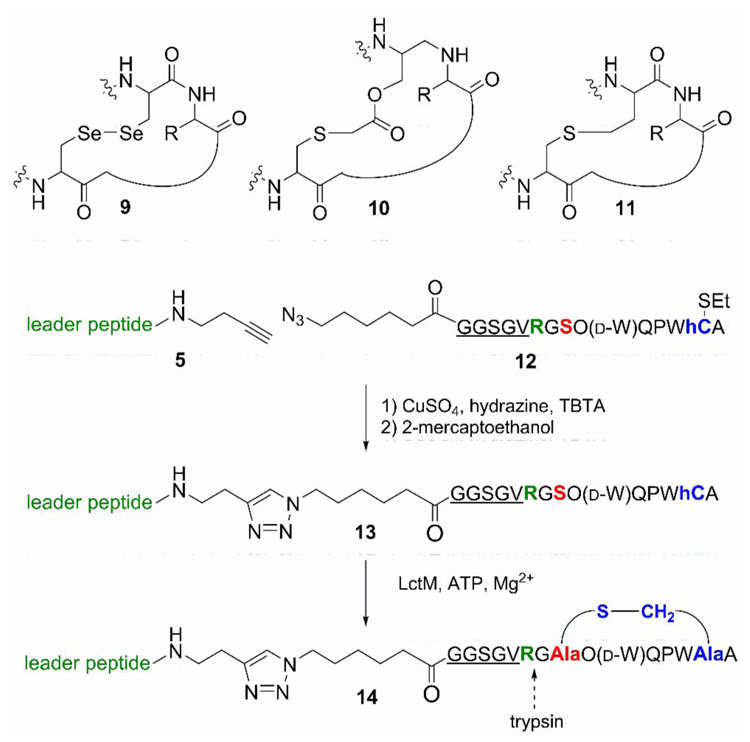
Enzymatic preparation of a thioether analog of contryphan-Sm.
The venom of the funnel web spider Agelenopsis aperta contains a d-serine residue that is incorporated by epimerization of an l-serine in the venom peptide. Tanner and coworkers reported a dehydroalanine containing peptide 3 as a potent inhibitor of the epimerase, prepared by oxidative elimination of a phenylselenocysteine precursor.11,38 Based on our previous work, we anticipated that the dehydropeptide could be prepared rapidly using an enzymatic dehydration. Although the precursor peptide can be prepared using molecular biology techniques because unlike contryphans, it does not contain any nonproteinogenic amino acids, we again used a purely synthetic approach as this allows the possibility for introduction of non-natural residues and therefore more synthetic flexibility. The strategy to prepare peptide 3 followed the scheme depicted in Figure 6. As expected, peptide 15 was a substrate for LctM resulting in the dehydrated product 16. The target dehydropeptide inhibitor 3 was obtained following incubation with endoproteinase Lys-C and HPLC purification, which provided peptide 3 as determined by ESI MS (Supporting Information).
Figure 6.
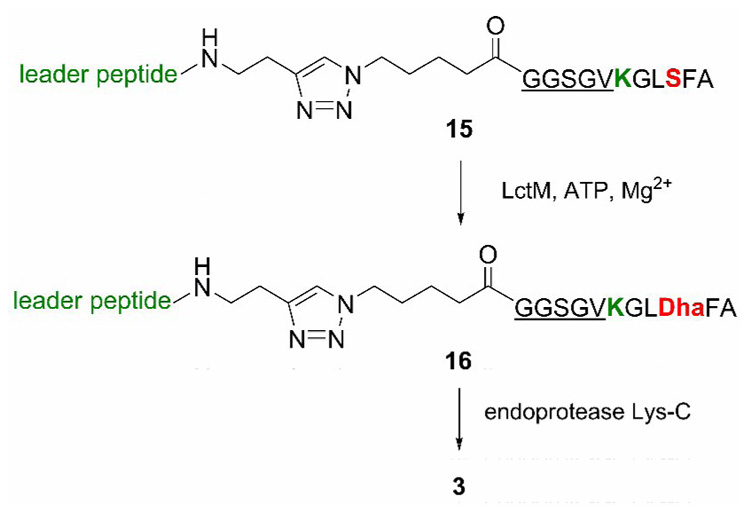
Enzymatic preparation of a snake venom epimerase inhibitor.
In a final application of the use of lacticin 481 synthetase, we focused on human tripeptidyl peptidase II from erythrocytes, a serine peptidase belonging to the subtilisin class. Previous studies have reported the dehydroalanine containing pentapeptide 4 as a potent inhibitor (Ki = 20 nM). The precursor peptide was again prepared by SPPS and Cu(I)-catalyzed ligation, and upon treatment with LctM in the presence of ATP and MgCl2, complete dehydration was observed (Supporting Information).
In summary, this work demonstrates the remarkable versatility of lacticin 481 synthetase. The enzyme efficiently catalyzed the dehydration of Ser residues that vary greatly in their flanking residues as well as the distance to the leader peptide. Furthermore the enzyme catalyzed the formation of thioether containing cyclic peptides, an approach that can be readily extended to the preparation of libraries of compounds. Although some of the products prepared in this study, notably peptides 3 and 4, could have been prepared more readily using straightforward peptide synthesis, the strength of the methodology described herein is in the preparation of long peptides that are still difficult to prepare by SPPS in non-specialized laboratories. Lantibiotic synthetases have been shown to process Ser/Thr residues as far as 42 residues C-terminal from the leader peptide in designed peptides.23 Therefore, these enzymes can be used to prepare peptides containing thioether rings and/or dehydro amino acids in large peptides that are not readily amenable to synthetic chemistry. The use of a protease cleavage site then allows the removal of the leader peptide and triazole linker. We note, that when a target peptide contains a Lys, the use of endoproteinase Lys-C or trypsin is prohibited, but we have previously demonstrated that other proteases with more defined recognition sites such as Factor Xa can be used as well.39 Therefore, the use of lantibiotic synthetases offers much potential for preparing designer peptides.
Supplementary Material
Supporting Information Available. Experimental procedures and mass spectrometric characterization of all peptides.
Acknowledgments
This work was supported by the National Institutes of Health (GM58822) and a Ruth-Kirschstein National Research Award T32 (GM008276 to MRL).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Matthew R. Levengood, Department of Chemistry, University of Illinois, 600 S. Mathews Avenue, Urbana, Illinois 61801.
Wilfred A. van der Donk, Department of Chemistry, University of Illinois, 600 S. Mathews Avenue, Urbana, Illinois 61801. vddonk@scs.uiuc.edu
References
- 1.Marx V. Chem. Eng. News. 2005;83:17. [Google Scholar]
- 2.Kessler H. Angew. Chem. Int. Ed. Engl. 1982;21:512. [Google Scholar]
- 3.Rew Y, Malkmus S, Svensson C, Yaksh TL, Chung NN, Schiller PW, Cassel JA, DeHaven RN, Goodman M. J. Med. Chem. 2002;45:3746. doi: 10.1021/jm020108k. [DOI] [PubMed] [Google Scholar]
- 4.Li P, Roller PP. Curr. Top. Med. Chem. 2002;2:325. doi: 10.2174/1568026023394209. [DOI] [PubMed] [Google Scholar]
- 5.Tugyi R, Mezo G, Fellinger E, Andreu D, Hudecz F. J. Pept. Sci. 2005;11:642. doi: 10.1002/psc.669. [DOI] [PubMed] [Google Scholar]
- 6.Paul M, van der Donk WA. Minirev. Org. Chem. 2005;2:23. [Google Scholar]
- 7.Cobb SL, Vederas JC. Org. Biomol. Chem. 2007;5:1031. doi: 10.1039/b618178c. [DOI] [PubMed] [Google Scholar]
- 8.Bregant S, Tabor AB. J. Org. Chem. 2005;70:2430. doi: 10.1021/jo048222t. [DOI] [PubMed] [Google Scholar]
- 9.Narayan RS, Vannieuwenhze MS. Org. Lett. 2005;7:2655. doi: 10.1021/ol0507930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou H, van der Donk WA. Org. Lett. 2002;4:1335. doi: 10.1021/ol025629g. [DOI] [PubMed] [Google Scholar]
- 11.Okeley NM, Zhu Y, van der Donk WA. Org. Lett. 2000;2:3603. doi: 10.1021/ol006485d. [DOI] [PubMed] [Google Scholar]
- 12.Burrage S, Raynham T, Williams G, Essex JW, Allen C, Cardno M, Swali V, Bradley M. Chem.-Eur. J. 2000;6:1455. doi: 10.1002/(sici)1521-3765(20000417)6:8<1455::aid-chem1455>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 13.Chatterjee C, Paul M, Xie L, van der Donk WA. Chem. Rev. 2005;105:633. doi: 10.1021/cr030105v. [DOI] [PubMed] [Google Scholar]
- 14.Xie L, van der Donk WA. Curr. Opin. Chem. Biol. 2004;8:498. doi: 10.1016/j.cbpa.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 15.Chatterjee C, Miller LM, Leung YL, Xie L, Yi M, Kelleher NL, van der Donk WA. J. Am. Chem. Soc. 2005;127:15332. doi: 10.1021/ja0543043. [DOI] [PubMed] [Google Scholar]
- 16.Xie L, Miller LM, Chatterjee C, Averin O, Kelleher NL, van der Donk WA. Science. 2004;303:679. doi: 10.1126/science.1092600. [DOI] [PubMed] [Google Scholar]
- 17.Levengood MR, Patton GC, van der Donk WA. J. Am. Chem. Soc. 2007;129:10314. doi: 10.1021/ja072967+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chatterjee C, Patton GC, Cooper L, Paul M, van der Donk WA. Chem. Biol. 2006;13:1109. doi: 10.1016/j.chembiol.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 19.Zhang X, Ni W, van der Donk WA. Org. Lett. 2007;9:3343. doi: 10.1021/ol071301h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang X, van der Donk WA. J. Am. Chem. Soc. 2007;129:2212. doi: 10.1021/ja067672v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kluskens LD, Kuipers A, Rink R, de Boef E, Fekken S, Driessen AJ, Kuipers OP, Moll GN. Biochemistry. 2005;44:12827. doi: 10.1021/bi050805p. [DOI] [PubMed] [Google Scholar]
- 22.Rink R, Kuipers A, de Boef E, Leenhouts KJ, Driessen AJ, Moll GN, Kuipers OP. Biochemistry. 2005;44:8873. doi: 10.1021/bi050081h. [DOI] [PubMed] [Google Scholar]
- 23.Rink R, Wierenga J, Kuipers A, Kluskens LD, Driessen AJM, Kuipers OP, Moll GN. Appl. Environ. Microbiol. 2007;73:1792. doi: 10.1128/AEM.02350-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rink R, Kluskens LD, Kuipers A, Driessen AJ, Kuipers OP, Moll GN. Biochemistry. 2007;46:13179. doi: 10.1021/bi700106z. [DOI] [PubMed] [Google Scholar]
- 25.Wang Q, Chan TR, Hilgraf R, Fokin VV, Sharpless KB, Finn MG. J. Am. Chem. Soc. 2003;125:3192. doi: 10.1021/ja021381e. [DOI] [PubMed] [Google Scholar]
- 26.Pitts KE, Summers AO. Biochemistry. 2002;41:10287. doi: 10.1021/bi0259148. [DOI] [PubMed] [Google Scholar]
- 27.Li B, Yu J-PJ, Brunzelle JS, Moll GN, van der Donk WA, Nair SK. Science. 2006;311:1464. doi: 10.1126/science.1121422. [DOI] [PubMed] [Google Scholar]
- 28.Toogood PL. Tetrahedron Lett. 1993;34:7833. [Google Scholar]
- 29.Polinsky A, Cooney MG, Toy-Palmer A, Osapay G, Goodman M. J. Med. Chem. 1992;35:4185. doi: 10.1021/jm00100a026. [DOI] [PubMed] [Google Scholar]
- 30.Craik DJ, Adams DJ. ACS Chem. Biol. 2007;2:457. doi: 10.1021/cb700091j. [DOI] [PubMed] [Google Scholar]
- 31.Armishaw CJ, Daly NL, Nevin ST, Adams DJ, Craik DJ, Alewood PF. J. Biol. Chem. 2006;281:14136. doi: 10.1074/jbc.M512419200. [DOI] [PubMed] [Google Scholar]
- 32.Bondebjerg J, Grunnet M, Jespersen T, Meldal M. ChemBioChem. 2003;4:186. doi: 10.1002/cbic.200390030. [DOI] [PubMed] [Google Scholar]
- 33.Jimenez EC, Olivera BM, Gray WR, Cruz LJ. J. Biol. Chem. 1996;271:28002. doi: 10.1074/jbc.271.45.28002. [DOI] [PubMed] [Google Scholar]
- 34.Jacobsen R, Jimenez EC, Grilley M, Watkins M, Hillyard D, Cruz LJ, Olivera BM. J. Pept. Res. 1998;51:173. doi: 10.1111/j.1399-3011.1998.tb01213.x. [DOI] [PubMed] [Google Scholar]
- 35.Jimenez EC, Watkins M, Juszczak LJ, Cruz LJ, Olivera BM. Toxicon. 2001;39:803. doi: 10.1016/s0041-0101(00)00210-5. [DOI] [PubMed] [Google Scholar]
- 36.Hansson K, Ma X, Eliasson L, Czerwiec E, Furie B, Furie BC, Rorsman P, Stenflo J. J. Biol. Chem. 2004;279:32453. doi: 10.1074/jbc.M313825200. [DOI] [PubMed] [Google Scholar]
- 37.Massilia GR, Schinina ME, Ascenzi P, Polticelli F. Biochem. Biophys. Res. Commun. 2001;288:908. doi: 10.1006/bbrc.2001.5833. [DOI] [PubMed] [Google Scholar]
- 38.Murkin AS, Tanner ME. J. Org. Chem. 2002;67:8389. doi: 10.1021/jo0204653. [DOI] [PubMed] [Google Scholar]
- 39.McClerren AL, Cooper LE, Quan C, Thomas PM, Kelleher NL, van der Donk WA. Proc. Nat. Acad. Sci. USA. 2006;103:17243. doi: 10.1073/pnas.0606088103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information Available. Experimental procedures and mass spectrometric characterization of all peptides.


