Abstract
TANK-binding kinase 1 (TBK1) is of central importance for the induction of type-I interferon (IFN) in response to pathogens. We identified the DEAD-box helicase DDX3X as an interaction partner of TBK1. TBK1 and DDX3X acted synergistically in their ability to stimulate the IFN promoter, whereas RNAi-mediated reduction of DDX3X expression led to an impairment of IFN production. Chromatin immunoprecipitation indicated that DDX3X is recruited to the IFN promoter upon infection with Listeria monocytogenes, suggesting a transcriptional mechanism of action. DDX3X was found to be a TBK1 substrate in vitro and in vivo. Phosphorylation-deficient mutants of DDX3X failed to synergize with TBK1 in their ability to stimulate the IFN promoter. Overall, our data imply that DDX3X is a critical effector of TBK1 that is necessary for type I IFN induction.
Keywords: DDX3X, innate immunity, interferon, phosphorylation, TBK1
Introduction
Innate immune responses are stimulated when microbial molecules referred to as pathogen-associated molecular patterns (PAMPs) bind to a cognate pattern recognition receptor (PRR) (Medzhitov and Janeway, 1997). PRRs can be expressed at the cell surface, the endosomal compartment or in the cytoplasm of mammalian cells (Akira, 2006). Three major families of PRRs have been identified: the Toll-like receptors (TLRs), the RIG-I-like RNA helicases and the NOD-like receptors (NLRs). A total of 11 different TLRs interact with lipid, protein or nucleic acid PAMPs either at the cell surface or the endosome (Akira, 2006). By contrast, the RIG-I-like helicases exclusively detect cytosolic RNA of viral origin (Yoneyama and Fujita, 2007). The NLRs bind to a large variety of PAMPs or danger-associated molecular patterns that include peptides, small molecules and nucleic acids (Kanneganti et al, 2007; Petrilli et al, 2007). Cytosolic DNA, a PAMP resulting from infection with DNA-containing intracellular pathogens, is recognized by a yet unknown number of DNA receptors. One component of the DNA recognition machinery is DAI (DLM-1/ZBP1), a protein that does not belong to either of the three major PRR families (Ishii et al, 2006, 2008; Takaoka et al, 2007; Wang et al, 2008).
PRR engagement triggers several intracellular signalling cascades that culminate in the production and secretion of pro-inflammatory cytokines, chemokines and type-I interferons (IFNs) (predominantly IFN-α and -β collectively termed IFN-I (Pestka et al, 2004; Kawai and Akira, 2007a, 2007b)). These secreted mediators, in turn, activate a gene expression programme and orchestrate a protective immune response against the invading pathogen.
Interferons are the most important antiviral cytokines and loss of IFN-I signalling leads to a severe immunodeficiency towards viral infection (Muller et al, 1994). As a primary response to infection, IFN-β production can be triggered by several PRRs, most notably TLR3 and TLR4, RIG-I and MDA5 and the cytoplasmic DNA receptors. TLR-induced signalling is initiated by the recruitment of an adaptor called TRIF and proceeds through the ubiquitin ligase TRAF3 and the kinases TANK-binding kinase 1 (TBK1)/IKK-i to phosphorylate and activate the transcription factors IFN-regulatory factors (IRF) 3 and 7 (Akira, 2006; Honda and Taniguchi, 2006; He et al, 2007). RIG-I and MDA5 signalling uses a different adaptor called MAVS (IPS-1, VISA, Cardif; Kawai et al, 2005; Meylan et al, 2005; Xu et al, 2005; Sun et al, 2006), but also involves TRAF3, TBK1/IKK-i and IRF3/IRF7 (Saha et al, 2006; Yoneyama and Fujita, 2007). DNA-mediated signalling initiated by DAI (DLM-1/ZBP1) and/or additional cytoplasmic receptors is poorly understood, but similar to the other PRRs it funnels into the TBK1/IKK-i–IRF3 pathway to activate the IFN-β gene (Ishii et al, 2006; Takaoka et al, 2007).
A plethora of studies thus converge to emphasize the role of TBK1 as an activator of IRF3, hence as a central regulator of IFN-I genes in response to many different PAMPs. TBK1-mediated phosphorylation of IRF3 is believed to occur in the C-terminal domain of IRF3 and triggers a conformational change that allows for the dimerization and subsequent nuclear translocation of IRF3 (Honda and Taniguchi, 2006; Hiscott, 2007). In some cells, TBK1 function is shared by the highly related IKK-i (or IKKɛ) kinase, whereas in others TBK1 has a non-redundant function as an IRF3 kinase (Hemmi et al, 2004; McWhirter et al, 2004; Matsui et al, 2006).
The Gram-positive bacterium Listeria monocytogenes has adapted to an intracellular lifestyle by virtue of its ability to escape from endosomal or phagosomal compartments to the cytoplasm of the host cell (Portnoy et al, 2002; Hamon et al, 2006). Disruption of the endosomal/phagosomal membrane results from the activity of the major Listeria virulence protein, a haemolysin called Listeriolysin O (Schnupf and Portnoy, 2007). Intracellular infection with L. monocytogenes stimulates robust expression of IFN-I genes and this was shown to require TBK1 as well as IRF3 (O'Connell et al, 2004, 2005; Stockinger et al, 2004; Decker et al, 2005). The nature of the PAMP delivered to the cytoplasm by L. monocytogenes, although speculatively DNA (Stetson and Medzhitov, 2006), requires final clarification. None of the known PRR adapters are essential to connect the Listeria ligand and TBK1 (Stockinger et al, 2004; Soulat et al, 2006; Sun et al, 2006). The L. monocytogenes ligand and cytoplasmic DNA pathways thus share the lack of known players to address and activate TBK1.
To start bridging the gap between the L. monocytogenes ligand and the IRF3 pathway, we sought to identify TBK1-interacting proteins from macrophages. We report here the surprising finding that a DEAD-box helicase not closely related to, and with properties clearly distinct from, the RIG-I family is phosphorylated by TBK1 to enhance expression of IFN-I genes.
Results
Identification of DDX3X as a novel TBK1 target
To identify novel interactors of TBK1, we generated stable RAW264.7 cell lines that express a GS-TAP-tagged version of TBK1 and purified the TBK1 protein complex by tandem affinity purification (Figure 1A and B) (Burckstummer et al, 2006). Mass spectrometry analysis identified TBK1 along with the core complex components TRAF family member-associated NF-kappa-B activator (TANK; 17 peptides), TBK-binding protein 1 (TBKBP1, also referred to as SINTBAD; 10 peptides) and TBKBP2 (also referred to as NAP1 or AZI2; 15 peptides), indicating that this native purification was efficient and in agreement with previously published data on the TBK1 core complex (Bouwmeester et al, 2004). In addition, we identified the DEAD-box helicase DDX3X (RefSeq NP_034158) with five peptides. Immunoprecipitation experiments using tagged TBK1 suggested that its interaction with DDX3X and the transcription factor IRF3 are significantly weaker than the interaction between TBK1 and TANK and therefore not detected by coimmunoprecipitation under stringent conditions (Supplementary Figure 1A). Likewise, we could not detect any association of DDX3X with IRF3 (Supplementary Figure 1B).
Figure 1.
The DEAD-box helicase DDX3X is a target of TBK1. (A) Schematic representation of the tandem affinity purification protocol: GS-TAP-tagged TBK1 expressed in RAW264.7 cells was purified using rabbit immunoglobulin G (IgG) agarose and eluted by tobacco etch virus (TEV) protease cleavage. Next, the remaining complex was purified using streptavidin agarose and eluted by boiling in SDS sample buffer. (B) The final TAP eluate was separated on an SDS–PAGE and stained by silver staining. Protein complex composition was analysed by LC-MSMS. The position of several complex components is depicted next to the region where it was identified.
Requirement of DDX3X for IFN-β production
We addressed the functional relevance of the TBK1/DDX3X interaction by reducing DDX3X expression and measuring the impact on type-I IFN production. siRNA transfection led to a decrease in DDX3X or TBK1 expression by at least 50% (Figure 2A and B). The cells were infected with L. monocytogenes and IFN-β mRNA production was evaluated by quantitative RT–PCR. As expected, TBK1 was required for IFN-β mRNA transcription (Figure 2C). Strikingly, we observed the same level of reduction for the cells in which DDX3X expression was reduced by siRNA, suggesting that DDX3X is necessary for IFN-β induction. Similar results were obtained for Listeria-mediated induction of RANTES (Figure 2C), another IRF3-dependent gene. To verify that the DDX3X knockdown does not have a general effect on the response to L. monocytogenes, we measured the induction of TNF-α after infection. TNF-α is strongly induced upon L. monocytogenes infection (data not shown), but it does not rely on the TBK1/IRF3 axis (Doyle et al, 2002). In agreement with this, we did not observe any defect in the TNF-α induction neither when TBK1 nor when DDX3X expression was reduced by RNA interference (Figure 2C).
Figure 2.
DDX3X is required for IFN-β induction. Expression of TBK1 or DDX3X was targeted in RAW264.7 macrophages using specific siRNAs. Expression levels of TBK1 (A) or DDX3X (B) were monitored by immunoblotting. Pan-ERK was monitored as a loading control. (C) siRNA-treated RAW264.7 cells were infected with L. monocytogenes for 4 h. Induction of IFN-β mRNA (left panel), RANTES mRNA (middle panel) and TNF-α mRNA (right panel) were measured by quantitative RT–PCR. (D) Similar cells were transfected with poly(I:C) (left panel), treated with LPS (middle panel) or transfected with poly(dA:dT) (right panel). Induction of IFN-β mRNA was measured by quantitative RT–PCR. (E) siRNA-treated RAW264.7 cells were infected with L. monocytogenes or treated with IFN-β for 4 h. Induction of IRF7 and Mx2 was measured by quantitative RT–PCR. Sets of data were analysed using the paired Student's t-test (two-tailed, equal variance). Statistical significance was assessed based on the P-value: *P<0.05, **P<0.01 and ***P<0.001.
Next, we wanted to address whether DDX3X has a specific role during L. monocytogenes infection or a more general role in IFN-β induction. To this end, we stimulated the siRNA-treated cells with different PAMPs such as LPS, transfected poly(I:C) or transfected poly(dA:dT) targeting, respectively, TLR4, MDA5 or the cytosolic DNA receptor. In all of these experimental conditions, reduction of DDX3X expression caused a decrease in IFN-β induction (Figure 2D), confirming that DDX3X is necessary for type-I IFN induction in general.
TBK1/IRF3-mediated production of IFN-β triggers a positive feedback loop to fortify IFN production (Honda et al, 2005). Newly produced IFN-β engages the type-I IFN receptor (IFNAR) which, in turn, stimulates the production of IRF7. Subsequently, IRF7 cooperates with IRF3 to cause a second wave of type-I IFN production. To address whether DDX3X is also involved in this second loop of signalling initiated by IFNAR engagement, we decided to analyse the impact of reduced DDX3X expression on IRF7 transcription. We observed a diminished IRF7 induction upon TBK1 or DDX3X knockdown in cells infected with L. monocytogenes (Figure 2E) and this reduction was similar to the one observed for induced expression of the IFN-β gene (Figure 2C). By contrast, there was no effect of the TBK1 or DDX3X knockdown on the IFN-β-stimulated IRF7 expression (Figure 2E). Similar results were obtained for Mx2, another downstream target of the IFNAR (Figure 2E), indicating that DDX3X does not have a function in IFN-induced gene expression. Overall, these experiments suggest that DDX3X is required for TBK1/IRF3-mediated IFN-β production.
Impact of TBK1 on DDX3X function
The function of DDX3X that may be best documented so far is its role in the nuclear export of human immunodeficiency virus-1 (HIV-1) mRNA. To test whether this function is affected by TBK1 activity, we used an established experimental set-up involving a Rev reporter plasmid (Yedavalli et al, 2004). This assay monitors the DDX3X-dependent nuclear export of a Rev response element-containing RNA encoding the CAT gene (Figure 3A). We found that DDX3X by itself has only a moderate impact on Rev-mediated nuclear export (Figure 3B). Coexpression of TBK1 strongly increased the activity of DDX3X. This effect was neither seen with an inactive mutant of TBK1 (K38M) (Fitzgerald et al, 2003) nor with an ATPase-deficient mutant of DDX3X (K230A) (Yedavalli et al, 2004), implying that both the kinase activity of TBK1 and the helicase activity of DDX3X are required for this function of DDX3X.
Figure 3.
TBK1 has an impact on Rev/DDX3X-dependent nuclear RNA export. (A) Schematic representation of the Rev reporter construct pDM128. In the absence of Rev-mediated nuclear mRNA export, the CAT gene is spliced and therefore not translated. If Rev is present, the mRNA is exported out of the nucleus, splicing is prevented and the CAT mRNA is translated. (B) HEK293 cells were transfected with the reporter plasmid pDM128 and constant amounts of Rev DDX3X wt or K230A and TBK1 wt or K38M were added as indicated. CAT expression was measured 24 h post-transfection by ELISA (Roche).
Synergistic activation of the IFN-β promoter by TBK1 and DDX3X
Is DDX3X sufficient to activate the IFN promoter? We decided to address this question using an IFN-β reporter plasmid in HEK293 cells. MAVS and, to a lesser extent, TBK1 induced the IFN-β promoter, whereas DDX3X by itself did not (Figure 4A). Nevertheless, coexpression of TBK1 with DDX3X led to a synergistic promoter activation that was also observed for the ATPase-deficient mutant of DDX3X (Figure 4A), indicating that the ATPase and the helicase functions of DDX3X are dispensable for its synergy with TBK1. DDX3X did not synergize with TBK1 with regard to NF-κB promoter induction (Figure 4B), sustaining the specific role of DDX3X in the IFN pathway.
Figure 4.
DDX3X and TBK1 led to synergistic activation of the IFN-β promoter. Cells were transfected with a firefly luciferase reporter plasmid and a Renilla luciferase plasmid. Firefly luciferase activity was measured after 24 h and normalized to Renilla luciferase activity. (A) HEK293 cells were transfected with the IFN-β reporter plasmid and 1 μg of plasmids encoding MAVS, DDX3X or DDX3X-K230A and 500 ng of a TBK1 expression plasmid as indicated. (B) HEK293 cells were transfected with the NF-κB reporter plasmid and 1 μg of plasmids encoding HA-tagged MAVS, DDX3X or DDX3X-K230A and 500 ng of a TBK1 expression plasmid as indicated. (C) MEFs from wt mice (black bars), TBK1-deficient mice (grey bars) or IRF3-deficient mice (white bars) were transfected with the IFN-β reporter plasmid and 1 μg of plasmids encoding HA-tagged MAVS, DDX3X, TBK1 and TBK1-S172A as indicated.
Next, we performed epistasis analyses to position DDX3X in the IRF3 pathway. To this end, we used mouse embryonic fibroblasts (MEFs) from TBK1- or IRF3-deficient mice along with wild-type controls. Again, MAVS was a strong activator of IFN-β and MAVS-mediated activation depended more on the presence of IRF3 than on TBK1 (Figure 4C). This result can be attributed to a certain degree of redundancy between TBK1 and IKK-i (Hemmi et al, 2004). When TBK1 was expressed at low levels (compare Supplementary Figure 2), TBK1 by itself was only a weak inducer of IFN-β. In contrast, DDX3X and TBK1 together gave a strong synergistic activation of the IFN-β promoter (Figure 4C). Importantly, this effect was also seen in TBK1−/− MEFs, but not in IRF3−/− MEFs, confirming that the DDX3X effect on IFN requires TBK1 and does not bypass the requirement for IRF3. Noteworthy, this synergy depends on kinase activity of TBK1 as inactive TBK1 S172A cannot synergize with DDX3X (Figure 4C). Overall, these data support that DDX3X has a positive role in the IFN pathway and is therefore required for efficient IFN production.
How does DDX3X affect IFN-β gene expression? Our data are consistent with the possibility that DDX3X impinges on IFN-β promoter activity directly. We therefore asked whether DDX3X can be recruited to the IFN promoter. To address this question, we used chromatin immunoprecipitation (ChIP) and amplified the enhanceosome-binding region of the IFN-β promoter by quantitative PCR (Figure 5A). IRF3, absent under non-stimulated conditions, was recruited to the IFN promoter upon infection with L. monocytogenes (Figure 5B). The specificity of this signal was ascertained using a control serum for ChIP. GS-TAP-tagged DDX3X, isolated by affinity purification using rabbit IgG agarose, was also recruited to the enhanceosome region upon L. monocytogenes infection (Figure 5C). This recruitment was specific because a control region (Figure 5A) could not be amplified by PCR under these conditions. This suggests that DDX3X exerts a direct effect on the IFN-β promoter.
Figure 5.
DDX3X is recruited to the enhanceosome-binding site on the IFN-β promoter. (A) Schematic representation of the IFN-β promoter. Enhanceosome-binding site (white box) is flanked by specific primers, whereas a 1 kb upstream region is flanked by control primers. (B) RAW264.7 macrophages were infected with L. monocytogenes for 0–3 h. Quantitative PCR was realized on ChIP samples treated with either an IRF3-specific antiserum (black bar) or an unrelated serum (white bar). (C) RAW264.7 NTAP(GS)-DDX3X was infected with L. monocytogenes for 0–3 h. Quantitative PCR was realized on samples immunoprecipitated with IgG beads with primers specific for either the enhanceosome-binding site (black bar) or the control region (white bar). Sets of data were analysed using the paired Student's t-test (two-tailed, equal variance). Statistical significance was assessed based on the P-value: *P<0.05 and **P<0.01.
To address whether DDX3X recruitment to the IFN-β promoter depends on IRF3, we synthesized a biotinylated double-stranded DNA fragment corresponding to the IFN-β enhancer (Panne et al, 2007), along with two variants in which the IRF3-binding site was either mutated or deleted (Supplementary Figure 3A). We used these oligos as affinity reagents to monitor IRF3 or DDX3X engagement using lysates from untransfected RAW264.7 cells. We found that the apparent majority of IRF3 strongly associated with the wild type, but only weakly with the mutated enhancer sequence (Supplementary Figure 3B). On the other hand, a minor fraction of DDX3X associated with both the wild-type and the mutated enhancer sequence, suggesting that its binding is independent of IRF3.
DDX3X as a TBK1 kinase substrate
Having established the importance of DDX3X in the TBK1-dependent IRF3 pathway, we further investigated the molecular nature of the relationship between TBK1 and DDX3X. Given the weak affinity of DDX3X for TBK1, we considered the possibility of a kinase–substrate interaction. We observed that coexpression of TBK1 with DDX3X is associated with a slight shift in DDX3X migration on an SDS–PAGE (Supplementary Figure 1). To investigate whether this shift in migration was dependent on TBK1 kinase activity, we generated different inactive mutants of TBK1, mutating either a putative phosphorylation site in the activation loop (S172A), the ATP-binding site (K38M) or the catalytic base (D135N). Activity of the TBK1 mutants was monitored by an in vitro kinase assay or by immunoblotting using an antibody that was raised against phosphoserine 172 in the activation loop of TBK1. Whereas TBK1 wt was active in the kinase assay (Supplementary Figure 4) and phosphorylated on S172 (Figure 6A, middle panel), all mutants were inactive and unphosphorylated on S172, suggesting that the antibody is a valuable tool to monitor the activation status of TBK1 in vivo.
Figure 6.
DDX3X is phosphorylated by TBK1. (A, B) HEK293 cells were transiently transfected with Myc- or HA-tagged constructs as indicated. Cells were treated with 10 μM staurosporine 1 h before lysis as indicated. Cells were lysed 48 h post transfection. Cell extracts were treated with 5 U CIP for 1 h as indicated. Cell extracts were analysed by immunoblotting using either anti-Myc (Rockland), anti-HA (Covance Research) or an antiserum against phosphoserine 172 in TBK1. (C) GST-tagged IRF3, DDX3X and Grb2 were expressed in E. coli and purified by HisTrap chromatography. Purity was assessed by Coomassie staining of three different dilutions of each protein (right panel). TBK1-mediated phosphorylation was assessed by incubating the three potential substrates (IRF3, DDX3X and Grb2) with TBK1 in the presence of [γ-32P]ATP. Lane 1 contains TBK1 without substrate, whereas lanes 2, 6 and 10 contain the substrates without TBK1 (left panel).
How does loss of TBK1 kinase activity affect DDX3X migration? The aforementioned shift in migration was strictly dependent on TBK1 kinase activity, as it was not seen when different inactive mutants of TBK1 (TBK1 S172A, K38M or D135N) were coexpressed with DDX3X (Figure 6A). To unequivocally prove that the shift in migration reflects a phosphorylation event, we added the non-selective kinase inhibitor staurosporine and showed that the TBK1-induced shift was strongly diminished in the presence of the drug. Likewise, treatment of the cell extract with calf intestinal phosphatase led to a decreased shift in DDX3X migration, providing strong evidence that coexpression of TBK1 causes DDX3X phosphorylation. In parallel, we compared DDX3X to IRF3 in this assay (Figure 6A). Strikingly, the phosphorylation pattern observed for IRF3 mirrored the one obtained for DDX3X. These results indicate that DDX3X and IRF3 are very similar in terms of their TBK1 substrate quality.
To demonstrate whether the shift in DDX3X migration was specific for TBK1, we coexpressed DDX3X with a number of kinases including IKK-i, the closest relative of TBK1. Interestingly, IKK-i could also phosphorylate DDX3X, but DDX3X phosphorylation was not observed when the kinases IKKα, IRAK1, Abl or FAK2 were coexpressed (Figure 6B). This suggests that DDX3X is rather specifically phosphorylated by TBK1 and IKK-i.
So far, the data presented supported the possibility that DDX3X is a novel TBK1 substrate, although we could not formally exclude the presence of an intermediate kinase activated by TBK1. To rule out any indirect effect, we expressed His-tagged IRF3, DDX3X and Grb2 in Escherichia coli and purified them by HisTrap chromatography. Equal amounts of purified proteins were then used for an in vitro kinase assay using TBK1 purified from HEK293 cells. Noteworthy, TBK1 alone (lane 1) or the substrates alone (lanes 2, 6 and 10) did not produce any signal. IRF3 was phosphorylated by TBK1 whereas Grb2 was not, suggesting that the assay was performed under conditions that warrant substrate specificity (Figure 6C). The signal obtained for DDX3X was equivalent to the one obtained for IRF3, implying that DDX3X is also a TBK1 substrate in vitro.
DDX3X phosphorylation site mapping
To map the phosphorylation site systematically, we decided for a peptide array that displays 73 peptides containing all serine and threonine residues of DDX3X (Supplementary Figure 5). As a control, we included a peptide derived from the TBK1 activation loop that contains putative autophosphorylation site S172 (Figure 6A). As another control, we included a peptide derived from the C-terminus of IRF3 that contains S396, one of the TBK1 target sites (Servant et al, 2003). As the control peptides contained several serines and threonines, we sequentially exchanged these for alanines to map which serine or threonine within these peptides was actually phosphorylated. Incubation of the peptide array with purified TBK1 produced a number of phosphorylation signals (Figure 7A). The TBK1 control peptide was phosphorylated by TBK1, suggesting that the activation loop is indeed an autophosphorylation site. Mutagenesis indicates that the phosphorylation occurs predominantly at S172, highlighting the relevance of the antibody that recognizes phospho-S172 in TBK1 (Figure 6A). The peptide derived from IRF3 was efficiently phosphorylated as well, but unexpectedly, the phosphorylation occurred exclusively at S402. Analysing the remaining hits in the peptide array, we identified 11 potential phosphorylation sites in 9 DDX3X-derived peptides (Figure 7A). Four of these target sites (S181, S183, S240 and S269) were found in the DEAD domain that contains the ATPase activity of DDX3X. The seven remaining sites were scattered throughout the helicase domain (S429, T438, S442, S456, S520, T542 and S543). We used the information gathered in the peptide array to derive a TBK1 consensus phosphorylation site (Figure 7B). Strikingly, TBK1 showed a strong preference for serine over threonine. In fact, none of the 12 peptides displayed on the array that contain exclusively threonines was phosphorylated. Furthermore, we identified several preferences of TBK1 in terms of amino acids adjacent to the actual site of phosphorylation, the most striking being a preference for a hydrophobic amino acid (L or F) in the +1 position as also seen in the two control peptides. It is interesting to note that S386, which has been identified as critical for IRF3 activation (Mori et al, 2004), also contains a Leu in the +1 position.
Figure 7.
Mapping of the TBK1 phosphorylation site in DDX3X. (A) Peptides derived from DDX3X or control peptides derived from TBK1 or IRF3 along with alanine mutants were incubated with TBK1 in the presence of [γ-32P]ATP. Each array contained a total of 82 peptides spotted in triplicate. (B) Phosphorylation sites obtained from the peptide array (see also text) were used to build a TBK1 phosphorylation consensus sequence. (C) Putative DDX3X phosphorylation sites were mapped onto the structure of a truncated version of DDX3X.
To test which of those putative phosphorylation sites are surface-exposed and therefore accessible to TBK1 in the context of the folded protein, we mapped them onto the structure of a truncated version of DDX3X that was recently published (Hogbom et al, 2007). All sites except T542/S543 mapped to the surface of DDX3X (Figure 7C), implying that the vast majority of these sites are bona fide TBK1 phosphorylation sites in vivo.
Phosphorylation of DDX3X required for synergy with TBK1
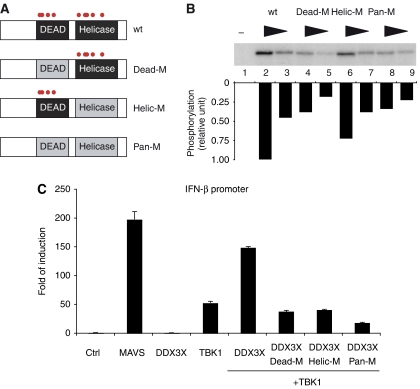
To obtain phosphorylation-deficient mutants for functional experiments, we created three DDX3X mutants. We mutated either all potential phosphorylation sites collectively (Pan-M) or the phosphorylation sites in the DEAD domain (Dead-M) or those in the helicase domain separately (Helic-M) (Figure 8A). Mutagenesis did not include T542 or S543 as those residues were predicted to disrupt the secondary structure of DDX3X. Again, we purified those DDX3X mutants from E. coli and analysed TBK1-mediated phosphorylation in vitro. DDX3X wt was strongly phosphorylated (Figure 8B). In contrast, phosphorylation of Pan-M was strongly diminished as was the case for the Dead-M mutant. The Helic-M mutant was also reduced in terms of TBK1 phosphorylation, but the effect was less pronounced. On the basis of these results, we conclude that both the DEAD domain and the helicase domain contain at least one TBK1 phosphorylation site and that the predominant phosphorylation event occurs in the DEAD domain. We tested these DDX3X mutants for their ability to synergize with TBK1 in the IFN-β reporter assay. Although DDX3X wt showed a strong synergy with TBK1, all the phosphosite mutants failed to enhance activity of the IFN-β promoter (Figure 8C). This establishes a causal link between TBK1-mediated phosphorylation of DDX3X and the ability of the two proteins to stimulate IFN-β expression.
Figure 8.
Phosphorylation-deficient DDX3X mutants fail to synergize with TBK1. (A) Phosphorylation sites obtained from the peptide array were mutated as follows: Dead-M (S181A, S183A, S240A and S269A), Helic-M (S429A, T438A, S442A, S456A and S520A) or Pan-M (S181A, S183A, S240A, S269A, S429A, T438A, S442A, S456A and S520A). (B) Mutated DDX3X proteins were purified from E. coli and analysed in the in vitro kinase assay as described in Figure 6B. (C) HEK293 cells were transiently transfected with the IFN-β reporter plasmid and MAVS, DDX3X wt, different DDX3X mutants (Dead-M, Helic-M and Pan-M) and TBK1 as indicated. Reporter activity was quantified as described in Figure 4.
Discussion
As far as current knowledge goes, TBK1 is arguably the most important kinase in the IFN pathway, yet we know of only two substrates, IRF3 and IRF7 (Pichlmair and Reis, 2007). In this study, we show that the DEAD-box helicase DDX3X is required for IFN production. DDX3X is found to be a TBK1 substrate in vitro and in vivo and phosphorylation by TBK1 is necessary for DDX3X to stimulate the IFN-β promoter. ChIP suggests that DDX3X can be recruited to the IFN promoter, positioning DDX3X downstream of TBK1 at the level of IRF3.
During these studies, we were also able to advance the biochemical characterization of TBK1: first, we identified and characterized a set of inactive mutants (Figure 6). Second, we developed a phospho-specific TBK1 antibody that can be used to monitor TBK1 activity in vivo (Figure 6). Third, we investigated the properties of TBK1 in vitro, using both recombinant proteins and peptides as substrates. On the basis of these analyses, we are providing what we believe may be the first consensus sequence for a TBK1 substrate, which could be helpful for the future identification of TBK1 target sites.
DEAD-box helicases constitute a large family of proteins that comprises at least 38 members in humans (Linder, 2006). These enzymes are characterized by the presence of nine conserved motifs organized in two highly conserved domains: a DEAD domain (named after the characteristic amino-acid sequence D-E-A-D) and a helicase domain. Their nucleic acid unwinding activity requires the hydrolysis of ATP through Walker motifs present in the DEAD domain. Although DEAD-box helicases are usually classified as RNA helicases, it has been suggested that some of them, including DDX3X, can also bind to DNA (Franca et al, 2007). Genetic and biochemical experiments mainly realized in yeast suggested that DEAD-box helicases could be involved at many steps of RNA metabolism, including the initiation of the transcription, RNA splicing, nuclear export, control of mRNA translation and the regulation of RNA stability.
DDX3X is an X chromosome-encoded DEAD-box helicase that has been implicated in nuclear export of HIV-1 RNA through Rev/Crm1 (Yedavalli et al, 2004) and transcriptional regulation of the p21waf1/cip1 promoter (Chao et al, 2006). Both of these activities require access to the nuclear compartment, which is achieved by the means of a nuclear localization signal and a nuclear export signal. Incubation of cells with nuclear export inhibitor leptomycin B traps DDX3X in the nucleus (Yedavalli et al, 2004), confirming that it is shuttling between nucleus and cytoplasm.
DDX3X belongs to the same family of helicases as RIG-I and MDA5 and yet, its mode of action in innate immunity seems to be distinct: unlike RIG-I and MDA5, DDX3X does not exert an effect as a PRR, but is situated rather downstream in the signalling cascade that controls IFN-β production. Furthermore, unlike RIG-I and MDA5, the DDX3X function in the IRF pathway does not rely on its ATPase activity, implying that it may rather exert an effect as a scaffold than as a helicase. A similar scenario has been described for other transcriptional co-activators that exert an effect as bridging factors to build the transcriptional activator complex (Valineva et al, 2005).
Our data suggest that DDX3X is required for the function of the TBK1/IRF3 module and that DDX3X exerts at least part of this function at the promoter level. We would like to propose that activated TBK1 phosphorylates DDX3X which, in the nucleus, gets recruited to the IFN promoter where it stimulates transcription of the IFN-β gene. How this effect is brought about is currently unclear and will be the objective of future studies. DDX3X has been reported to promote transcription by direct interaction with transcription factors (Chao et al, 2006). In our case, however, we fail to observe a direct interaction between IRF3 and DDX3X and the association of DDX3X with the IFN-β enhancer appeared to be independent of the IRF3-binding site. This is reminiscent of the direct association of DDX3X with the E-Cadherin promoter (Botlagunta et al, 2008). On the basis of available literature, however, it is likely that the bulk of the DDX3X molecular functions involve its RNA binding and helicase function. So far, our attempts to find evidence for a post-transcriptional role of DDX3X in the IFN pathway have failed. We cannot rule out the possibility that DDX3X may affect IFN-β production at other stages of gene regulation as well. For instances, DDX3X has been reported to bind to the cap-binding protein complex (Shih et al, 2008). As capping is a cotranscriptional event required for quality control of nascent transcripts (Orphanides and Reinberg, 2002), one could speculate that DDX3X modulates transcriptional elongation of the IFN-β gene. Additionally, there is the possibility of DDX3X having an effect on nuclear export or translation of the IFN-β mRNA. In fact, DDX3X has been implied in the nuclear export of HIV-1 RNA (Yedavalli et al, 2004) on top of cap-dependent mRNA translation (Shih et al, 2008), overall suggesting that DDX3X may indeed interfere with IFN induction at different stages (transcription, nuclear export and mRNA translation) to allow for a fine-tuning or fail saving of the process. This may also explain why TBK1 seems to rely on at least two substrates (IRF3 and DDX3X) to regulate IFN-β production.
The fact that DDX3X is necessary for IFN production implies that it has an antiviral function. In apparent contradiction to this notion, hepatitis C virus (HCV) and HIV-1 seem to exploit DDX3X and use it to their advantage (Yedavalli et al, 2004; Ariumi et al, 2007): DDX3X is needed to sustain the viral life cycle of HIV-1 by facilitating the nuclear export of HIV-1 RNA (Yedavalli et al, 2004). Likewise, DDX3X binds to the HCV core protein and supports viral replication (Ariumi et al, 2007). This obvious contradiction, however, may be resolved by interpreting it in light of the constant evolutionary struggle between host and pathogen: DDX3X may have evolved as an antiviral protein, but viruses then, in turn, developed strategies to hijack DDX3X function and use it for their own benefit, thereby counteracting the antiviral effect of DDX3X. On the basis of this, we postulate that additional viral DDX3X antagonists may exist that inactivate DDX3X either by marking it for proteasome-mediated degradation, by direct proteolytic cleavage or by altering its phosphorylation pattern.
Materials and methods
Antibodies
The TBK1-specific antiserum was obtained from Cell Signaling. The IRF3-specific antiserum was obtained from Invitrogen. The DDX3X-specific antiserum was generated by immunizing rabbits using a recombinant purified domain of DDX3X. Immunizations were carried out by Eurogentec (Belgium).
Plasmids
All ESTs were obtained from RZPD (Berlin, Germany). Genes were based on the following RefSeq IDs: Human DDX3X (NM_001356), human IRF3 (NM_001571). HIV-1 Rev and the reporter plasmid pDM128 were kind gifts of Dr Thomas Hope (University of California, San Diego) and Dr Bryan Cullen (Duke University). Mutagenesis of DDX3 was performed by custom gene synthesis (Genscript Corporation, Piscataway, NJ, USA; www.genscript.com).
Tandem affinity purification
Tandem affinity purification using the GS-TAP cassette was performed as described previously (Burckstummer et al, 2006). Elution was performed by boiling the sample in SDS sample buffer.
siRNA treatment of RAW264.7 cells
siRNAs for TBK1 and DDX3X were purchased from Dharmacon. RAW264.7 cells (2 × 104) were transfected with 25 nM siRNA complexed with HiPerFect (Qiagen) according to the manufacturer's instructions. Transfection was repeated once after 48 h. At 72 h after the first transfection, 5 × 105 cells were seeded onto 6 cm dish to be stimulated 24 h later.
RAW264.7 stimulation
siRNA-treated RAW264.7 cells were infected with L. monocytogenes LO28 at a multiplicity of infection of 10 for 4 h. In parallel, cells were transfected with 30 μg/ml poly(I:C) (Amersham Biosciences) complexed with HiPerFect (Qiagen) for 3 h or 10 μg/ml poly(dA:dT) (Sigma) complexed with PolyFect (Qiagen) for 4 h according to manufacturer's instructions. Additionally, cells were treated with 100 ng/ml LPS derived from Salmonella minnesota (Alexsis) or with 500 U/ml IFN-β (Calbiochem, La Jolla, CA) for 4 h.
RNA isolation, cDNA synthesis and quantitative PCR
After stimulation, total RNA was extracted from RAW264.7 cells using Nucleospin RNA II kit (Macherey & Nagel). The RNA was then reverse transcribed with oligo(dT)18 primer and RevertAid M-MuLV Reverse Transcriptase (MBI Frementas) according to the manufacturer's instructions. The cDNA was then analysed by quantitative PCR using the iCycler IQ machine (Bio-Rad). The target genes (IFN-β, RANTES, TNFα, IRF7 and Mx2) were quantified using the standard curve method and normalized to the endogenous control GAPDH. The PCR was performed by using SybrGreen (Molecular Probes) and the Taq polymerase (MBI Fermentas) and the following primers: GAPDH for, 5′-CATGGCCTTCCGTGTTCCTA-3′; GAPDH rev, 5′-GCGGCACGTCAGATCCA-3′; IFN-β for, 5′-TCAGAATGAGTGGTGGTTGC-3′; IFN-β rev, 5′-GACCTTTCAAATGCAGTAGATTCA-3′; TNFα for, 5′-CAAAATTCGAGTGACAAGCCTG-3′; TNFα rev,5′-GAGATCCATGCCGTTGCC-3′; IRF7 for, 5′-CTGGAGCCATGGGTATGCA-3′; IRF7 rev, 5′-AAGCAGAAGCCCAGACTGCT-3′; Mx2 for, 5′-CCAGTTCCTCTCAGTCCCAAGATT-3′; Mx2 rev, 5′-TACTGGATGATCAAGGGAACGTGG-3′; RANTES for, 5′-CTCACCATCATCCTCACTGC-3′; RANTES rev, 5′-ACTTGGCGGTTCCTTCG-3′.
Rev reporter assay
HEK293 cells were seeded at 5 × 105 cells per well in six-well plates. Cells were transfected with 500 ng of pDM128 (Rev reporter plasmid), 30 ng of Rev expression plasmid, 300 ng of DDX3X expression plasmid and 500 ng of TBK1 expression plasmid. All samples were normalized for DNA content using an empty plasmid. Cells were harvested 24 h post-transfection and CAT levels were analysed by ELISA (Roche) according to the manufacturer's instructions.
Generation of point mutant of TBK1 and DDX3X
QuikChange® Multi Site-Directed Mutagenesis Kit (Stratagene) was used to generate point mutants with appropriate primers regarding the sequence to target.
Luciferase assay
HEK293 and MEF cells were transfected for 24 h with the IFN-β reporter (firefly luciferase) plasmid (pTA-Luc-IFN-β) or the NF-κB firefly luciferase plasmid (pTA-Luc-NF-κB) together with the Renilla luciferase control plasmid (phRG-TK) in complex with ExGen500 (MBI Fermentas) according to the manufacturer's instructions. As target vectors, N-HA-MAVS or N-HA-DDX3X plasmid (or derived mutants) and/or myc-TBK1 (or derived mutants) plasmid were used as indicated. Empty HA vector was used to equalize the DNA content of each transfection. At 24 h after the transfection, the cells were lysed and the extracts were analysed by using the Dual-Luciferase Reporter Assay System (Promega) and the luminometer Lumat (LB9501; Berthold). For quantification, the firefly luciferase activity was then normalized to the Renilla luciferase activity.
ChIP
A total of 3 × 107 RAW264.7 wt or NTAP(GS)-DDX3X cells were used per time point. ChIP assays were performed as recently described (Zupkovitz et al, 2006) using 3 μg of IRF3-specific antibody (Zymed, Invitrogen) followed by a protein A pulldown or using rabbit IgG agarose for NTAP(GS)-DDX3X. For the analysis of the enhanceosome-binding site of ifnb1 promoter, the following primers were used: forward, 5-caggatgagcagctactctgc-3, and reverse, 5-ctgctacctgcaagatgagg-3. For the 1 kb upstream control region, the following primers were used: forward, 5-gccaatgatgtggttcagc-3, and reverse, 5-gagacctgtctgctgttagtgg-3.
Generation of the antiserum directed against phosphoserine 172 in TBK1
The phospho-specific antiserum was custom-synthesized by Eurogentec using a phosphorylated peptide derived from the activation loop of TBK1 (DDEQFVpSLYGTEE) (Eurogentec).
Transient transfection of HEK293 and preparation of cell extracts
HEK293 cells were transiently transfected using PolyFect (Qiagen) according to the manufacturer's instructions. Cells were harvested 24 or 48 h post-transfection and lysed in Frackelton buffer (10 mM Tris–HCl pH 7.5, 50 mM NaCl, 30 mM sodium pyrophosphate, 1% Triton X-100, 1 mM DTT, 100 μM sodium orthovanadate, 50 μM NaF and protease inhibitors).
Kinase assays
For peptide-based assays, Myc-TBK1 was isolated from HEK293 cells by immunoprecipitation using anti-Myc agarose (Sigma). Precipitated TBK1 was incubated with a biotinylated peptide derived from IRF3 (DLHISNSHPLSLC) in the presence of 50 μM ATP, 5 μCi [γ-32P]ATP and kinase buffer (40 mM Tris–HCl pH 7.5, 10 mM MgCl2 and 1 mM DTT) for 30 min at 30°C. The reaction was terminated by addition of guanidinium chloride (2.5 M final concentration) and the reaction was spotted onto a SAM2 Biotin Capture membrane (Promega) and further treated according to the manufacturer's instructions. Kinase activity was then measured by scintillation counting.
For those kinase assays where recombinant proteins were used as substrates, we purified those substrates from E. coli using HisTrap chromatography (Amersham Biosciences). Recombinant TBK1 was expressed in HEK293 cells and purified by affinity chromatography (data not shown). Purified TBK1 was incubated with three dilutions of each purified substrate in the presence of 50 μM ATP and 5 μCi [γ-32P]ATP using kinase buffer (40 mM Tris–HCl pH 7.5, 10 mM MgCl2 and 1 mM DTT) at 30°C. In some instances, the ATP concentration was lowered to 10 μM to increase substrate specificity. The reaction was terminated by boiling in SDS sample buffer after 5–15 min. Samples were separated by SDS–PAGE, the gels were dried and analysed by autoradiography.
Peptide array
The peptide array was custom synthesized by Jerini Peptide Technologies (Berlin, Germany). The peptide sequences that were spotted on the array in triplicate are depicted in Supplementary Figure 5. Recombinant TBK1 was expressed in HEK293 cells and purified by affinity chromatography (data not shown). The peptide array was incubated with TBK1 in the presence of 50 μM ATP and 100 μCi [γ-32P]ATP using kinase buffer (40 mM Tris–HCl pH 7.5, 10 mM MgCl2 and 1 mM DTT) at 30°C. After 30 min, the reaction was terminated by addition of 0.1 M phosphoric acid. The slides were then washed with 0.1 M phosphoric acid for five times and with deionized water for five times. After a final wash with methanol, the slides were dried and analysed by autoradiography.
Structural mapping of phosphorylation sites and building of the TBK1 phosphorylation consensus
The structure of DDX3 was downloaded from the Protein Database (PDB) (accession number 2i4i). Phosphorylation sites were highlighted using PyMOL (www.pymol.org). The TBK1 phosphorylation consensus was built by manual inspection of the TBK1 target peptides taking into consideration only those amino acids that were seen more than three times (out of 13 positive peptides). Amino-acid sizes reflect the relative abundance of a given amino acid in that particular position (e.g. Leu in position +1 was found in 5 out of 13 peptides and has therefore been represented as 5/13 of the size of the S/T peak).
Statistical analysis
Sets of data were analysed using the paired Student's t-test (two-tailed, equal variance). Statistical significance was assessed based on the P-value: *P<0.05, **P<0.01 and ***P<0.001. For the siRNA experiments (Figure 2), we calculated the standard deviation and the statistical significance (t-test) of the percentage of reduction based on a minimum of three independent experiments. For reporter gene assays (Figures 3, 4 and 8), we chose to show the absolute level of induction (versus relative). The difference in transfection efficiency across different experiments precludes the calculation of standard deviations based on absolute levels. We therefore decided to show one representative experiment and hereby state that we have replicated each result at least twice, with a statistically significant difference in all three cases.
Supplementary Material
Supplementary Figures 1 and 5
Acknowledgments
We are grateful to Thomas Hope and Bryan Cullen for providing the plasmids for the Rev reporter assay. We thank Hannah Jahn for technical assistance, Melanie Planyavsky for preparation of the TAP samples for analysis by mass spectrometry, Thomas Stranzl for help with the TBK1 consensus site and Andreas Pichlmair, Christoph Baumann and the entire Superti-Furga laboratory for helpful discussions. TB is supported by a research fellowship (BU 2180/1-1) from the German Research Foundation (DFG). DS was supported for a year by a fellowship from the French Foundation for Medical Research (FRM). SW is supported by the University of Vienna through Initiativkolleg ‘Symbiotic Interactions'. CeMM is supported by the Austrian Academy of Sciences. Work described here was funded by the Austrian Proteomics Platform-II of the GenAU Program of the Austrian Ministry of Education and Research, by a grant from the Austrian National Bank and by a grant (P20522-B05) from the Austrian Science Foundation (FWF). Research at the Max F Perutz laboratories was supported by the Austrian Science Foundation (FWF) through grants AP17859, P20522-B05 and SFB 28 to TD.
References
- Akira S (2006) TLR signaling. Curr Top Microbiol Immunol 311: 1–16 [DOI] [PubMed] [Google Scholar]
- Ariumi Y, Kuroki M, Abe K, Dansako H, Ikeda M, Wakita T, Kato N (2007) DDX3 DEAD-box RNA helicase is required for hepatitis C virus RNA replication. J Virol 81: 13922–13926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botlagunta M, Vesuna F, Mironchik Y, Raman A, Lisok A, Winnard P Jr, Mukadam S, Van Diest P, Chen JH, Farabaugh P, Patel AH, Raman V (2008) Oncogenic role of DDX3 in breast cancer biogenesis. Oncogene; doi:10.1038/onc.2008.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwmeester T, Bauch A, Ruffner H, Angrand PO, Bergamini G, Croughton K, Cruciat C, Eberhard D, Gagneur J, Ghidelli S, Hopf C, Huhse B, Mangano R, Michon AM, Schirle M, Schlegl J, Schwab M, Stein MA, Bauer A, Casari G et al. (2004) A physical and functional map of the human TNF-alpha/NF-kappa B signal transduction pathway. Nat Cell Biol 6: 97–105 [DOI] [PubMed] [Google Scholar]
- Burckstummer T, Bennett KL, Preradovic A, Schutze G, Hantschel O, Superti-Furga G, Bauch A (2006) An efficient tandem affinity purification procedure for interaction proteomics in mammalian cells. Nat Methods 3: 1013–1019 [DOI] [PubMed] [Google Scholar]
- Chao CH, Chen CM, Cheng PL, Shih JW, Tsou AP, Lee YH (2006) DDX3, a DEAD box RNA helicase with tumor growth-suppressive property and transcriptional regulation activity of the p21waf1/cip1 promoter, is a candidate tumor suppressor. Cancer Res 66: 6579–6588 [DOI] [PubMed] [Google Scholar]
- Decker T, Muller M, Stockinger S (2005) The yin and yang of type I interferon activity in bacterial infection. Nat Rev Immunol 5: 675–687 [DOI] [PubMed] [Google Scholar]
- Doyle S, Vaidya S, O'Connell R, Dadgostar H, Dempsey P, Wu T, Rao G, Sun R, Haberland M, Modlin R, Cheng G (2002) IRF3 mediates a TLR3/TLR4-specific antiviral gene program. Immunity 17: 251–263 [DOI] [PubMed] [Google Scholar]
- Fitzgerald KA, McWhirter SM, Faia KL, Rowe DC, Latz E, Golenbock DT, Coyle AJ, Liao SM, Maniatis T (2003) IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol 4: 491–496 [DOI] [PubMed] [Google Scholar]
- Franca R, Belfiore A, Spadari S, Maga G (2007) Human DEAD-box ATPase DDX3 shows a relaxed nucleoside substrate specificity. Proteins 67: 1128–1137 [DOI] [PubMed] [Google Scholar]
- Hamon M, Bierne H, Cossart P (2006) Listeria monocytogenes: a multifaceted model. Nat Rev Microbiol 4: 423–434 [DOI] [PubMed] [Google Scholar]
- He JQ, Oganesyan G, Saha SK, Zarnegar B, Cheng G (2007) TRAF3 and its biological function. Adv Exp Med Biol 597: 48–59 [DOI] [PubMed] [Google Scholar]
- Hemmi H, Takeuchi O, Sato S, Yamamoto M, Kaisho T, Sanjo H, Kawai T, Hoshino K, Takeda K, Akira S (2004) The roles of two IkappaB kinase-related kinases in lipopolysaccharide and double stranded RNA signaling and viral infection. J Exp Med 199: 1641–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiscott J (2007) Triggering the innate antiviral response through IRF-3 activation. J Biol Chem 282: 15325–15329 [DOI] [PubMed] [Google Scholar]
- Hogbom M, Collins R, van den Berg S, Jenvert RM, Karlberg T, Kotenyova T, Flores A, Hedestam GB, Schiavone LH (2007) Crystal structure of conserved domains 1 and 2 of the human DEAD-box helicase DDX3X in complex with the mononucleotide AMP. J Mol Biol 372: 150–159 [DOI] [PubMed] [Google Scholar]
- Honda K, Taniguchi T (2006) IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat Rev Immunol 6: 644–658 [DOI] [PubMed] [Google Scholar]
- Honda K, Yanai H, Takaoka A, Taniguchi T (2005) Regulation of the type I IFN induction: a current view. Int Immunol 17: 1367–1378 [DOI] [PubMed] [Google Scholar]
- Ishii KJ, Coban C, Kato H, Takahashi K, Torii Y, Takeshita F, Ludwig H, Sutter G, Suzuki K, Hemmi H, Sato S, Yamamoto M, Uematsu S, Kawai T, Takeuchi O, Akira S (2006) A Toll-like receptor-independent antiviral response induced by double-stranded B-form DNA. Nat Immunol 7: 40–48 [DOI] [PubMed] [Google Scholar]
- Ishii KJ, Kawagoe T, Koyama S, Matsui K, Kumar H, Kawai T, Uematsu S, Takeuchi O, Takeshita F, Coban C, Akira S (2008) TANK-binding kinase-1 delineates innate and adaptive immune responses to DNA vaccines. Nature 451: 725–729 [DOI] [PubMed] [Google Scholar]
- Kanneganti TD, Lamkanfi M, Nunez G (2007) Intracellular NOD-like receptors in host defense and disease. Immunity 27: 549–559 [DOI] [PubMed] [Google Scholar]
- Kawai T, Akira S (2007a) Antiviral signaling through pattern recognition receptors. J Biochem 141: 137–145 [DOI] [PubMed] [Google Scholar]
- Kawai T, Akira S (2007b) Signaling to NF-kappaB by Toll-like receptors. Trends Mol Med 13: 460–469 [DOI] [PubMed] [Google Scholar]
- Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S (2005) IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol 6: 981–988 [DOI] [PubMed] [Google Scholar]
- Linder P (2006) Dead-box proteins: a family affair—active and passive players in RNP-remodeling. Nucleic Acids Res 34: 4168–4180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui K, Kumagai Y, Kato H, Sato S, Kawagoe T, Uematsu S, Takeuchi O, Akira S (2006) Cutting edge: role of TANK-binding kinase 1 and inducible IkappaB kinase in IFN responses against viruses in innate immune cells. J Immunol 177: 5785–5789 [DOI] [PubMed] [Google Scholar]
- McWhirter SM, Fitzgerald KA, Rosains J, Rowe DC, Golenbock DT, Maniatis T (2004) IFN-regulatory factor 3-dependent gene expression is defective in Tbk1-deficient mouse embryonic fibroblasts. Proc Natl Acad Sci USA 101: 233–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R, Janeway CA Jr (1997) Innate immunity: the virtues of a nonclonal system of recognition. Cell 91: 295–298 [DOI] [PubMed] [Google Scholar]
- Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J (2005) Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437: 1167–1172 [DOI] [PubMed] [Google Scholar]
- Mori M, Yoneyama M, Ito T, Takahashi K, Inagaki F, Fujita T (2004) Identification of Ser-386 of interferon regulatory factor 3 as critical target for inducible phosphorylation that determines activation. J Biol Chem 279: 9698–9702 [DOI] [PubMed] [Google Scholar]
- Muller U, Steinhoff U, Reis LF, Hemmi S, Pavlovic J, Zinkernagel RM, Aguet M (1994) Functional role of type I and type II interferons in antiviral defense. Science 264: 1918–1921 (issn: 0036-8075) [DOI] [PubMed] [Google Scholar]
- O'Connell RM, Saha SK, Vaidya SA, Bruhn KW, Miranda GA, Zarnegar B, Perry AK, Nguyen BO, Lane TF, Taniguchi T, Miller JF, Cheng G (2004) Type I interferon production enhances susceptibility to Listeria monocytogenes infection. J Exp Med 200: 437–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell RM, Vaidya SA, Perry AK, Saha SK, Dempsey PW, Cheng G (2005) Immune activation of type I IFNs by Listeria monocytogenes occurs independently of TLR4, TLR2, and receptor interacting protein 2 but involves TANK-binding kinase 1. J Immunol 174: 1602–1607 [DOI] [PubMed] [Google Scholar]
- Orphanides G, Reinberg D (2002) A unified theory of gene expression. Cell 108: 439–451 [DOI] [PubMed] [Google Scholar]
- Panne D, Maniatis T, Harrison SC (2007) An atomic model of the interferon-beta enhanceosome. Cell 129: 1111–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestka S, Krause CD, Walter MR (2004) Interferons, interferon-like cytokines, and their receptors. Immunol Rev 202: 8–32 [DOI] [PubMed] [Google Scholar]
- Petrilli V, Dostert C, Muruve DA, Tschopp J (2007) The inflammasome: a danger sensing complex triggering innate immunity. Curr Opin Immunol 19: 615–622 [DOI] [PubMed] [Google Scholar]
- Pichlmair A, Reis ESC (2007) Innate recognition of viruses. Immunity 27: 370–383 [DOI] [PubMed] [Google Scholar]
- Portnoy DA, Auerbuch V, Glomski IJ (2002) The cell biology of Listeria monocytogenes infection: the intersection of bacterial pathogenesis and cell-mediated immunity. J Cell Biol 158: 409–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha SK, Pietras EM, He JQ, Kang JR, Liu SY, Oganesyan G, Shahangian A, Zarnegar B, Shiba TL, Wang Y, Cheng G (2006) Regulation of antiviral responses by a direct and specific interaction between TRAF3 and Cardif. EMBO J 25: 3257–3263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnupf P, Portnoy DA (2007) Listeriolysin O: a phagosome-specific lysin. Microbes Infect 9: 1176–1187 [DOI] [PubMed] [Google Scholar]
- Servant MJ, Grandvaux N, tenOever BR, Duguay D, Lin R, Hiscott J (2003) Identification of the minimal phosphoacceptor site required for in vivo activation of interferon regulatory factor 3 in response to virus and double-stranded RNA. J Biol Chem 278: 9441–9447 [DOI] [PubMed] [Google Scholar]
- Shih JW, Tsai TY, Chao CH, Wu Lee YH (2008) Candidate tumor suppressor DDX3 RNA helicase specifically represses cap-dependent translation by acting as an eIF4E inhibitory protein. Oncogene 27: 700–714 [DOI] [PubMed] [Google Scholar]
- Soulat D, Bauch A, Stockinger S, Superti-Furga G, Decker T (2006) Cytoplasmic Listeria monocytogenes stimulates IFN-beta synthesis without requiring the adapter protein MAVS. FEBS Lett 580: 2341–2346 [DOI] [PubMed] [Google Scholar]
- Stetson DB, Medzhitov R (2006) Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity 24: 93–103 [DOI] [PubMed] [Google Scholar]
- Stockinger S, Reutterer B, Schaljo B, Schellack C, Brunner S, Materna T, Yamamoto M, Akira S, Taniguchi T, Murray PJ, Muller M, Decker T (2004) IFN regulatory factor 3-dependent induction of type I IFNs by intracellular bacteria is mediated by a TLR- and Nod2-independent mechanism. J Immunol 173: 7416–7425 [DOI] [PubMed] [Google Scholar]
- Sun Q, Sun L, Liu HH, Chen X, Seth RB, Forman J, Chen ZJ (2006) The specific and essential role of MAVS in antiviral innate immune responses. Immunity 24: 633–642 [DOI] [PubMed] [Google Scholar]
- Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T, Lu Y, Miyagishi M, Kodama T, Honda K, Ohba Y, Taniguchi T (2007) DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature 448: 501–505 [DOI] [PubMed] [Google Scholar]
- Valineva T, Yang J, Palovuori R, Silvennoinen O (2005) The transcriptional co-activator protein p100 recruits histone acetyltransferase activity to STAT6 and mediates interaction between the CREB-binding protein and STAT6. J Biol Chem 280: 14989–14996 [DOI] [PubMed] [Google Scholar]
- Wang Z, Choi MK, Ban T, Yanai H, Negishi H, Lu Y, Tamura T, Takaoka A, Nishikura K, Taniguchi T (2008) Regulation of innate immune responses by DAI (DLM-1/ZBP1) and other DNA-sensing molecules. Proc Natl Acad Sci USA 105: 5477–5482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu LG, Wang YY, Han KJ, Li LY, Zhai Z, Shu HB (2005) VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol Cell 19: 727–740 [DOI] [PubMed] [Google Scholar]
- Yedavalli VS, Neuveut C, Chi YH, Kleiman L, Jeang KT (2004) Requirement of DDX3 DEAD box RNA helicase for HIV-1 Rev-RRE export function. Cell 119: 381–392 [DOI] [PubMed] [Google Scholar]
- Yoneyama M, Fujita T (2007) Function of RIG-I-like receptors in antiviral innate immunity. J Biol Chem 282: 15315–15318 [DOI] [PubMed] [Google Scholar]
- Zupkovitz G, Tischler J, Posch M, Sadzak I, Ramsauer K, Egger G, Grausenburger R, Schweifer N, Chiocca S, Decker T, Seiser C (2006) Negative and positive regulation of gene expression by mouse histone deacetylase 1. Mol Cell Biol 26: 7913–7928 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures 1 and 5