Abstract
Protein glutathionylation is a post-translational modification that may account for a broad mechanism of redox signaling. The caspase family of cysteine proteases represent a potential target for regulation by glutathionylation. To examine this, caspase proteins, derived from HL-60 cells after activation with actinomycin D, were incubated with GSSG. Total protein glutathionylation was enhanced and caspase-3 activity was inhibited in a dose and time dependent manner by GSSG. Caspase inhibition was reversible by thiol-specific reducing reagents. Proteolytic activation of caspases was also affected, as the activation of procaspase-3 and procaspase-9 in HL-60 cell extracts induced by cytochrome c and dATP was inhibited by preincubation with GSSG. When biotin labeled GSSG was incubated with recombinant caspase-3, biotin label was found associated with both p12 and p17 subunits of active caspase-3 by non-reducing SDS-PAGE. Caspase-3 glutathionylation was confirmed by matrix assisted laser desorption ionization (MALDI) mass spectrometric analysis of GSSG-treated recombinant caspase-3. Specific sites of glutathionylation were identified as Cys135 of the p17 protein (the active site) and Cys45 of the p12 protein. These results indicate that glutathionylation of caspase can occur at physiologically relevant concentrations of GSSG and results in the inhibition of caspase activation and activity.
Keywords: glutathionylation, glutathione, caspase, thionylation, oxidative stress, apoptosis
1. Introduction
Protein glutathionylation is an important redox sensitive post-translational protein modification in which glutathione forms mixed disulfide bonds with cysteine residues on target proteins [1]. During oxidative stress, GSH is oxidized to GSSG either enzymatically (eg. glutathione peroxidase) or nonenzymatically. As endogenous levels of GSSG increase, cysteine moieties of target proteins can be glutathiolated by thiol disulfide exchange. Alternatively, protein thiols can be directly oxidized to form thiyl radical (P-S) or sulfenic acid (P-SOH), which can subsequently react with GSH to form the mixed disulfide (P-SSG). While the role of protein glutathionylation in vivo remains to be elucidated, it has been implicated in the stabilization of extracellular proteins, protection of protein sulfhydryls against irreversible oxidation and regulation of enzyme activities. Enhanced protein glutathionylation has also been implicated as a potential factor in the pathogenesis of oxidative stress-related diseases and disorders such as HIV infection, hyperlipidemia, diabetes, cancer and aging [2–4]. To date, numerous proteins, such as carbonic anhydrase III [5], protein kinase C [6, 7], creatine kinase [8], tyrosine hydroxylase [9], NF-κB [10], c-Jun [11], actin [12], thioredoxin [13], cAMP-dependent protein kinase [14] and protein phosphatase 2A [15] have been shown to be regulated by protein glutathionylation.
Caspases (Cysteinyl Aspartate-Specific Proteases), a family of cysteine proteases responsible for the deliberate disassembly of a cell into apoptotic bodies during apoptosis, represent a likely target for protein glutathionylation. Caspases are present as inactive pro-enzymes (procaspases), most of which are activated by proteolytic cleavage during apoptosis. Caspases have a conserved pentapeptide sequence QACXG (X can be G, R or Q) at the active site and a reduced sulfhydryl group in the cysteine moiety is required for caspase activities. Modification of the active site sulfhydryl groups with thiol-blocking reagents such as iodoacetamide and N-ethylmaleimide completely inhibits caspase activity [16]. Studies indicating that recombinant caspase-3 is inactivated by low doses of hydrogen peroxide in a reversible fashion with dithiothreitol also support the role of sulfhydryl modification in the redox regulation of caspase activity and apoptosis [17]. Recent results in endothelial cells provide evidence that glutathionylation of caspases-3 inhibited its cleavage and that TNF-α-induced apoptosis in these cells was associated with glutaredoxin-induced deglutathionylation of caspase-3 [18].
In the present study, we provide direct evidence for caspase-3 inhibition by glutathionylation at specific cysteine residues. The effect of glutathionylation induced by incubation with GSSG on the activity of caspases derived from apoptotic cell HL-60 cell lysates pretreated with actinomycin D was examined. Also, we utilized a cell-free system for caspase activation to determine whether GSSG affected caspase activation. The formation of glutathiolated caspase-3 was confirmed by using biotin-labeled GSSG, mass spectrometric analysis, and isoelectric focusing/immunobloting methodologies.
2. Materials and Methods
2.1. Materials
Caspase substrates (Ac-VDVAD-pNA, Ac-DEVD-pNA, Ac-VEID-pNA, Ac-IETD-pNA and Ac-LEHD-pNA, utilized as substrates for caspase-2, -3, -6, -8, -9, respectively) were purchased from Alexis Biochemicals (San Diego, CA). Rabbit polyclonal antibodies against caspase-3 (H-277) and caspase-9 (H-170) were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). SuperSignal West Pico Chemiluminescent Substrate and Sulfo-NHS-biotin were obtained from Pierce (Rockford, IL). Human recombinant caspase-3 was obtained from Upstate Biotechnology (Lake Placid, NY). All other reagents were obtained from Sigma (St. Louis, MO) unless otherwise indicated.
2.2. Cell Culture
Human promyelocytic leukemia HL-60 cells were cultured in suspension culture in RPMI-1640 medium with glutamine (Mediatech, Herndon, VA), containing 10% heat-inactivated fetal bovine serum (Gemini Bioproducts, Woodland, CA), 100 unit/ml penicillin G, and 100 µg/ml streptomycin (Invitrogen/Gibco, Carlsbad, CA) and maintained at 37°C and 5% CO2. All experiments were conducted using cells during the exponential growth phase. In certain experiments, where specified, apoptosis was induced by addition of 0.5 µg/ml actinomycin D to the culture medium for 6 hours prior to harvesting. Optimal conditions for actinomycin D-induced caspase activation were established in preliminary experiments (data not shown).
2.3. Enhancement of Total Protein Glutathionylation by GSSG
HL-60 apoptotic cell lysates containing 2 mg protein were incubated with either 0 or 1 mM GSSG for 90 min at 37°C. Aliquots of the GSSG-treated samples were subsequently treated with either 0 or 1 mM DTT. All samples were subjected to analyses of protein bound glutathione as previously described [19]. Briefly, proteins were precipitated by 5% metaphosphoric acid (MPA). After washing 3 times by resuspension (in 5% MPA) and centrifugation, the pellets were resuspended in 8 M urea/1 mM EDTA and incubated for 10 min at 40°C. Potassium borohydride was added to a final concentration of 35 mg/ml and the solutions were incubated for 45 min at 40°C. A few drops of octanol were added to reduce foaming. The proteins in solutions were precipitated by 20% MPA for 15 min on ice. The mixtures were then centrifuged at 14,000 × g for 15 min. The supernatant fractions were analyzed for glutathione by an enzymatic-recycling method as previous described [20, 21].
2.4. Caspase Activity Assay
After treated with actinomycin D, HL-60 cells were collected by centrifugation at 1000 × g and washed 3 times with phosphate buffered saline (PBS). Cell pellets were resuspended in caspase assay buffer (50 mM HEPES, pH 7.4, 100 mM NaCl, 1 mM EDTA, 10% glycerol, 0.1% CHAPS). Cell extracts were prepared by freeze/thawing, followed by centrifugation for 15 minutes at 14,000 × g. All procedures were conducted at 4°C. Protein concentrations of the supernatant fractions were measured using the Bio-Rad Protein Assay Kit (Bio-Rad Laboratories, Hercules, CA). Aliquots of cell extracts containing 100 µg protein pretreated with different concentrations of GSSG (0 – 1 mM) were incubated with 200 µM colorimetric caspase substrates in 96-well plates (at a final volume of 200 µl per well) at room temperature. Absorbance at 405 nm was measured every 10 minutes for 1 hour in a 96-well MR-5000 microplate reader (Dynatech Laboratories, Chantilly, VA). Caspase activities were expressed as units/mg protein (1 unit=1 ΔO.D./ hr ).
2.5. Caspase Activation Assay in Cell-free System
Untreated HL-60 cells were collected by centrifugation at 1000 × g and washed 3 times with PBS. Cell pellets were resuspended in caspase activation buffer (25 mM HEPES, pH 7.4, 5 mM MgCl2, 1 mM EGTA). Cellular extracts were prepared by homogenization with an all-glass Ten Broeck homogenizer followed by centrifugation for 15 min at 14,000 × g. All procedures were conducted at 4°C. Protein concentrations of cell extracts were measured as described above. Aliquots containing 100 µg proteins were preincubated with different concentrations of GSSG (0 – 1 mM) in caspase activation buffer in 96-well microplates (at a final volume of 100 µl per well) for 90 min at room temperature. To initiate activation, cytochrome c (final concentration of 10 µM) and dATP (final concentration of 1 mM) were added and samples were incubated at 37°C for 60 min. Caspase activation was assessed by subjecting the reaction mixtures to western blot analysis as described below. Alternatively, for measurement of caspase activity, 80 µl caspase assay buffer and 20 µl caspase-9 or caspase-3 substrate stock solution (2 mM in caspase assay buffer) were added to each well (at the final volume of 200 µl per well) and activities were measured at room temperature as described above. Activation of procaspases occurs in caspase activation buffer but not caspase assay buffer due to differences in different ionic strength between these two buffer systems.
2.6. Western Blot Analysis
Proteins were separated using 12% SDS-PAGE, transferred to nitrocellulose membranes and probed with rabbit anti-caspase-3 (1:800) or anti-caspase-9 (1:1000) polyclonal antibodies, followed by horseradish peroxidase (HRP)-conjugated goat anti-rabbit antibody (1:3000). After incubation with SuperSignal West Pico Chemiluminescent Substrate for 5 min according to instructions, membranes were exposed to x-ray film for 0.5 – 5 min.
2.7. Biotinylation of GSSG
Biotinylation of GSSG was based on a modification of a method previously described for the biotinylation of glutathione ethyl ester [22]. GSSG was reacted with sulfo-NHS-biotin, a biotinylation reagent selective for primary amines, at a 1:2 molar ratio in 50 mM NaHCO3, pH 8.5 for 1 hr at room temperature. The reaction was terminated by the addition of a 5-fold molar excess of NH4HCO3 relative to the starting sulfo-NHS-biotin amount. The resulting solution was analyzed for unlabeled GSSG by HPLC with electrochemical detection [23]. Results indicated that >90% of GSSG was biotinylated under these conditions.
2.8. Glutathionylation of Native Caspase-3 by Biotinylated GSSG
Cell lysate proteins (10 mg) derived from actinomycin D treated HL60 cells were incubated with 50 µM biotinylated GSSG in caspase assay buffer for 90 min at 37°C. After dialysis against Tris-NaCl buffer (50 mM Tris, 150 mM NaCl, pH 8.0) for 24 hours, samples were incubated on a rotating mixer with 500 µl streptavidin-agarose beads for 12 hours at 4°C. The beads were subsequently washed two times with Tris-NaCl buffer for 5 minutes and three-times with high salt Tris-NaCl buffer (50 mM Tris, 500 mM NaCl, pH 8.0) for 5 minutes. Bound proteins were eluted with 50 mM DTT and desalted and concentrated using ultrafiltration on Microcon Centrifugal Filter Devices (YM-10, Millipore, Milford, MA). Resulting proteins were subjected to western blot analysis for caspase-3.
2.9. Glutathionylation of Recombinant Caspase-3 with Biotinylated GSSG
Human recombinant caspase-3 (300 ng) was incubated with increasing concentrations of biotinylated GSSG (0–1 mM) in caspase assay buffer for 90 min at 37°C. Aliquots of reaction mixtures (150 ng) was subjected to 15% SDS-PAGE using either reducing (with 20 mM DTT in loading buffer) or non-reducing conditions. Proteins were transferred to nitrocellulose membranes which were subsequently blocked by 5% milk Tris buffered saline (TBS) for 1 hr at room temperature. Membranes were probed for glutathiolated caspase by avidin peroxidase (1:40000) (Sigma, St. Louis, Mo.). After incubation with SuperSignal West Pico Chemiluminescent Substrate for 5 min according to instructions, membranes were exposed to x-ray film for 15 – 60 sec.
2.10. Detection of Glutathiolated Recombinant Caspase-3 by Mass Spectrometry
Human recombinant caspase-3 (2 µg) was incubated with either 50 µM or 1 mM GSSG in caspase assay buffer at 37°C for 90 min. An aliquot of the 1 mM GSSG-treated sample was subsequently reduced with 1 mM DTT at 37°C for 30 min. All samples were desalted and concentrated to 20 µl by ultrafiltration as described above. The samples were subjected to mass spectrometric analysis. The samples were further desalted and concentrated using C4 ZipTip (Millipore, Billerica, MA), mixed with 0.5 µl of 10 mg/ml α-cyano-4-hydroxysuccinnamic acid in a mixture solvent of formic acid/water/isopropanol (1:3:2), and applied onto a MALDI plate. MALDI mass spectra were recorded with a PerSeptive Voyager-DE STR MALDI-TOF mass spectrometer (PerSeptive Biosysystems, Framingham, MA) operated in the linear mode. The mass measurement accuracy with internal calibration was, in general, better than 100 ppm. The remaining enzyme was subjected to trypsin digestion, followed by mass mapping. Tryptic digestion was started with the addition of 25 ng/µl Sequence Grade Modified Trypsin (Promega, Madison, WI) in ammonium bicarbonate buffer. The protein was digested for at least 16 hr at 37°C with agitation. The digestion products were cleaned and concentrated using micro-C18 ZipTips (Millipore, Billerica, MA) mixed with 0.5 µl of 10 mg/ml 2, 5-dihydroxybenzoic acid in 50% acetonitrile and 0.1% (v/v) trifluoroacetic acid, and applied onto a MALDI plate. MALDI mass spectra were recorded with a PerSeptive Voyager-DE STR MALDI-TOF mass spectrometer operated in the reflection mode.
2.11. Detection of Glutathionylated Caspase-3 in Rat Liver by Isoelectric Focusing
Liver samples were obtained from male 5–6-week old F344 rats (Charles River Labs, Wilmington, MA). Animals were sacrificed by CO2 asphyxiation and livers were removed immediately, rinsed in ice-cold saline, blotted dry and weighed. Samples of liver (~200 mg) were homogenized in metaphosphoric acid and analyzed for total glutathiolated proteins and GSH and GSSG as described previously [4]. Liver pieces (~ 1 g) were homogenized at 4°C in buffer containing 20 mM HEPES, pH 7,4, 5 mM EDTA and 5 mM EGTA. Homogenates were centrifuged at 3,000 g at 4°C for 10 min and the resulting supernatants were centrifuged at 14,000 g for 15 min. Resulting supernatants were assessed for protein content (Bio-Rad) and frozen at −80°C until analysis. Prior to analysis, some samples was incubated with or without 20 mM DTT for 30 min at 37°C to confirm the thiol-specific reversibility of the PI shift. In some cases, samples were incubated with 1 mM GSSG or 1 mM cystine at 37°C for 90 min either before or after DTT treatment.
Tissue extracts were fractionated by isoelectric focusing (IEF) before immunodetection [24] using a vertical polyacrylamide minigel system (Mini-Protecan III, Bio-Rad Laboratory, Hercules, CA). IEF gels contained 5% (w/v) acrylamide, 0.16% (w/v) bisacrylamide, 12% (v/v) glycerol, and 2% (v/v) ampholytes (pH range 5–7). Extracts containing 50–100 µg protein were mixed with 50% (v/v) glycerol prior to loading on gels. IEF was initiated at a constant 150 V for 1 hr followed by a constant 250V for 1 hr and a constant 500 V for 30 min. After equilibrating with ice-cold 0.7% (v/v) acetic acid for 15 min, gels were transferred to nitrocellulose membranes (placed on the cathode side of the gel) at a constant 20V for 30 min (Bio-Rad Trans- Blot semi-dry Transfer Cell, Bio-Rad Laboratories, Hercules, CA). Immunoblotting was performed as described above.
3. Results
3.1. Inhibition of Caspase Activity in HL-60 Cell Lysates by GSSG
Incubation of HL-60 cells with 0.5 µg/ml actinomycin D induced caspase-3 activity based on the cleavage of a synthetic tetrapeptide substrate Ac-DEVD-pNA. Optimal caspase-3 activities were reached at 6 hr incubation (data not shown). Thus, for all future experiments requiring active caspases, cell extracts were obtained from HL-60 cells pretreated with 0.5 µg/ml actinomycin D for 6 hr.
GSSG was utilized as a glutathionylating reagent and induction of protein glutathionylation was confirmed by analysis of protein-bound glutathione. Total glutathiolated protein levels in HL-60 cell lysates were significantly enhanced from <0.4 to 3.22 ± 0.13 (mean ± SEM) nmol/mg by incubation with 1 mM GSSG for 90 min at 37°C. This increase of protein glutathionylation was completely reversible by the subsequent addition of 1 mM DTT (data not shown).
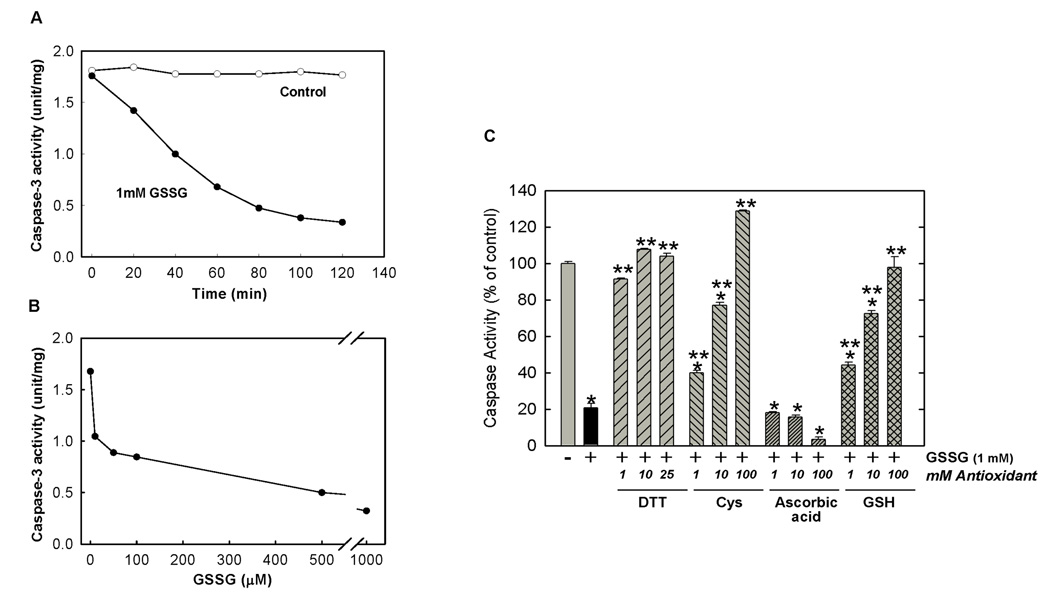
Incubation of HL-60 cell lysates with GSSG inhibited caspase-3 activity in a time and dose dependent manner (Figure 1A & B). Maximum inhibition by GSSG was achieved at 80–100 minutes (Figure 1A). For all subsequent experiments, an incubation period of 90 min was used. Caspase inhibition was enhanced with increasing concentrations of GSSG (0 – 1 mM) reaching maximal inhibition of 80% at 1 mM GSSG (IC50=~75 µM) (Figure 1B). A substantial 30% inhibition was obtained even at 10 µM, the lowest concentration of GSSG examined.
Figure 1.
Time Course, Dose Response and Reversibility of Inhibition of Caspase-3 Activity by GSSG. (A) Time course of caspase-3 Inhibition by GSSG. HL-60 cell lysates (100 µg protein) were incubated with (solid circle) or without (open circle) 1 mM GSSG at 37°C for various times (0–120 min). Caspase-3 activity was measured according to Experimental Procedures. 1 unit=1 ΔO.D./min. (B) Dose Response of Caspase-3 Inhibition by GSSG. HL-60 cell lysates (100 µg protein) were incubated with GSSG (0–1 mM) for 90 min at 37°C. Caspase-3 activity was measured according to Experimental Procedures. (C) Reversibility of Caspase Inhibition by GSSG is Thiol Specific. HL-60 apoptotic cell lysates (100 µg protein) were preincubated with 1 mM GSSG for 90 min at 37°C, then incubated with various concentrations of DTT, cysteine, ascorbic acid and GSH for 30 min. Caspase-3 activity was measured as described in Experimental Procedures. Results are presented as means ± SEM (n=3). * Significantly different from untreated control, (P<0.01). ** Significantly different from GSSG-treated control, (P<0.01).
The inhibition of caspase-3 by GSSG was completely reversible by subsequent addition of thiol-specific reducing agents (Figure 1C). DTT was most effective with nearly 100% recovery of caspase-3 activity obtained at a concentration of 1 mM. In addition to DTT, other thiol-specific antioxidants including cysteine and GSH were also effective at reversing the GSSG-induced inhibition, although higher concentrations were required. Ascorbic acid was ineffective at reversing the GSSG-induced inhibition at concentrations up to 100 mM.
Similar levels of inhibition by GSSG and recovery by DTT were observed when other caspase substrates (Ac-VDVAD-pNA, Ac-VEID-pNA, Ac-IETD-pNA, Ac-LEHD-pNA) were used to assess activity (data not shown). While these different substrates show specificity for different caspases (caspases 2, 6, 8 and 9, respectively) and are routinely used to assess caspase-specific activities, a significant level of cross-reactivity with caspase-3 was observed (data not shown). As a result, it is likely that caspase-3 was primarily responsible for the observed activities with all caspase substrates.
3.2. Inhibition of Caspase Activation by GSSG in a Cell-free System
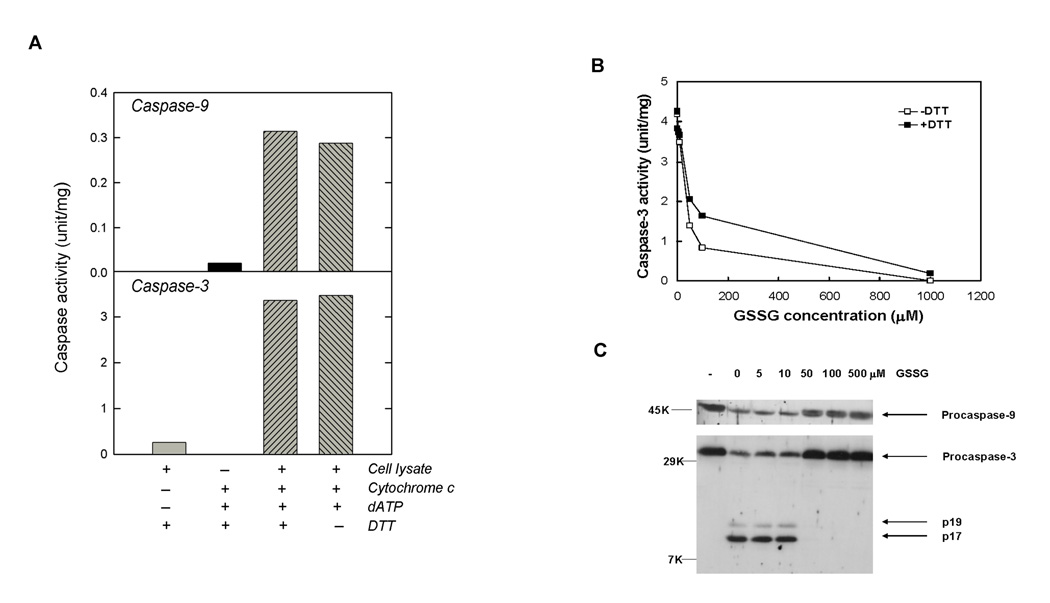
To examine if GSSG incubation could affect the proteolytic activation of caspases, we utilized a cell-free system first described by Liu et al [25]. Using extracts from HL-60 cells without prior treatment with actinomycin D, proteolytic activation of caspase-9 and caspase-3 was accomplished by incubation with cytochrome c and dATP (Figure 2A). As expected, very little activity was observed for both caspase-3 and caspase-9 in untreated cell lysates. Activation of both caspases occurred after addition of 10 µM cytochrome c and 1 mM dATP as previously described [25]. In addition, optimal caspase activities were not dependent on the addition of DTT to the reaction mixture. Since this cell-free caspase activation system was stable at room temperature but not stable when cell extracts were preincubated at 37°C (data not shown), all subsequent preincubations with GSSG in this cell-free system were conducted at room temperature.
Figure 2.
Inhibition of Caspase-9 and Caspase-3 by GSSG in a Cell-Free System. (A) Activation of Caspase-9 and Caspase-3 in HL-60 Cytosolic Cell Lysates by Cytochrome c and dATP. HL-60 cytosolic cell lysates (100 µg protein) were incubated with +/− 10 µM cytochrome c, +/− 1 mM dATP and +/− 1 mM DTT for 60 min at 37°C. Reactions were stopped by addition of caspase activity buffer. Activities of caspase-9 and caspase-3 were measured using synthetic substrates Ac-LEHD-pNA and Ac-DEVD-pNA, respectively. (B) Inhibition of caspase-3 activation by GSSG. HL-60 cytosolic cell lysates (100 µg protein) were preincubated with different concentrations of GSSG for 90 min at room temperature. Cytochrome c and dATP were added to activate caspases. After 60 min at 37°C, reactions were stopped by addition of caspase activity buffer. Caspase-3 activity was measured in the presence (solid square) or absence of 5 mM DTT (open square) using the synthetic tetrapeptide substrate. (C) Inhibition of caspase cleavage by GSSG. Resulting reaction solutions (100 µl) after caspase activation (above) were directly subjected to western blot analysis for caspase-3 and caspase-9 using rabbit anti-caspase-3 and anti-caspase-9, respectively. Lane 1 (labeled “-“) contained no cytochrome C and dATP.
Preincubation of cell extracts with increasing concentrations of GSSG resulted in a dose dependent inhibition in caspase-3 activity (Figure 2B). At 50 µM GSSG, caspase-3 activities were only 33% of the control value. After activation with cytochrome c and dATP in caspase activation buffer, cell lysates were treated with or without 5 mM DTT in caspase assay buffer. With addition of 5 mM DTT, only partial recovery of caspase-3 activity was observed in GSSG treated lysates. Recoveries were only 22%, 23% and 4% for GSSG concentrations of 50, 100, 1000 µM, respectively. No additional improvement in recovery was observed with higher concentrations of DTT (data not shown). Thus, the observed GSSG-induced inhibition was not primarily due to the inhibition of active caspase-3 as described above. To examine if inhibition was a result of decreased caspase proteolytic activation, western blot analyses of caspase-3 and caspase-9 were performed (Figure 2C). In untreated cell lysates, only procaspases were observed (lane 1). When cell lysates were treated with cytochrome c and dATP (lane 2), reductions in both procaspase-9 and procaspase-3 were observed together with the appearance of active forms of caspase-3 indicating the proteolytic activation of the caspases. Banding patterns for cell lysates preincubated with GSSG concentrations of 50 µM or greater prior to treatment with cytochrome c and dATP were identical with cell lysates without addition of cytochrome c and dATP (lane 1) indicating that GSSG at these concentrations inhibited the proteolytic activation of both caspase-9 and caspase-3.
3.3. Induction of Caspase-3 Glutathionylation by GSSG
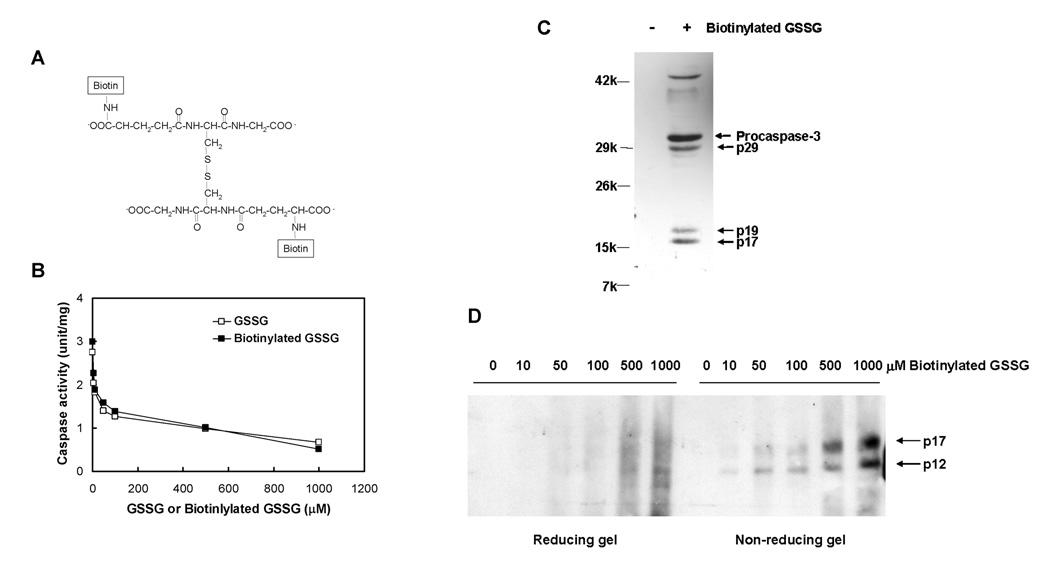
To examine if caspase glutathionylation was responsible for GSSG-induced inhibition of caspase activity and caspase activation, biotin-labeled GSSG was used to confirm protein binding. Biotinylation of amine groups on the γ-glutamyl moieties of GSSG was accomplished using sulfo-NHS-biotin (Figure 3A). Biotinylation of GSSG did not alter its ability to inhibit caspase-3 as identical dose response relationships were observed for both GSSG and biotinylated GSSG (Figure 3B). When cell extracts derived from actinomycin D-treated HL-60 cells were incubated with 50 µM biotinylated GSSG, western analysis of resulting biotin-labeled proteins isolated with streptavidin-agarose beads, revealed the presence of both active caspase-3 and procaspase-3 (Figure 3C, lane 2), suggesting the formation of both glutathiolated active caspase-3 and procaspase-3 under these conditions. When the samples were analyzed in a similar fashion without prior treatment with biotinylated GSSG, few proteins were recovered and no caspase-3 bands were observed by Western blot analysis (Figure 3C, lane 1). To determine if GSSG-induced glutathionylation occurs with recombinant caspase-3, human recombinant enzyme was incubated with biotinylated GSSG. Biotin was found associated with both the p12 and p17 subunits of active caspase-3 in a dose dependent fashion by non-reducing SDS-PAGE (Figure 3D). When samples were analyzed by SDS-PAGE under reducing conditions (with 20 mM DTT), substantially diminished levels of caspase-3-associated biotin were observed, suggesting that this protein-associated biotin was a result of glutathionylation. The apparent glutathionylation of p12 occurred at GSSG concentrations lower than that observed for p17, where the active site is located.
Figure 3.
Glutathionylation of Native and Recombinant Caspase-3 by Biotinylated GSSG. (A) Structure of biotinylated GSSG. (B) Inhibition of Caspase-3 activity in HL-60 Cell Lysate by GSSG (open square) and Biotinylated GSSG (solid square). (C) Glutathionylation of Native Caspase-3 in HL-60 Cell Lysates. HL-60 apoptotic cell lysates containing 10 mg protein were incubated with or without 50 µM biotinylated GSSG for 90 min. Proteins were dialyzed to remove unbound biotinylated GSSG and then incubated with streptavidin-agarose beads. The bound proteins were eluted by addition of 50 mM DTT, then subjected to western blot analysis and probed with rabbit anti-caspase-3 antibody. (D) Glutathionylation of Recombinant Caspase-3 by Biotinlyated GSSG. Human recombinant caspase-3 (150 ng) was incubated with increasing concentrations of biotinylated GSSG for 90 min. Western blot analysis of caspase-3 was performed under reducing and non-reducing conditions using SDS-PAGE and probed with avidin peroxidase.
Mass spectrometry was also utilized to confirm glutathionylation of caspase-3. Human recombinant active caspase-3 was incubated with GSSG for 90 min at 37°C and analyzed by MALDI-TOF mass spectrometry. Two protein ions were observed at m/z = 12960 and 16614 Da, corresponding to p12 and p17, respectively in non-GSSG treated samples (Figure 4A). The observed mass of p12 in the mass spectrum was higher than the theoretical mass calculated from sequence data (MW=11895 Da) due to the presence of a histidine tag and a linker on the recombinant enzyme. Incubation with 50 µM GSSG led to the appearance of two new ions at 13265 and 16919, which were approximately 305 Da greater than the original p12 and p17 ions, respectively (Figure 4B). The mass differences of 305 Da are equivalent to that of a glutathione moiety attached to the protein. MALDI is capable of dissociating non-covalent complexes at high laser fluence and in a “hot” matrix. The MALDI mass spectrum from reaction products of caspase-3 with GSSG showed that the product ions at m/z = 13265 and 16919 were dominant in the spectrum even at the high laser fluence, consistent with the formation of a covalent bond in the products. Similar results were obtained with GSSG at 1 mM concentration (Figure 4C). GSSG-induced mass shifts were reversed by incubation of GSSG-treated recombinant caspase-3 with 1 mM DTT prior to analysis, further confirming the presence of glutathiolated caspase-3 (Figure 4D).
Figure 4.
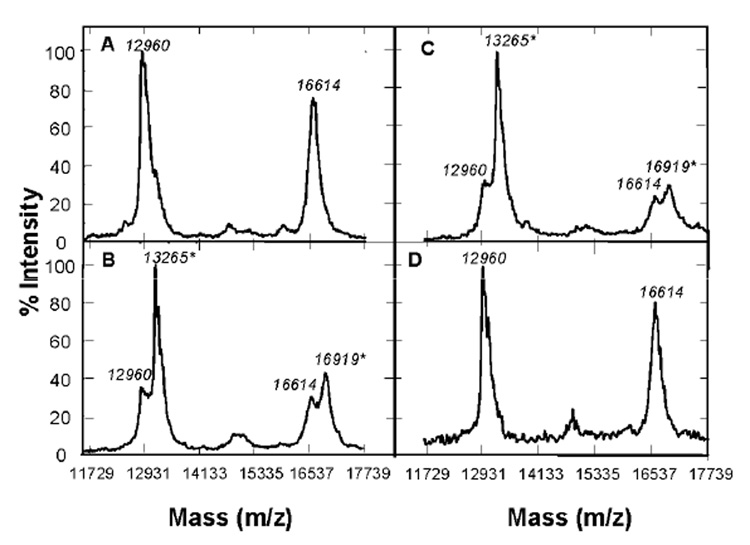
Mass Spectrometric Analysis of GSSG-treated Recombinant Caspase-3. Human recombinant caspase-3 (2 µg) was treated with (A) 0 µM GSSG, (B) 50 µM GSSG, (C) 1 mM GSSG, or (D) 1 mM GSSG and subsequently with 1 mM DTT at 37°C. The exposure times for GSSG and DTT treatments were 90 and 30 min, respectively. All samples were subjected to mass spectrometric analyses as described in Experimental Procedures.
To identify the modification sites, untreated and 50 µM GSSG-treated recombinant enzymes were digested by trypsin. The peptide products were analyzed by MALDI-TOF-MS. The comparison of the mass spectra of tryptic peptides from untreated and GSSG-treated caspase-3 resulted in the identification of two peaks at 1268.636 Da and 1903.817 Da, which were only observed in the mass spectrum of the GSSG-treated sample (Table 1). Masses of these two peaks corresponded to attachment of a glutathione moiety (305 Da) to the tryptic peptide L(129–136)R (963.545 Da) of p17 and D (36–49) K (1598.770 Da) of p12, respectively. These results demonstrate that Cys45 on p12 and Cys135 on p17 (active site) were glutathiolated. These cysteine residues correspond to Cys220 and Cys163 in procaspase-3 sequence, respectively.
Table 1.
Identification of Glutathiolated Cysteine Residues on Caspase-3 by MALDI-TOF
| Peptide fragment | Control | 50 µM GSSG | ||
|---|---|---|---|---|
| MW (Da) | Intensity | MW (Da) | Intensity | |
| p17 L(129–136)R | 963.5 | 2623 | 963.5 | 439 |
| 1268.6 | 1158 | |||
| p12 D(36–49)K | 1598.8 | 500 | ||
| 1903.8 | 389 | |||
3.4. Identification of Glutathionylated Procaspase-3 in Liver by IEF
To examine for the presence of glutathionylated caspases in protein extracts derived from liver samples from male F344 rats, samples were separated by IEF and analyzed by immunoblot with anti-caspase-3 antibody. Based on the addition of a negatively charged GSH residue, glutathionylated caspases are expected to appear as additional bands at a lower pH on the IEF gel. For each untreated liver sample analyzed, multiple anti-caspase-3 antibody reactive bands were present (Figure 5, lane 1). The band exhibiting the highest pI corresponded to authentic procaspase-3. Adjacent bands running at a slightly lower pH suggested the presence of multiple forms of procaspase-3. In order to determine if these adjacent bands represented glutathionylated forms of the protein, aliquots of the protein extracts were treated with 20 mM DTT prior to IEF analysis (Figure 5, lane 2). In the DTT treated samples, only the highest pI band remained corresponding to the parent procaspase-3. The multiple putative glutathionylated procaspase-3 bands likely represent different levels of glutathionylation (eg. monoglutathionylation, biglutathionylation and triglutathionylation). To further confirm the identity of these bands, liver extracts that had been previously reduced by DTT were dialyzed overnight to remove DTT and then treated with either 1 mM GSSG or 1 mM cysteine and subjected to IEF (Figure 5, lane 3). The banding pattern of the GSSG-treated sample was identical to that observed with the original untreated protein sample. However, only the high pH procaspases-3 band was present in the cystine-treated samples (Lane 4). This is consistent with cysteinylation as the addition of cysteine does not introduce an additional negative charge to the protein. Levels of glutathionylated procaspases-3 were quantitated by densitometric scanning of corresponding procaspases-3 bands and compared with levels of GSSG and total glutathionylated proteins in liver samples from six rats. Levels of glutathionylated procaspases-3 varied >2-fold and where significantly correlated with both total glutathionylated proteins (r=0.77) and GSSG levels (r=0.86) (data not shown).
Figure 5.
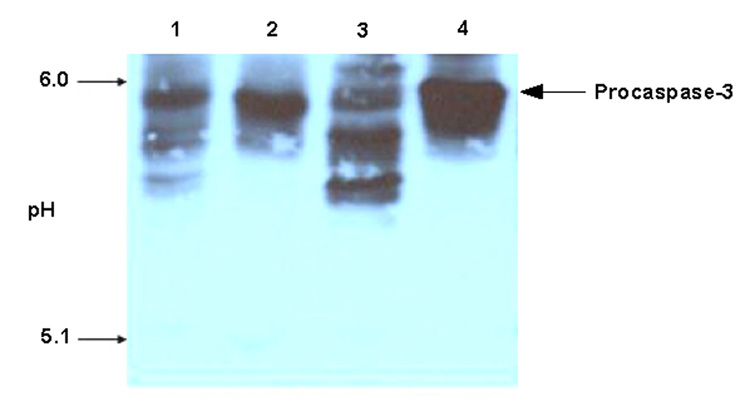
Glutathionylation of Procaspase-3 in Liver. Protein extracts from rat liver were prepared as described in Methods. Portions were reduced with 20 mM DTT for 30 min at 37°C and then dialyzed over night. Portions of the DTT-treated samples were incubated with either 1 mM GSSG or 1 mM cystine at 37°C for 90 min. All samples were subjected to IEF and immunoblot. Lane 1, the original liver extract; lane 2, reduced liver extract; lane 3, GSSG-treated liver extract; lane 4, cystine-treated liver extract.
4. Discussion
The results from these studies indicate caspase-3, and perhaps other caspases, can be regulated by glutathionylation. Protein glutathionylation, resulting from the oxidation of the major cellular antioxidant GSH, has received much recent attention as an important redox-sensitive post-translational modification involved in the functional regulation of many proteins and as a general marker for oxidative stress [26]. The progressive glutathionylation of key proteins is proposed as a molecular switch by which cells respond in an immediate and reversible fashion to oxidative stress and caspases may serve as one such group of proteins. In studies using endothelial cells in culture, deglutathionylation of procaspases-3 by glutaredoxin was shown to be an important step leading to TNF-α induced apoptosis [18]. However, in these studies, the specific cysteine residues subject to glutathionylation were not identified.
In the present study, incubation of HL-60 apoptotic cell lysates with GSSG induced total levels of protein-bound glutathione and concomitantly inhibited the activity of caspase-3 in a dose and time dependent manner. The inhibition of caspase-3 activity was fully recovered by addition of thiol reducing reagents such as DTT, GSH and cysteine, but not ascorbic acid, suggesting that the inhibition is due to oxidation specifically at thiol residues. When recombinant caspase-3 was incubated with various concentrations of biotinylated GSSG, the biotin label was found to be associated with both p12 and p17 subunits and the enzyme-associated biotin was largely removable by DTT suggesting that glutathionylation of both p12 and p17 occurred under these conditions. Mass spectrometric analysis confirmed the glutathionylation of recombinant caspase-3 as increases of 305 Da (equivalent to the expected mass increase by GS-S-protein mixed disulfide formation) were observed for both p12 and p17 subunits after treatment with GSSG. This mass shift was also reversible by the subsequent addition of DTT.
In addition to active caspase-3, procaspases were also subjected to glutathionylation by incubation with GSSG. When HL-60 apoptotic cell lysates containing both active caspase-3 and procaspase-3 were incubated with biotinylated GSSG, both caspase-3 and procaspase-3 were among the glutathiolated proteins observed after isolation with streptavidin beads. Glutathionylation of procaspases was associated with an inhibition in their ability to be proteolytically activated as cytochrome c and dATP failed to activate caspase-9 and caspase-3 in HL-60 cell lysates pretreated with GSSG.
The evidence that GSSG can directly lead to the glutathionylation of both caspases and procaspases suggests that the mechanism involves thiol/disulfide exchange between GSSG and protein thiols. In addition to GSSG, several other methods have been reported to induce the glutathionylation of specific proteins. In a number of studies, GSH binding was induced by addition of the thiol specific oxidant diamide together with GSH [6]. However, since diamide can also directly oxidize proteins without addition of GSH, it can be difficult to distinguish between effects resulting from glutathionylation and those due to other oxidative mechanisms, especially for studies on enzymes that have a cysteine as the active site. Indeed, in our hands diamide was effective at inhibiting caspase-3 without requiring the addition of GSH (data not shown). In other studies, proteins were treated with GSH after being pre-activated by incubation with 5, 5′-dithiobis (2-nitrobenzoic acid) (DTNB) [27]. While this method likely involves the thiol/disulfide exchange of thiol-conjugated proteins with GSH, its physiological relevance is limited. In the present study, we utilized GSSG due to its direct physiological relevance and glutathionylation-specific mechanism of action.
Inhibition of both caspase activity and activation occurred at concentrations of GSSG that can be achieved physiologically as caspase-3 activity was inhibited ~30% by incubation with as little as 10 µM GSSG (IC50: 75 µM). Caspase-3 appears to be particularly sensitive to glutathionylation by GSSG as millimolar concentrations were required for glutathionylation of other proteins including protein kinase C (PKC) [12], creatine kinase [28] and thioredoxin [13]. Physiological levels of GSSG can vary greatly in different tissues ranging up to 120 µM [23] and transient increases of several-fold can occur as a result of acute oxidative stress in vivo.
We also demonstrate the presence of glutathionylated procaspases-3 in rat liver. Identification of glutathiolated forms were based on pI changes in IEF gels and reversal after DTT treatment. In a recent report, glutathionylated caspase-3 was detected in endothelial cells in culture using biotin-labeled, glutathione ethyl ester [18]. The use of IEF to detect glutathionylated caspases in vivo has several advantages to other potential methods in that it does not require the use of labeling procedures or extensive sample processing or purification and is sensitive, requiring only small amounts of protein (50–100 µg).
Mass spectrometric analysis revealed both p12 and p17 subunits of caspase-3 were glutathiolated upon incubation with GSSG. Further analysis of tryptic peptides derived from GSSG-treated caspase-3 revealed Cys45 (Cys220 in the procaspase-3 sequence) on p12 and Cys135 (Cys163 in the procaspase-3 sequence) on p17 were glutathiolated. Incorporation of biotinylated glutathione to the p12 subunit appeared to occur at a lower concentration than for the p17 subunit suggesting that the most sensitive site for glutathionylation is Cys45 on p12 and not the active site (Cys135 on p17), although at higher GSSG concentrations (≥ 500 µM) the band for p17 appears greater than that for p12. Hydrophobicity analysis of the primary sequence of procaspase-3 reveals that the two cysteine residues capable of being glutathiolated were located in or near hydrophilic domains, which suggests that these cysteine residues were on the protein surface in aqueous solution. Cys135 on p17 (active site) was found to be the primary site of modification by nitrosylation with NO [29] or alkylation with iodoacetamide [16], both processes are known to completely abolish caspase-3 activity. One explanation for the high reactivity for the active site Cys may be due to its low pKa, compared with other non-reactive cysteine residues. Like caspase-3, creatine kinase also has a reactive cysteine residue at the active site, which is prone to glutathionylation upon oxidative stress [8]. Another explanation for Cys135 being a target for glutathionylation may be related to the physical properties of the amino acids in its vicinity. On the primary sequence, Cys135 is next to a basic amino acid, Arg136, which might stabilize the adduct by interacting electrostatically with the carboxyl group of the γ-glutamyl portion of GSH. Similar stabilizing interactions between GSH and basic amino acids were shown to occur in the glutathionylation of c-Jun [11] and thioredoxin [13]. Stabilization may also result from amino acids in vicinity in the three-dimensional structure but not in the primary sequence as observed for carbonic anhydrase III [30]. However, this is difficult to assess for caspase-3 because the three-dimensional structure of active caspase-3 is unavailable. Overall, it is difficult to explain how glutathionylation of Cys45 on p12 affects caspase-3 activity. While modification of this cysteine residue has not been reported previously, cysteine-to-serine mutation at this site, as well as at Cys135, was associated with increased cleavage compared to wildtype procaspases-3 [18]. Glutathionylation of this cysteine residue may prevent substrate access to the active site, or may change the confirmation of the enzyme, or may completely disrupt the enzyme by dissociating the two subunits.
Oxidative stress can have differential effects on cells in tissues including both induction of apoptosis as well as cell proliferation depending upon numerous factors including dose and type of oxidant and cell type [31, 32]. Our results suggest that oxidative stress-induced caspase-3 glutathionylation may lead to an inhibition of apoptosis. Although oxidative stress induces apoptosis in numerous cell types, such as hepatocytes [33], retinal pigment epithelial cells [34] and cancer cell lines (eg. HL60) [35], other studies suggest that excessive oxidative stress may inhibit caspase activity and apoptosis. For example, hydrogen peroxide at low concentrations (50 µM) but not high concentrations (200–500 µM) was effective at inducing caspase activation and apoptosis in Jurkat T-lymphocytes [36]. It is possible that the differential effects of the type and extend of oxidative stress on apoptosis is, in part, driven by differential effects on caspase glutathionylation. It should also be noted that caspase-independent oxidative stress-inducible pathways such as the release of Apotosis-Inducing Factor (AIF) from the mitochondria [37] may be playing a role in the induction of apoptosis under conditions where caspase-dependant pathways are inhibited by glutathionylation.
In summary, we report on a novel protein modification of caspases, protein glutathionylation that regulates caspases at two different levels: caspase activities and caspase proteolytic activation. Future experiments are needed to investigate the relationship between caspase glutathionylation and apoptosis. The addition of caspases and procaspases to the growing list of critical cellular proteins regulated by glutathionylation, adds further support for the importance of protein glutathionylation as a cell regulator.
Acknowledgements
We thank Dr. Wei Dai (New York Medical College) and Dr. C.V. Rao (Institute for Cancer Prevention) for their support and advice and also Dr. Despina Komninou and Wayne Kleinman for helpful discussion and technical support. This research was supported by NIH grants DE13222 and CA68384.
Abbreviations used
- dATP
2′-deoxyadenosine 5′-triphosphate
- DTNB
5, 5′-dithiobis (2-nitrobenzoic acid)
- DTT
dithiothreitol
- HRP
horseradish peroxidase
- MALDI
matrix assisted laser desorption ionization
- MPA
metaphosphoric acid
- PBS
phosphate buffered saline
- PKC
protein kinase C
- TBS
Tris buffered saline
- TOF
time of flight.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Klatt P, Lamas S. Regulation of protein function by S-glutathiolation in response to oxidative and nitrosative stress. Eur J Biochem. 2000;267:4928–4944. doi: 10.1046/j.1432-1327.2000.01601.x. [DOI] [PubMed] [Google Scholar]
- 2.Dalle-Donne I, Milzani A, Gagliano N, Colombo R, Giustarini D, Rossi R. Molecular Mechanisms and Potential Clinical Significance of S-Glutathionylation. Antioxidants & redox signaling. 2007 doi: 10.1089/ars.2007.1716. [DOI] [PubMed] [Google Scholar]
- 3.Giustarini D, Rossi R, Milzani A, Colombo R, Dalle-Donne I. S-glutathionylation: from redox regulation of protein functions to human diseases. J Cell Mol Med. 2004;8:201–212. doi: 10.1111/j.1582-4934.2004.tb00275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang Z, Komninou D, Kleinman W, Pinto JT, Gilhooly EM, Calcagnotto A, et al. Enhanced levels of glutathione and protein glutathiolation in rat tongue epithelium during 4-NQO-induced carcinogenesis. Int J Cancer. 2007;120:1396–1401. doi: 10.1002/ijc.22525. [DOI] [PubMed] [Google Scholar]
- 5.Cabiscol E, Levine RL. The phosphatase activity of carbonic anhydrase III is reversibly regulated by glutathiolation. Proc Natl Acad Sci U S A. 1996;93:4170–4174. doi: 10.1073/pnas.93.9.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ward NE, Stewart JR, Ioannides CG, O'Brian CA. Oxidant-induced S-glutathiolation inactivates protein kinase C-alpha (PKC-alpha): a potential mechanism of PKC isozyme regulation. Biochemistry. 2000;39:10319–10329. doi: 10.1021/bi000781g. [DOI] [PubMed] [Google Scholar]
- 7.Ward NE, Pierce DS, Chung SE, Gravitt KR, O'Brian CA. Irreversible inactivation of protein kinase C by glutathione. J Biol Chem. 1998;273:12558–12566. doi: 10.1074/jbc.273.20.12558. [DOI] [PubMed] [Google Scholar]
- 8.Reddy S, Jones AD, Cross CE, Wong PS, Van Der Vliet A. Inactivation of creatine kinase by S-glutathionylation of the active-site cysteine residue. Biochem J. 2000;347(Pt 3):821–827. [PMC free article] [PubMed] [Google Scholar]
- 9.Borges CR, Geddes T, Watson JT, Kuhn DM. Dopamine biosynthesis is regulated by S-glutathionylation. Potential mechanism of tyrosine hydroxylast inhibition during oxidative stress. J Biol Chem. 2002;277:48295–48302. doi: 10.1074/jbc.M209042200. [DOI] [PubMed] [Google Scholar]
- 10.Pineda-Molina E, Klatt P, Vazquez J, Marina A, Garcia de Lacoba M, Perez-Sala D, et al. Glutathionylation of the p50 subunit of NF-kappaB: a mechanism for redox-induced inhibition of DNA binding. Biochemistry. 2001;140:14134–14142. doi: 10.1021/bi011459o. [DOI] [PubMed] [Google Scholar]
- 11.Klatt P, Lamas S. c-Jun regulation by S-glutathionylation. Methods Enzymol. 2002;348:157–174. doi: 10.1016/s0076-6879(02)48635-1. [DOI] [PubMed] [Google Scholar]
- 12.Wang J, Boja ES, Tan W, Tekle E, Fales HM, English S, et al. Reversible glutathionylation regulates actin polymerization in A431 cells. J Biol Chem. 2001;276:47763–47766. doi: 10.1074/jbc.C100415200. [DOI] [PubMed] [Google Scholar]
- 13.Casagrande S, Bonetto V, Fratelli M, Gianazza E, Eberini I, Massignan T, et al. Glutathionylation of human thioredoxin: a possible crosstalk between the glutathione and thioredoxin systems. Proc Natl Acad Sci U S A. 2002;99:9745–9749. doi: 10.1073/pnas.152168599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Humphries KM, Juliano C, Taylor SS. Regulation of cAMP-dependent protein kinase activity by glutathionylation. J Biol Chem. 2002;277:43505–43511. doi: 10.1074/jbc.M207088200. [DOI] [PubMed] [Google Scholar]
- 15.Rao RK, Clayton LW. Regulation of protein phosphatase 2A by hydrogen peroxide and glutathionylation. Biochem Biophys Res Commun. 2002;293:610–616. doi: 10.1016/S0006-291X(02)00268-1. [DOI] [PubMed] [Google Scholar]
- 16.Nicholson DW, Ali A, Thornberry NA, Vaillancourt JP, Ding CK, Gallant M, et al. Identification and inhibition of the ICE/CED-3 protease necessary for mammalian apoptosis. Nature. 1995;376:37–43. doi: 10.1038/376037a0. [DOI] [PubMed] [Google Scholar]
- 17.Borutaite V, Brown GC. Caspases are reversibly inactivated by hydrogen peroxide. FEBS Lett. 2001;500:114–118. doi: 10.1016/s0014-5793(01)02593-5. [DOI] [PubMed] [Google Scholar]
- 18.Pan S, Berk BC. Glutathiolation regulates tumor necrosis factor-alpha-induced caspase-3 cleavage and apoptosis: key role for glutaredoxin in the death pathway. Circulation research. 2007;100:213–219. doi: 10.1161/01.RES.0000256089.30318.20. [DOI] [PubMed] [Google Scholar]
- 19.Kleinman WA, Komninou D, Leutzinger Y, Colosimo S, Cox J, Lang CA, et al. Protein glutathiolation in human blood. Biochem Pharmacol. 2003;65:741–746. doi: 10.1016/s0006-2952(02)01560-5. [DOI] [PubMed] [Google Scholar]
- 20.Richie JP, Jr, Skowronski L, Abraham P, Leutzinger Y. Blood glutathione concentrations in a large-scale human study. Clin Chem. 1996;42:64–70. [PubMed] [Google Scholar]
- 21.Tietze F. Enzymic method for quantitative determination of nanogram amounts of total and oxidized glutathione: applications to mammalian blood and other tissues. Anal Biochem. 1969;27:502–522. doi: 10.1016/0003-2697(69)90064-5. [DOI] [PubMed] [Google Scholar]
- 22.Sullivan DM, Wehr NB, Fergusson MM, Levine RL, Finkel T. Identification of oxidant-sensitive proteins: TNF-alpha induces protein glutathiolation. Biochemistry. 2000;39:11121–11128. doi: 10.1021/bi0007674. [DOI] [PubMed] [Google Scholar]
- 23.Kleinman WA, Richie JP., Jr Determination of thiols and disulfides using high-performance liquid chromatography with electrochemical detection. J Chromatogr B Biomed Appl. 1995;672:73–80. doi: 10.1016/0378-4347(94)00194-a. [DOI] [PubMed] [Google Scholar]
- 24.Robertson EF, Dannelly HK, Malloy PJ, Reeves HC. Rapid isoelectric focusing in a vertical polyacrylamide minigel system. Anal Biochem. 1987;167:290–294. doi: 10.1016/0003-2697(87)90166-7. [DOI] [PubMed] [Google Scholar]
- 25.Liu X, Kim CN, Yang J, Jemmerson R, Wang X. Induction of apoptotic program in cell-free extracts: requirement for dATP and cytochrome c. Cell. 1996;86:147–157. doi: 10.1016/s0092-8674(00)80085-9. [DOI] [PubMed] [Google Scholar]
- 26.Muscat JE, Kleinman W, Colosimo S, Muir A, Lazarus P, Park J, et al. Enhanced protein glutathiolation and oxidative stress in cigarette smokers. Free Radic Biol Med. 2004;36:464–470. doi: 10.1016/j.freeradbiomed.2003.11.018. [DOI] [PubMed] [Google Scholar]
- 27.Dalle-Donne I, Giustarini D, Rossi R, Colombo R, Milzani A. Reversible S-glutathionylation of Cys 374 regulates actin filament formation by inducing structural changes in the actin molecule. Free Radic Biol Med. 2003;34:23–32. doi: 10.1016/s0891-5849(02)01182-6. [DOI] [PubMed] [Google Scholar]
- 28.Lenton KJ, Therriault H, Wagner JR. Analysis of glutathione and glutathione disulfide in whole cells and mitochondria by postcolumn derivatization high-performance liquid chromatography with ortho-phthalaldehyde. Anal Biochem. 1999;274:125–130. doi: 10.1006/abio.1999.4258. [DOI] [PubMed] [Google Scholar]
- 29.Zech B, Wilm M, van Eldik R, Brune B. Mass spectrometric analysis of nitric oxide-modified caspase-3. J Biol Chem. 1999;274:20931–20936. doi: 10.1074/jbc.274.30.20931. [DOI] [PubMed] [Google Scholar]
- 30.Mallis RJ, Poland BW, Chatterjee TK, Fisher RA, Darmawan S, Honzatko RB, et al. Crystal structure of S-glutathiolated carbonic anhydrase III. FEBS Lett. 2000;482:237–241. doi: 10.1016/s0014-5793(00)02022-6. [DOI] [PubMed] [Google Scholar]
- 31.Sykes MC, Mowbray AL, Jo H. Reversible glutathiolation of caspase-3 by glutaredoxin as a novel redox signaling mechanism in tumor necrosis factor-alpha-induced cell death. Circulation research. 2007;100:152–154. doi: 10.1161/01.RES.0000258171.08020.72. [DOI] [PubMed] [Google Scholar]
- 32.England K, Cotter TG. Direct oxidative modifications of signalling proteins in mammalian cells and their effects on apoptosis. Redox Rep. 2005;10:237–245. doi: 10.1179/135100005X70224. [DOI] [PubMed] [Google Scholar]
- 33.Li J, Huang CY, Zheng RL, Cui KR, Li JF. Hydrogen peroxide induces apoptosis in human hepatoma cells and alters cell redox status. Cell Biol Int. 2000;24:9–23. doi: 10.1006/cbir.1999.0438. [DOI] [PubMed] [Google Scholar]
- 34.Jin GF, Hurst JS, Godley BF. Hydrogen peroxide stimulates apoptosis in cultured human retinal pigment epithelial cells. Curr Eye Res. 2001;22:165–173. doi: 10.1076/ceyr.22.3.165.5517. [DOI] [PubMed] [Google Scholar]
- 35.Matsura T, Kai M, Fujii Y, Ito H, Yamada K. Hydrogen peroxide-induced apoptosis in HL-60 cells requires caspase-3 activation. Free Radic Res. 1999;30:73–83. doi: 10.1080/10715769900300081. [DOI] [PubMed] [Google Scholar]
- 36.Hampton MB, Orrenius S. Dual regulation of caspase activity by hydrogen peroxide: implications for apoptosis. FEBS Lett. 1997;414:552–556. doi: 10.1016/s0014-5793(97)01068-5. [DOI] [PubMed] [Google Scholar]
- 37.Boujrad H, Gubkina O, Robert N, Krantic S, Susin SA. AIF-mediated programmed necrosis: a highly regulated way to die. Cell cycle (Georgetown, Tex. 2007;6:2612–2619. doi: 10.4161/cc.6.21.4842. [DOI] [PubMed] [Google Scholar]