Abstract
Intermodular aminoacyl transfer is the fundamental bond forming reaction in the biosynthesis of polypeptides by ribosomes and nonribosomal peptide synthetases (NRPS). Here we report the design and functional characterizations of short synthetic α-helical peptides that mimic the aminoacyl loading and intermodular aminoacyl transfer steps of NRPS with aminolysis rate enhancements in neutral aqueous solutions of up to 5400-fold (kcat/kuncat). The catalysts operate as noncovalently associated peptide assemblies with composite active sites fashioned at the interface between helical subunits. Following the substrate loading at the active site cysteine, the juxtaposition of the resulting aminoacyl thiolester and the nucleophilic amine of the acyl acceptor moiety gives rise to high effective concentrations (up to 54 M) that facilitate interhelical aminoacyl transfer with rates typically exceeding 10−4 sec−1. Moreover, studies based on homo- and heteromeric assemblies, active site amino acid substitutions, kinetic analysis, and reaction modeling indicate that the de novo designed supramolecular catalysts reported herein exhibit some of the basic characteristics of natural enzymes, including precise positioning and p Ka modulation of active site residues, covalent catalysis, and multiple product turnovers.
Intermodular aminoacyl transfer is the fundamental bond forming reaction in the biosynthesis of polypeptides by ribosomes and nonribosomal peptide synthetases (NRPS).1 Here we report the design and functional characterizations of short α-helical peptides that mimic the aminoacyl loading and intermodular aminoacyl transfer steps of NRPS with aminolysis rate enhancements in neutral aqueous solutions of up to 5400-fold (kcat/kuncat). The catalysts operate as noncovalently associated assemblies with composite active sites fashioned at the interface between helical subunits. Following substrate loading at the active site cysteine, the juxtaposition of the resulting aminoacyl thiolester and the amine of the acyl acceptor moiety gives rise to high effective concentrations (up to 54 M) that facilitate interhelical aminoacyl transfer. Moreover, studies based on homo- and heteromeric assemblies, active site substitutions, kinetic analysis, and reaction modeling indicate that the supramolecular catalysts exhibit some basic characteristics of natural enzymes, including precise positioning and pKa modulation of active site residues, covalent catalysis, and product turnover.2,3
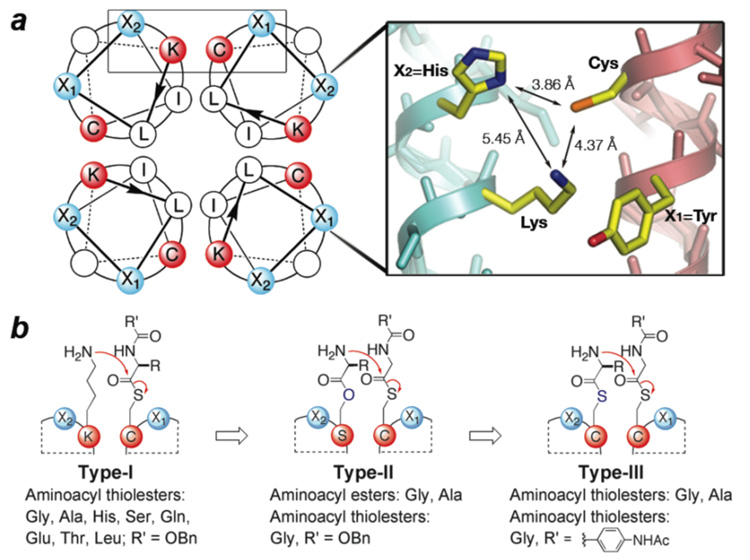
In the present study, our primary objective was to mimic the two fundamental chemical steps of NRPS: aminoacyl substrate anchoring and intermodular aminoacyl transfer. To bring about efficient acyl transfer, we sought to exploit primarily principles of catalysis by approximation4 using 26-residue coiled-coil peptides5 to assemble noncovalently the aminoacyl donor and acceptor moieties into productive complexes (Fig. 1, S1). Because parallel coiled-coil homotetramers are pseudo-four-fold symmetric, each complex contains four putative active sites (Figure 1a). We designed and evaluated three active site variations consisting of Cys for substrate loading and one of the following acyl acceptors: a lysine residue (Type-I); an aminoacyl ester tethered at a serine residue (Type-II); and a second aminoacyl thiolester tethered at a cysteine residue (Type-III) (Figure 1b).
Figure 1.
Representations of the coiled-coil scaffold and the composite aminoacyl transfer active sites. a) (left) Helical wheel diagram of the homotetrameric coiled-coil illustrating the four symmetry-related active sites juxtaposing an aminoacyl donor (cysteine for covalent substrate anchoring via transthiolesterification), an aminoacyl acceptor (amine from lysine or a covalently tethered amino acid), and X1 and X2 residues potentially providing electrostatic or general acid-base catalysis. (right) The 2.17 Å crystal structure of a designed homotetramer6 illustrating the juxtaposition of the putative active site residues. b) Schematic illustration of the three active site designs, highlighting aminoacyl transfer from an aminoacyl-donor to the -acceptor moiety located on an adjacent helix. For clarity, only one of the four symmetry-related active sites is shown (boxed region in the helical wheel diagram). For peptide sequences see Table S1.
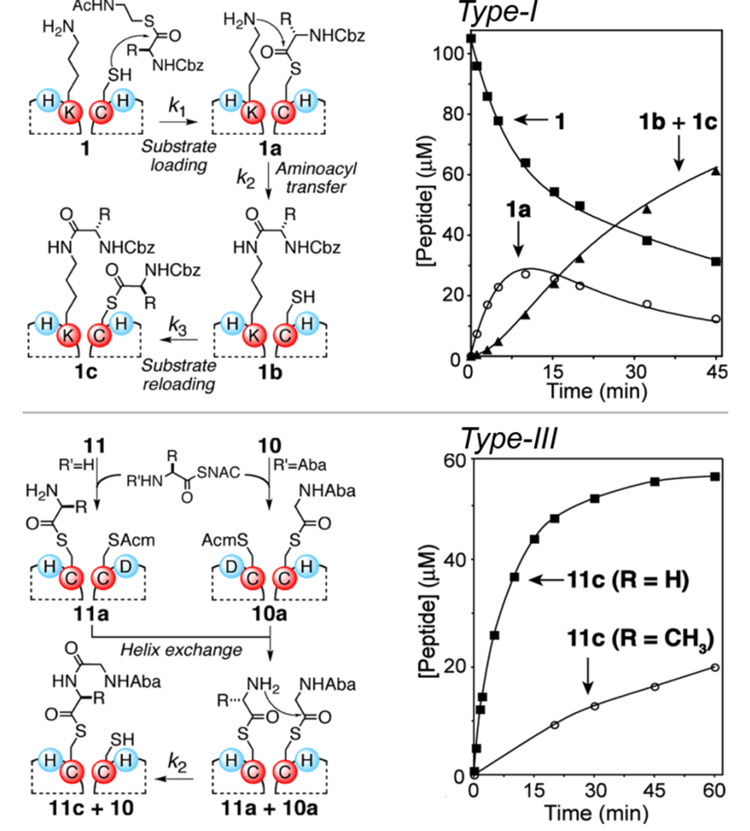
We initially evaluated Type-I active sites (peptides 1–4) using 100-fold excess Cbz-protected N-acetylcysteamine glycyl-thiolester as substrate. Under the neutral aqueous conditions, 1 underwent substrate loading at the active site Cys to form 1a, subsequent interhelical aminoacyl transfer to produce 1b, and reloading to yield product 1c (Figure 2, Figure S3, Table 1). Increases in the aminolysis rate of 5400-fold were observed for 1 relative to the background aminoacylation of Lys in control tripeptide 7. The interhelical mode of acyl transfer was supported by mixing purified 1c with 1, which resulted in rapid aminoacyl transfer to yield approximately two equivalents of 1b (see Fig. S2 for details and an additional proof using labeled peptides). The rate of aminoacyl transfer for 1 was reduced by >300-fold in 6 M Gnd.HCl as would be expected due to the partial unfolding of the coiled coil. Intermolecular rates of lysine aminoacylation in the context of the folded scaffold were determined by assaying 8 and 9, which lack an active site Cys residue. Interestingly, the Lys residues in folded peptides 8 and 9 were acylated 50- and 10-fold faster than Lys in unstructured tripeptide 7, respectively. Preliminary results suggest that this enhanced reactivity is due to a depression of the Lys active site pKa.2a,f Substrate generality was assessed using a representative set of L-aminoacyl thiolesters (Table S2). Although substitutions of the α-substituent had little effect on the aminoacyl transfer rates (k2 varied by ~3-fold), side chain β-substitution resulted in 7 to 13-fold slower aminoacyl loading rates. To probe the influence of active site histidines (X1 and X2) on the aminoacyl loading and transfer steps, 2–4 were employed in which either or both residues were substituted with alanine. While reductions in the acyl transfer rate of up to 13-fold resulted, the observed acyl transfer efficiency of 4 discounts the possibility of acylimidazolyl intermediates in the reaction.2c,3
Figure 2.
Reactions schemes and product formation in time for Type-I and -III active sites. For Type-I, reactions contained ~100 µM peptide, 10 mM Cbz-Gly-SNAC, 10 mM tris-carboxyethyl phosphine (TCEP) as reducing agent, 285 mM Hepes pH 7.0, and ~200 µM acetamidobenzoic acid (Aba) as internal concentration standard. Curve fits shown are from reaction modeling using SIMFIT7. Type-III reactions were initiated under similar conditions by combining 10a (~70 µM) with 11a (~560 µM).
Table 1.
Aminoacyl loading and transfer rate constants.a
The potential for aminoacyl transfer between two proximally tethered amino acids was assessed using Type-II and Type–III active site designs (Fig. 1b). Encouragingly, 5 and 6 having a serine-anchored aminoacyl ester as the acyl acceptor (Type-II) underwent substrate loading and aminoacyl transfer with rates similar to the analogous reaction for 1 (Fig. S5, Table 1), although the reduced transfer rate for 6 likely results from the increased steric influence of the acyl acceptor α-substituent. The possibility of aminoacyl transfer in the Type-III active site was studied using pre-loaded aminoacyl donor (10a) and acceptor (11a, R = H, Me) modules (Figure 2). In this system Cys8 in 10a and Cys13 in 11a are disabled by Acm side chain protection. However, combining 10a and 11a allows the formation of heterotetrameric assemblies which brings together aminoacyl-donor and –acceptor moieties to create competent active sites (Fig. 2c, S4). Indeed, reaction mixtures containing 10a and 11a resulted in aminoacyl transfer with rates similar to those observed for Type-I and -II active sites (Fig. 2), suggesting fast helix exchange followed by efficient intermodular aminoacyl transfer.
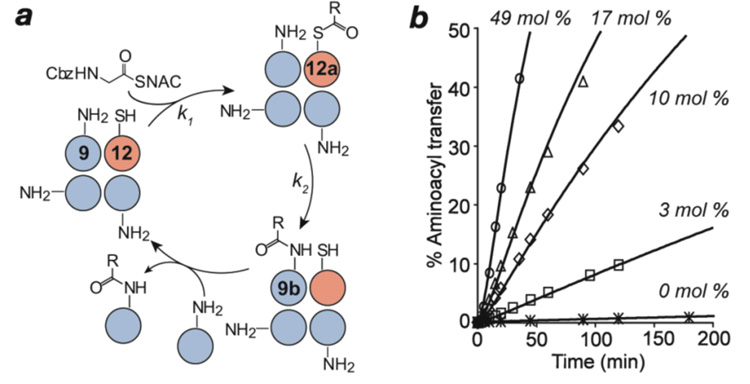
We next examined the viability of catalytic turnover by exploiting heterotetrameric peptide assemblies. Combining 9, which contains an active site aminoacyl-acceptor (Lys) but no – donor (Cys), with 12, having an aminoacyl-donor but no – acceptor, makes possible the formation of heterotetrameric assemblies in which Cys and Lys residues are brought into proximity to form composite Type-I active sites (Figure 3a). As expected, the observed rates of aminoacylation depended markedly on the concentration of 12 present (Figure 3b), reflecting catalyst participation in the reaction. Multiple product turnovers were observed, suggesting that helix subunit exchange rates are faster than the rate of intermodular aminoacyl transfer.
Figure 3.
a) Schematic representation of catalytic aminoacyl loading and transfer cycles based on heterotetrameric assemblies (Type-I active site). Colored circles represent α-helices for the donor peptide 12 (pink) and acceptor peptide 9 (blue). For clarity, we depict only the statistically most predominant catalytically-competent heterotetrameric coiled-coils. b) Aminoacylation of 9 (~360 µM) in reactions containing catalyst 12 at varying concentrations. The curves shown represent best fits of the data to a minimal reaction model, yielding rate constants of k1 = 1.4 × 10−3 sec−1 and k2 = 11.0 × 10−4 sec−1. The catalyst shows multiple turnovers (~1 turnover per hour for the first 20 hours in the 3 mol % reaction).
The studies reported here establish that two fundamental steps of NRPS can be effectively mimicked by appropriately designed self-assembling peptides. However, the hallmark of NRPS lies in their ability based on the logic of their domain organization to instruct the formation of specific peptide sequences.1 It remains to be seen whether the supramolecular approach described here can be further advanced toward programmed peptide synthesis by exploiting the sequence-dependent selective coiled-coil assembly recently demonstrated in the design of complex networks.8
Supplementary Material
Acknowledgment
We thank NIH (GM57690) for financial support and NSF for a predoctoral fellowship (L.J.L.).
Footnotes
Supporting Information Available: Experimental details, biophysical characterizations, tables S1–S2, and figures S1–S4. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.For recent NRPS reviews see:Fischbach MA, Walsh CT. Chem. Rev. 2006;106:3468–3496. doi: 10.1021/cr0503097.Sieber SA, Marahiel MA. Chem. Rev. 2005;105:715–738. doi: 10.1021/cr0301191.Khosla C, Harbury PB. Nature. 2001;409:247–252. doi: 10.1038/35051723.Cane DE, Walsh CT, Khosla C. Science. 1998;282:63–68. doi: 10.1126/science.282.5386.63.
- 2.For examples of catalyst designs based on helical peptide assemblies see:Johnsson K, Allemann RK, Widmer H, Benner SA. Nature. 1993;365:530–532. doi: 10.1038/365530a0.Lee DH, Granja JR, Martinez JA, Severin K, Ghadiri MR. Nature. 1996;382:525–528. doi: 10.1038/382525a0.Yao S, Ghosh I, Zutshi R, Chmielewski J. Nature. 1998;396:447–450. doi: 10.1038/24814.Broo KS, Nilsson H, Nilsson J, Flodberg A, Baltzer L. J. Am. Chem. Soc. 1998;120:4063–4068.Allert M, Baltzer L. Chem. Eur. J. 2002;8:2549–2560. doi: 10.1002/1521-3765(20020603)8:11<2549::AID-CHEM2549>3.0.CO;2-X.Andersson LK, Caspersson M, Baltzer L. Chem. Eur. J. 2002;8:3687–3697. doi: 10.1002/1521-3765(20020816)8:16<3687::AID-CHEM3687>3.0.CO;2-8.Kaplan J, DeGrado WF. Proc. Nat. Acad. Sci. 2004;101:11566–11570. doi: 10.1073/pnas.0404387101.
- 3.Peptide-based acylation catalysts:Miller SJ. Acc. Chem. Res. 2004;37:601–610. doi: 10.1021/ar030061c.Blank JT, Miller SJ. Biopolymers. 2005;84:38–47. doi: 10.1002/bip.20392.
- 4.For designs exploiting proximity to bring about aminoacyl transfer see:Wieland T, Bokelmann E, Bauer L, Lang HU, Lau H, Schafer W. Justus Liebigs Ann. Chem. 1953;583:129–149.Kemp DS. Biopolymers. 1981;20:1793–1804.Sasaki S, Shionoya M, Koga K. J. Am. Chem. Soc. 1985;107:3371–3372.Gennari C, Molinari F, Piarulli U, Bartoletti M. Tetrahedron. 1990;46:7289–7300.Dawson PE, Muir TW, Clark-Lewis I, Kent SB. Science. 1994;266:776–779. doi: 10.1126/science.7973629.Offer J, Boddy CNC, Dawson PE. J. Am. Chem. Soc. 2002;124:4642–4646. doi: 10.1021/ja016731w.Tamura K, Schimmel P. Proc. Nat. Acad. Sci. 2003;100:8666–8669. doi: 10.1073/pnas.1432909100.Ishiwata A, Ichiyanagi T, Takatani M, Ito Y. Tetrahedron Lett. 2003;44:3187–3190.Leleu S, Penhoat M, Bouet A, Dupas G, Papamicael C, Marsais F, Levacher V. J. Am. Chem. Soc. 2005;127:15668–15669. doi: 10.1021/ja054775p.Snyder TM, Liu DR. Angew. Chem. Int. Ed. 2005;44:7379–7382. doi: 10.1002/anie.200502879.Chen G, Warren JD, Chen J, Wu B, Wan Q, Danishefsky SJ. J. Am. Chem. Soc. 2006;128:7460–7462. doi: 10.1021/ja061588y.
- 5.Harbury PB, Zhang T, Kim PS, Alber T. Science. 1993;262:1401. doi: 10.1126/science.8248779. [DOI] [PubMed] [Google Scholar]
- 6.Yadav MK, Redman JE, Leman LJ, Alvarez-Gutierrez JM, Zhang Y, Stout CD, Ghadiri MR. Biochemistry. 2005;44:9723–9732. doi: 10.1021/bi050742a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sievers D, von Kiedrowski G. Chem. Eur. J. 1998;4:629–641. [Google Scholar]
- 8.Ashkenasy G, Jagasia R, Yadav M, Ghadiri MR. Proc. Nat. Acad. Sci. 2004;101:10872–10877. doi: 10.1073/pnas.0402674101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.