Abstract
The type 4 adenylyl cyclase, Adcy4, is the least abundant of five different adenylyl cyclase isoforms expressed in the Y1 mouse adrenocortical cell line and is deficient in a Y1 mutant with impaired steroidogenic factor 1 (SF1) activity. This study examines the contributions of SF1 and other DNA promoter/regulatory elements to Adcy4 expression in the Y1 cell line and its derivative Adcy4-deficient mutant. Primer extension and in silico analyses indicate that Adcy4 transcription initiates from multiple sites just downstream of a GC-rich sequence. Luciferase reporter gene assays identify a 124-bp sequence, situated 19 bp upstream of the major transcription start site and highly conserved among several mammalian species, as the major determinant of Adcy4 expression in Y1 cells and as a site of compromised activity in the Adcy4-deficient mutant. EMSAs using competitor nucleotides and specific antibodies indicate that this conserved region contains three specificity protein (Sp)-1/Sp3-binding sites and one SF1-binding site. As determined by site-specific mutagenesis, the 5′-most Sp1/Sp3-site enhances promoter activity, whereas the middle Sp1/Sp3 and SF1 sites each repress Adcy4 promoter activity. In the Adcy4-deficient mutant, mutating the SF1 site restores Adcy4 promoter activity and knocking down SF1 with small interfering RNAs increases Adcy4 expression, confirming the contribution of SF1 to the mutant phenotype. These studies demonstrate roles for Sp1/Sp3 and SF1 in Adcy4 expression in Y1 cells and establish a repressor function for SF1 in certain promoter contexts.
THE NINE MEMBRANE-BOUND adenylyl cyclase (Adcy) isoform types identified in mammalian cells are distinguished by their tissue-specific patterns of distribution and by their responses to G protein subunits, calcium, protein kinases A and C, and forskolin, properties that contribute to the orchestrated actions of a diverse group of hormones and on their respective target tissues (see Refs. 1, 2 for review). Relatively little is known, however, about the tissue-specific functions of the Adcy isoforms or the factors that govern their expression and contribute to their tissue-selective patterns of distribution. The type-3 enzyme (Adcy3) has a critical role in olfaction (3) and its expression in olfactory neurons is regulated by a transcription factor (Olf-1) that participates in the coordinate regulation of a set of neuronal cell-specific olfactory genes (4); Adcy1 has an important role in aspects of synaptic plasticity and memory (5), and its expression in neurons and pinealocytes requires a 15-bp compound promoter regulatory element comprised of a nuclear receptor binding motif and an E-box (6); Adcy8 has critical roles in synaptic plasticity and memory (5), in modulation of anxiety (7) and in Ca2+-stimulated cAMP accumulation in the parotid (8), and its expression in neuronal tissue is regulated via a cAMP/cAMP response element-binding protein-dependent pathway (9). Although Adcy has long been regarded as an important effector of ACTH action on the adrenal cortex (10,11), the factors governing the expression of the Adcy isozymes in the adrenal cortex and their contributions to adrenocortical function have not been studied extensively. Based on in situ hybridization studies, Adcy9 is the major isozyme expressed in the rat adrenal cortex and its expression is increased after ACTH stimulation (12). Immunohistochemical studies and experiments involving pharmacological manipulation of Adcy activity indicate that the human adrenal cortex expresses Adcy 1-6 and that ACTH action is mediated primarily by the Ca2+-sensitive type 5/6 and to a lesser extent by the β/γ-sensitive type 2/4 isoforms (13).
We investigated the factors that govern the expression of the type-4 isozyme, Adcy4, using the Y1 mouse adrenocortical tumor cell line as a model expression system. Adcy4 is expressed most highly in brain but has been detected at lower levels in a variety of other tissues, suggesting that Adcy4 expression is influenced by both tissue-specific and widely expressed transcription factors (1). Adcy4 is the least abundant of the five Adcy isoforms expressed in the Y1 adrenal cell line and is deficient in a family of mutants that we had isolated from the Y1 adrenal cell line previously. The other Adcy isoforms expressed in Y1 adrenal cells, Adcy1, Adcy3, Adcy5, and Adcy6, are not affected in this mutant family (14). Previously we reported on the sequence of mouse Adcy4 cDNA, the intron-exon organization of the Adcy4 gene and the activity of 1920 bp of 5′-flanking DNA on reporter gene expression in transient transfection experiments (15). The activity of this flanking DNA, which extended from the translation start site into the 3′ end of the upstream Ripk3 gene, was impaired in the Adcy4-deficient Y1 adrenal cell mutants (15), suggesting that specific promoter regulatory sequences in this region contributed to the differential expression of the gene in parent and mutant cells. We thus postulated that determining the molecular basis for the differential expression of this gene in parent and mutant cells is likely to lead to the identification of regulatory factors critical for Adcy4 expression and to a better understanding of the factors that govern its expression in different tissues.
We also showed that the Adcy4-deficient mutants harbor a defect that impairs the ability of steroidogenic factor 1 (SF1) to interact with coactivators such as glucocorticoid receptor-interacting protein-1 without compromising its DNA binding activity (16,17). The defect does not reside in SF1 per se; rather the defect seems to reside in a gene that affects the transactivation function of SF1 (18,19). The loss of SF1 function results in a complex phenotype characterized not only by the loss of Adcy4 expression but also by deficiencies in other SF1-regulated genes such as Mc2r, Cyp11a1, Cyp11b1, and Star (17). These mutants also overexpress SF1 due to the presence of multiple copies of the gene on acentric chromosomal fragments from chromosome 2 (18,19,20). The coexistence of an SF1 defect with an Adcy4 deficiency in the mutant clones prompted us to suggest a regulatory role for SF1 in Adcy4 expression in cells from the adrenal cortex (15). In the present study, we determined the transcription start site of Adcy4, and identified a sequence upstream of the major transcription start site (spanning positions −135 to −19) that is conserved in the Adcy4 flanking DNA from several mammalian species. We have demonstrated that this conserved region is an important determinant of Adcy4 promoter activity in Y1 cells and its derivative mutants and that the transcription factors specificity protein (Sp)-1, Sp3, and SF1 contribute to Adcy4 promoter activity via interactions with this conserved region; the SF1 site in this region acts as a repressor of transcription and accounts, at least in part, for the differential activity of the Adcy4 promoter in parent and mutant cells. We also demonstrate an inhibitory effect of the first intron of Adcy4 on gene expression.
Materials and Methods
Cells and cell culture
The properties of the ACTH-responsive mouse adrenocortical cell line used in this study (clone Y1BS1) and the Adcy4-deficient derivative mutant 10r-6 have been described previously (21) as have the conditions of cell culture. Briefly, cells were grown at 36.5 C under a humidified atmosphere of 5% CO2 in air in nutrient mixture F10 supplemented with 15% heat-inactivated horse serum and 2.5% heat-inactivated fetal bovine serum and the antibiotics penicillin G sodium and streptomycin sulfate. Sera were obtained from Invitrogen Canada Inc. (Burlington, Ontario, Canada).
Plasmids
The reporter gene p-1459Adcy4Luc was prepared as described previously (15). An NcoI restriction fragment isolated from a 10-kb genomic Adcy4 clone (GenBank accession no. AZ420139) was ligated into the unique NcoI site of the luciferase vector pGL3-basic (Promega Corp., Madison, WI) in the correct orientation. This inserted fragment was truncated at the 5′ end by digestion with NheI, thereby generating a luciferase reporter gene containing 1459 bp of 5′-flanking DNA, exon I, intron I, and the noncoding portion of exon II from the Adcy4 gene. Constructs with 5′ deletions were generated from p-1459Adcy4Luc using available restriction sites. The p-404(Δ −135/−19)Adcy4Luc reporter gene was prepared by excision of a BglI/BsmI fragment from p-404Adcy4Luc followed by blunt-end ligation of the plasmid. The p-404(Δ intron)Adcy4Luc reporter gene was prepared by sequential PCR amplification of two fragments that overlapped at exon I as described (22). The first fragment was derived from p-404Adcy4Luc and extended from −404 to the 3′-end of exon I. The second fragment was derived from the Adcy4 cDNA clone BF782774 (15), and extended from the 5′ end of exon I through the NcoI site that marked the translation initiation ATG in exon II. The heteroduplex formed by hybridization of the homologous exon I regions from both fragments was amplified by PCR, and substituted into p-404Adcy4Luc using available restriction sites. The p-142/+92Adcy4Luc plasmid was constructed by PCR amplification of a −142/+92 fragment using p-404Adcy4Luc as template and appropriate oligonucleotides primers; the forward primer incorporated a KpnI restriction site at the 5′ end. The resultant PCR product was subcloned into the EcoRV site of pBluescript II KS+ (Stratagene, La Jolla, CA) by T/A cloning, excised using KpnI and HindIII and inserted into the reporter vector pTA-Luc (CLONTECH Laboratories, Inc., Mountain View, CA) replacing the minimal TATA promoter of the reporter gene with Adcy4 promoter/regulatory sequences but leaving the translation initiation signals of the vector intact. The p-142/+92Adcy4Luc plasmids harboring mutations that disrupted the Sp1A, Sp1B, and SF1 binding sites were generated from p-404Adcy4Luc by PCR using the oligonucleotide based mutation strategy described previously (22). The mutated −142/+92 fragments were inserted into the KpnI and HindIII sites of the pTA-Luc vector as described above for p-142/+92Adcy4Luc.
The small interfering RNA (siRNA) expression plasmid pRNAT-CMV3.1/Neo (Genscript Corp., Piscataway, NJ), was used to express hairpin siRNAs targeting mouse SF1. Three SF1 hairpin oligonucleotides were synthesized based on predictions of optimal effectiveness using siRNA Target Finder and siRNA Construct Builder software from Genscript Corp. The three siRNAs, targeted nucleotides 151/171 (SF1 siRNA no. 1), 565/585 (SF1 siRNA no. 2) and 1375/1395 (SF1 siRNA no. 3), respectively, in the mouse transcript (accession no. AF511594). A germ cell nuclear factor (GCNF) siRNA expression vector, targeting nucleotides 716/736 in GCNF (accession no. NM_010264) was prepared similarly. The identities of all PCR amplified fragments and siRNA cassettes were confirmed by DNA sequencing.
A functional His-tagged SF1 expression vector and its inactive antisense counterpart were prepared as described previously (17).
Primer extension and quantitative RT-PCR
Total RNA was extracted from Y1 mouse adrenocortical tumor cells using guanidine thiocyanate and purified by centrifugation through 5.7 m CsCl (23); Total RNA from B57Bl/6 mouse liver was isolated using a RNeasy minikit (QIAGEN Inc., Canada, Mississauga, Ontario, Canada). Poly(A)-enriched RNA was isolated by two rounds of affinity chromatography over oligo(deoxythymidine) cellulose (New England Biolabs Ltd., Pickering, Ontario, Canada).
Primer extension reactions were carried out using an oligonucleotide primer (5′-AAGAGATCTTCGCTGGGAGG-3′) complementary to the Adcy4 transcript 47 nt downstream of the initiator ATG as described (22). The primer was end labeled with [γ-32P]ATP (50 μCi; 3000 Ci/mmol) using T4 polynucleotide kinase (20 U; New England Biolabs), purified over a stacked column of DE53/AG50W-X8 ion exchange resin and hybridized with 10–50 μg of RNA. The labeled primer was extended by incubation with Superscript II reverse transcriptase (40 U) for 1 h at 42 C in the presence of deoxynucleotide triphosphates and first-strand buffer (Invitrogen Canada). The extended primer was separated by electrophoresis on a 6% acrylamide per 7 m urea gel and visualized by exposure to X-OMAT AR x-ray film (Kodak, Rochester, NY) with the aid of an intensifying screen. The lengths of the extended products were estimated using the same primer in a sequencing reaction with the mouse cDNA clone BF782774, a kidney-derived Adcy4 cDNA that includes 112 bp of 5′ untranslated sequence (15).
For quantitative RT-PCR assays, total RNA (5 μg), purified using an RNeasy minikit (QIAGEN), was reverse transcribed into cDNA using 200 U SuperScript II ribonuclease H− reverse transcriptase (Invitrogen Canada) and an oligo-(deoxythymidine) 18 primer (100 pmol) for 1 h at 42 C in a 20-μl reaction containing 50 mm Tris-HCl (pH 8.4), 75 mm KCl, 3 mm MgCl2, 10 mm dithiothreitol, and deoxynucleotide triphosphates (500 μm each). The cDNA [5 μl of a 1:5 dilution in 10 mm Tris-HCl (pH 8.5)] was analyzed for specific transcripts by quantitative real-time PCR in a 25-μl reaction containing 5 pmol of each primer per well using a Platinum SYBER Green qPCR SuperMix-UDG kit (Invitrogen Canada) and the ABI 7300 real-time PCR system (Applied Biosystems, Foster City, CA). Amplified products were quantitated from cycle threshold values generated using known amounts of cDNA for each gene in the quantitative PCR. Each sample was assayed in triplicate and experiments were repeated at least four times. As an internal control, transcripts were normalized to the amount of TKT transcript in each sample.
EMSAs
Protein binding to the Adcy4 promoter was evaluated in EMSAs essentially as described (22) using radioactive probes encompassing the Sp1A binding site (AACGGGAAGGGGCTGGG CTGTATCGC); the Sp1B and SF1 binding sites (GGGGAAGTGGGAGGGGGCCTTG GCC); and the Sp1A (AACGAGAGGTAGC GGGCTGTATCGC), Sp1B (GGGGAAGT GCTAGACGGCCTTGGCC), and SF1 (GG GGAAGTGGGAGGGGGCTCTTGCGC) sites mutated, respectively. Mutated bases are underlined. Each reaction contained 5 μg nuclear extract, 40 fmol (50,000 cpm/reaction) of double-stranded cDNA probe labeled by end-filling with [α-32P]dGTP (3000 Ci/mmol; PerkinElmer LAS Canada, Woodbridge, Ontario, Canada) and 2 μg double-stranded poly(dI-dC) in a total of 20 μl of binding buffer [20 mm HEPES (pH 7.9), 50 mm KCl, 1 mm EDTA, 1 mm dithiothreitol, and 5% glycerol]. Where indicated, competitor oligonucleotides (100-fold molar excess) or rabbit antibodies against Sp1, Sp3 (1 μg purified IgG/reaction; Upstate Cell Signaling Solutions, Lake Placid, NY) and SF1 (1 μl antiserum) were added. DNA-protein complexes were resolved on 4% nondenaturing polyacrylamide gels and visualized by exposure to x-ray film with the aid of intensifying screens.
Chromatin immunoprecipitation assays
Chromatin immunoprecipitation assays were carried out as described previously (24). Briefly, Y1 adrenal cells stably transfected with a plasmid encoding SF1 with tandem myc and hemagglutinin epitope tags were grown to saturation density, serum deprived for 48 h, and then treated with 2.5 μm α-amanitin (Sigma Chemical Co., St. Louis, MO) for 2 h followed by a 30-min washout to remove the α-amanitin. Cells were rinsed and chromatin was cross-linked by incubating the cells with formaldehyde at a final concentration of 1% at 37 C for 10 min. Cells were rinsed, nuclei were isolated and lysed in 150 μl of a buffer containing 1% sodium dodecyl sulfate, 10 mm EDTA, 50 mm Tris-HCl (pH 8.1), 1× protease cocktail (Sigma) per 2 × 106 cells. Samples were sonicated so as to shear chromosomal DNA to an average length of 500 bp, cleared by centrifugation and then immunocleared with 15 μl of preimmune serum, 2 μg of sheared salmon sperm DNA (Invitrogen), and 80 μl protein A-Sepharose, added as a 50% slurry. Immunoprecipitation was carried out overnight at 4 C and immune complexes were recovered by absorption to protein A-Sepharose beads for 1 h at 4 C. The beads then were washed extensively and immune complexes were extracted using 1% sodium dodecyl sulfate and 0.1 m NaHCO3. Extracts were heated overnight at 65 C to reverse cross-linking and DNA fragments were isolated by extraction with phenol-chloroform and then precipitated. DNA products were assayed for Adcy4 fragments in 25 μl PCR containing 0.5 mm MgCl2 and 25 pmol each of primer pairs spanning −199 to +4 (forward primer: CACCTGCTCATCGACATCG; reverse primer: CTTTGCTAGTGGCTCAGGG) or spanning a 205-bp region within intron 4 (forward primer: GAGCCAAGCATTGATAACACC; reverse primer: TCCATATTCATCTCCTGAGCC). PCRs were initiated by incubating samples at 94 C for 4 min and then were continued for 40 cycles: each cycle consisted of incubations at 94 C for 45 sec, 58 C for 30 sec, and 72 C for 30 C. Samples were then incubated at 72 C for 10 min and cooled to 4 C.
Luciferase reporter gene assays
Cells (2 × 105/dish) were replicate plated in 60-mm tissue culture dishes, grown for 3 d and then transferred to α-MEM supplemented with heat-inactivated horse and fetal bovine sera. Cells were transfected in triplicate with super-coiled plasmid DNA (0.41 pmol/dish) using a high-efficiency calcium phosphate precipitation technique (22) over 24 h, rinsed to remove the DNA, and incubated in fresh medium for 48 h. Cells then were harvested by scraping into a buffer containing 50 mm Tris.2-[N-morpholino]ethanesulfonic acid (pH 7.8), 1% Triton X-100, 4 mm EGTA, and 1 mm dithiothreitol; disrupted by vortexing; and clarified by centrifugation in a refrigerated microfuge. Luciferase activity in cell extracts was assayed by luminometry in a 250-μl cocktail containing 9 mm magnesium acetate, 3 mm Na2ATP, and 60 μm luciferin (see Ref. 18). To correct for variations in transfection efficiencies among experiments, separate dishes of cells from the same replicate plating were transfected with a control vector (pGL3 control; Promega) containing a luciferase reporter gene under control of the Simian virus 40 core promoter and enhancer (17). Results are expressed as a percentage of the activity achieved with the control vector.
Results
Identification of the transcription start site of mouse Adcy4
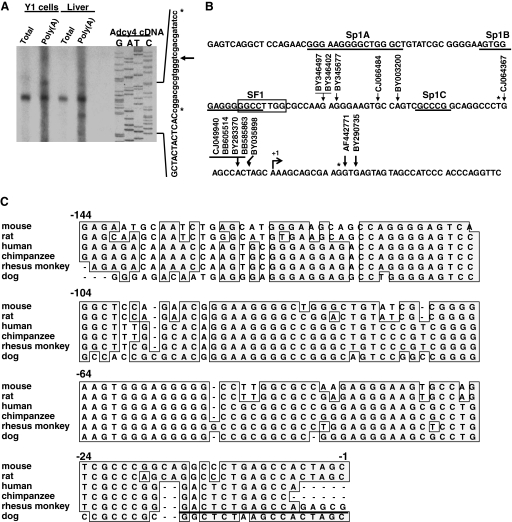
The transcription start site of the mouse Adcy4 gene was determined by primer extension using a radiolabeled Adcy4 oligonucleotide probe and RNA from either Y1 mouse adrenocortical tumor cells or from mouse liver as detailed in Materials and Methods. Total RNA from either source yielded a major product that extended 121 nt upstream of the translation initiator ATG, i.e. 12 nt longer than the mouse Adcy4 cDNA clone isolated and characterized previously (see accession no. AF42771 and NM_080435 and Ref. 15). Reactions using poly(A)-enriched RNA from both Y1 adrenal cells and mouse liver revealed additional, minor primer extension products, including a longer product, extending 133 nt upstream of the translation initiator ATG, and several shorter products (Fig. 1A).
Figure 1.
The proximal promoter region of Adcy4. A, The 5′ ends of Adcy4 transcripts were determined by primer extension using total and poly(A)-enriched RNA from Y1 cells and from mouse liver. The sizes of the extended fragments were determined by sequencing the complementary strand of an Adcy4 cDNA clone (accession no. AF442771) using the same primer. The sequences in capital letters are derived from the Adcy4 cDNA, the sequences in small letters are derived from the vector; the relative positions of the major (arrow) and minor (asterisks) transcripts on the sequencing ladder are indicated. B, The 5′ ends of the Adcy4 transcripts from Y1 cells and mouse liver were compared with those of several full-length RIKEN cDNA clones obtained from various origins. These ends were mapped to a region of the Adcy4 genomic clone (accession no. AC098877). The 5′-end of the major transcript identified in Y1 adrenal cells and C57BL/6 mouse liver is arbitrarily designated +1. Minor transcripts are denoted with asterisks. This region contains three putative Sp1 binding sites (underlined and labeled Sp1A, Sp1B and Sp1C) and a putative SF1 binding site that partially overlaps the Sp1B site (boxed) as identified by searching the Transfac Professional database (51) with the PATCH algorithm (formerly Patsearch) (52). C, The proximal 144 bp of DNA sequence flanking the mouse Adcy4 was compared with corresponding sequences flanking the Adcy4 from rat (accession no. NT_039606), human (accession no. NT_026437), chimpanzee (accession no. NW_001224596), rhesus monkey (accession no. NW_0011211199), and dog (accession no. NW_8876327). The sequences were aligned using a ClustalW alignment tool provided with MacVector 7.2 software (MacVector, Inc., Cary, NC); conserved sequences (boxed) and gaps introduced for optimal alignment (−) are indicated. Part of the sequence assigned to exon I of the rhesus monkey Adcy4 (underlined) is included to complete the comparison.
To further map the first exon of Adcy4 and to identify the transcription start site, full-length RIKEN cDNA clones with 5′-sequences homologous to mouse Adcy4 cDNA were retrieved from the mouse est database at the National Center for Biotechnology Information (search date: July 5, 2007) using BLAST (basic local alignment search tool) (25) and aligned with the mouse Adcy4 genomic sequence (accession no. AC098877) (15). As shown in Fig. 1B, the RIKEN clones had 5′ ends of different lengths and contained sequences that were contiguous with the sequence previously assigned to the first exon of Adcy4 (15). These sequences flanked the major and minor transcription start sites determined by primer extension of Y1 and mouse liver RNA. This in silico analysis confirms the previous assignment of Adcy4 exon I (15) and indicates that the 5′-end of this exon may extend by as much as 53 nt in some mouse tissues. The primary transcription start site in mouse liver and Y1 mouse adrenocortical cells, based on our primer extension data, is arbitrarily designated +1. The sequence in the upstream vicinity of the Adcy4 transcription start site does not contain an identifiable TATA box core promoter but is GC-rich with three Sp1-like motifs centered at −23, −57, and −86 nt and with an SF1-like motif that partially overlaps with the Sp1 site at −57 nt (Fig. 1B). These putative regulatory elements reside in a stretch of 144 nucleotides that is highly conserved (>75%) in the immediate upstream regions of the Adcy4 genes from rat, human, chimpanzee, rhesus monkey, and dog (Fig. 1C). Together these results suggest that transcription of the mouse Adcy4 may initiate at multiple sites within a GC rich region upstream of exon I and that Sp1- and SF1-like motifs may be important determinants of Adcy4 expression.
Deletion analysis of the mouse Adcy4 promoter
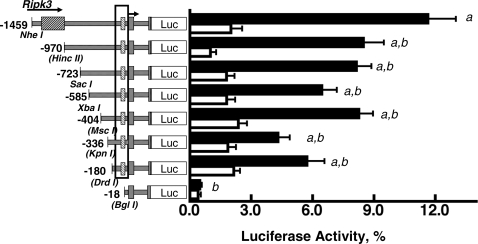
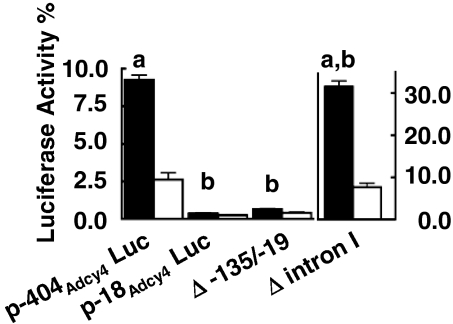
To identify important promoter regulatory elements in the 5′-flanking region of Adcy4, a series of luciferase reporter gene constructs, each containing exon I, intron I, the noncoding portion of exon II, and various lengths of Adcy4-flanking DNA (Adcy4Luc) was prepared and evaluated for luciferase activity after transfection into Y1 adrenal cells. The reporter gene p-1459Adcy4Luc containing 1459 bp of flanking DNA extending into the last intron of the upstream Ripk3 gene had 24-times the activity of p-18Adcy4Luc, a truncated product containing only 18 bp of 5′-flanking DNA (Fig. 2). Progressively truncating the flanking DNA from 1459 to 180 bp yielded plasmids with promoter activities that were significantly lower (30–50%) than p-1459Adcy4Luc but not significantly different from each other (Fig. 2). Further truncation of the 5′ end from 180 to 18 bp reduced activity 11-fold and yielded an activity that was not significantly different from that of the basic luciferase reporter vector devoid of Adcy4 sequences (data not shown). Extending the amount of flanking DNA upstream of the transcription start site to 4536 bp, i.e. into intron I of the Ripk3 gene, did not further enhance the activity of the luciferase reporter (data not shown). These results thus indicate that the regions between −1459 and −970 and between −180 and −18 in the DNA flanking Adcy4 contribute to its promoter activity. The latter region includes the 144 nt that are highly conserved in the Adcy4 promoters of several other species (see Fig. 1C). Deleting this conserved region, in the context of p-404Adcy4Luc, resulted in a promoter activity that was significantly decreased and not different from that obtained with p-18Adcy4Luc (Fig. 3). Surprisingly, deletion of intron I increased Adcy4 promoter activity 3.4-fold over that seen with p-404Adcy4Luc, indicating an inhibitory influence of the first intron on the reporter gene (Fig. 3).
Figure 2.
Effects of 5′ deletions on Adcy4 promoter activity in Y1 and mutant 10r-6 cells. Y1 (black bars) and mutant 10r-6 cells (white bars) were transfected with luciferase expression vectors (0.41 pmol/dish) containing exon I, intron I, and part of exon II up to the initiator ATG and varying lengths of DNA flanking the transcription start site (arrow). The last exon of the Ripk3 gene is indicated, as is the sequence between −135 and −19 that is conserved among species (boxed). The restriction sites used to progressively truncate the 5′ end of the Adcy4 promoter also are shown; the sites destroyed by blunt end ligation are indicated with brackets. Results are expressed as mean percentages of the activity obtained with the pGL3 control vector (i.e. a luciferase reporter gene under control of the Simian virus 40 core promoter and enhancer) ± sem. Depending on the plasmid, data were compiled from eight to 20 independent experiments in Y1 cells and from four to 11 independent experiments for 10r-6 cells. Statistically significant differences, determined by ANOVA followed by the Newman-Keuls multiple comparison test, are: a, different from p-18Adcy4Luc, P < 0.001; b, different from p-1459Adcy4Luc, P < 0.001. All of the constructs, except p-336Adcy4Luc and p-18Adcy4Luc, had activities that were significantly higher (P < 0.05) in parent Y1 cells than in mutant 10r-6 cells.
Figure 3.
Effects of internal deletions on Adcy4 promoter activity in Y1 and mutant 10r-6 cells. The activities of p-404Adcy4Luc and p-18Adcy4Luc were compared with the activities of p-404Adcy4Luc constructs containing either an internal deletion between −135 and −19 (Δ −135/−19) or a deletion of intron I (Δ intron I) after transfection into Y1 (black bars) and mutant 10r-6 cells (white bars) as described in Fig. 2. Results were compiled from three to six independent experiments, depending on the plasmid and cell line and are expressed as means ± sem. Note that the activities obtained using the Δ intron I construct are represented on a different scale. Bars (a), values significantly different from p-18Adcy4Luc, P < 0.001; bars (b), values significantly different from p-404Adcy4Luc, P < 0.001. Only p-404Adcy4Luc, and the Δ intron I construct had activities that were significantly different in parent Y1 cells and mutant 10r-6 cells, P < 0.001 (a and b).
To further test the contributions of the Adcy4 flanking DNA to Adcy4 expression, we evaluated the activities of the same deletion constructs in the Adcy4-deficient Y1 adrenal cell mutant, 10r-6. Each of the constructs tested yielded activities that were significantly lower in the mutant than in the Y1 parent, with the exceptions of p-18Adcy4Luc and p-336Adcy4Luc, which did not differ between parent and mutant cells (Figs. 2 and 3). As noted above, the activity of p-18Adcy4Luc did not differ from the promoterless vector. The apparent loss of differential expression of p-336Adcy4Luc in parent and mutant cells appeared to reflect a type II statistical error rather than the loss of an important regulatory element, because mutating the −404 to −320 region of the p-404Adcy4Luc vector did not reduce promoter activity in parent Y1 cells or abolish the differential expression of the reporter gene in parent and mutant cells (data not shown). In addition, truncating p-336Adcy4Luc to p-180Adcy4Luc yielded a plasmid with significantly greater activity in parent Y1 cells than in the 10r-6 mutant cells (Fig. 2).
Taken together, our data suggest that the conserved sequence in the proximal region of the Adcy4 promoter contains one or more important regulatory elements that drive gene expression in parent Y1 cells and have reduced activity in the 10r-6 mutant.
Identification of transcription factor binding sites in the Adcy4 promoter
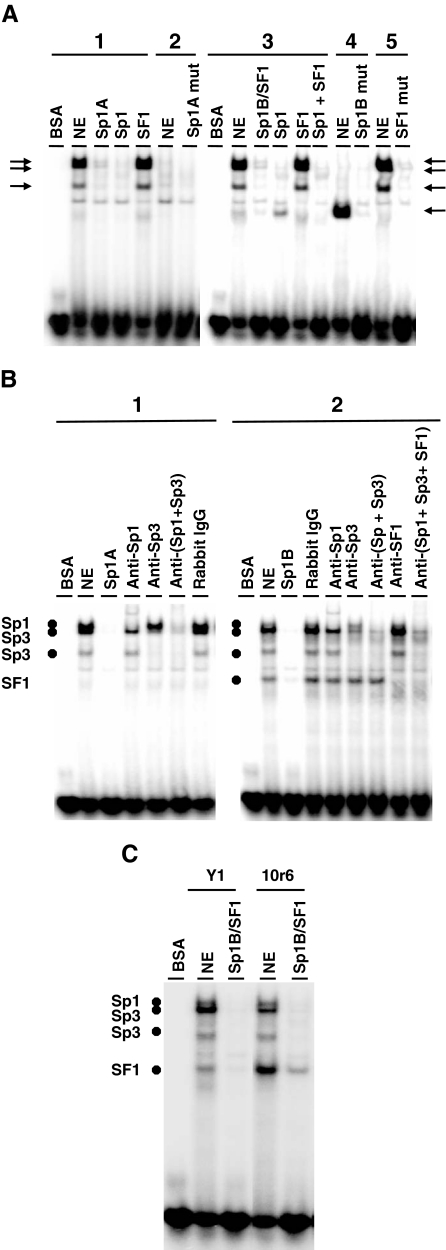
Seven overlapping labeled oligonucleotides were used in EMSAs with nuclear extracts from Y1 adrenocortical cells to identify transcription factor binding sites in the −135 to −19 region of the Adcy4 promoter. Only those oligonucleotides that included the putative Sp1 binding sites and the putative SF1 site (e.g. Fig. 1B) formed specific shifted complexes with Y1 nuclear extracts. Incubation of nuclear extracts from Y1 cells with a [32P]-labeled oligonucleotide encompassing the Sp1A binding site produced three shifted complexes that were competed away with an excess of the corresponding unlabeled oligonucleotide (Fig. 4A). Incubation of Y1 nuclear extracts with a labeled oligonucleotide that included the Sp1B and overlapping SF1 sites yielded the same three shifted complexes seen with the Sp1A probe plus an additional complex of faster mobility, all of which were competed away by an excess of the corresponding unlabeled oligonucleotide (Fig. 4A). To determine whether Sp1 and SF1 were associated with these shifted complexes, EMSAs were conducted in the presence of competitor oligonucleotides corresponding in sequence to a consensus Sp1 binding site (26) or to an authentic SF1 binding site (17). As shown in Fig. 4A, the three shifted complexes common to both oligonucleotide probes were competed away by the Sp1-binding oligonucleotide but not by the SF1-binding oligonucleotide. In addition, mutating the putative Sp1 binding sites in the corresponding labeled oligonucleotides prevented the formation of these complexes. Similar results were obtained using an oligonucleotide probe that included the Sp1C site (data not shown). Interestingly, the shifted complex unique to the labeled oligonucleotide containing the Sp1B and SF1 sites was enhanced in the presence of the Sp1-binding competitor oligonucleotide; this complex was also enhanced when using a labeled oligonucleotide with the Sp1 sites mutated. This latter complex was specifically competed away by the SF1-binding oligonucleotide and did not form when EMSAs were conducted using a labeled oligonucleotide with the SF1 binding site mutated (Fig. 4A). Antibodies to Sp1 partially disrupted and partially supershifted the complex of slowest mobility common to the labeled probes containing the Sp1A and Sp1B/SF1 sites; antibodies to Sp3 disrupted the other two complexes common to both probes; combining the Sp1 and Sp3 antibodies markedly diminished the assembly of all three complexes but did not affect the formation of the putative SF1 complex; an antibody to SF1 specifically disrupted the complex associated with the SF1 binding site (Fig. 4B). Combining anti-Sp1, -Sp3, and -SF1 antibodies markedly diminished all the specific complexes formed on the labeled Sp1B/SF1 oligonucleotide, whereas a control rabbit IgG did not affect complex formation (Fig. 4B). Nuclear extracts from the 10r-6 mutant produced the same shifted complexes as did extracts from parent Y1 cells using either the Sp1A-containing oligonucleotide (data not shown) or the Sp1B/SF1 containing oligonucleotide (Fig. 4C) as the labeled probe; however, the fastest migrating complex associated with the SF1 binding site was more prominent (Fig. 4C). This latter finding is consistent with our earlier observation that SF1 is overexpressed in the 10r-6 mutant (18).
Figure 4.
Sp1 and SF1 binding sites in the proximal promoter region of Adcy4. A, DNA-protein interactions were assessed using radiolabeleld double-stranded oligonucleotides that included the Sp1A site (1), the Sp1A site mutated (2), the overlapping Sp1B/SF1 sites (3), and the Sp1B and SF1 sites mutated (4 and 5, respectively). These radiolabeled oligonucleotides were evaluated for interaction with BSA or with nuclear extracts (NE) from Y1 adrenal cells by EMSA. Where indicated, binding reactions were carried out in the presence of a 100-fold molar excess of the corresponding unlabeled oligonucleotides, or bona fide Sp1 (ATTCGATCGGGGCGGGGCGAGC) and SF1 (CTTTACTCAAGGTGAGGATAAA)binding sites. The positions of specific shifted complexes (arrows) are indicated. B, Complex formation was determined as in panel A using double-stranded oligonucleotide probes containing the Sp1A site (1) or the overlapping Sp1B/SF1 sites (2). Where indicated, the corresponding unlabeled oligonucleotides (100-fold molar excess), a rabbit IgG control, IgG antibodies against Sp1 or Sp3 (1 μg), or SF1 antiserum (1 μl) were added to the reaction. The positions of the complexes displaced by the Sp1, Sp3, and SF1 antibodies are indicated. C, DNA-protein interactions on the overlapping Sp1B/SF1 site were assessed using nuclear extracts from either Y1 or 10r6 cells or BSA as indicated. Reactions carried out in the presence of 100-fold molar excess of the unlabeled SP1B/SF1 oligonucleotide were included as controls for specificity. The identities of the shifted complexes are based on their displacement with specific antibodies (B).
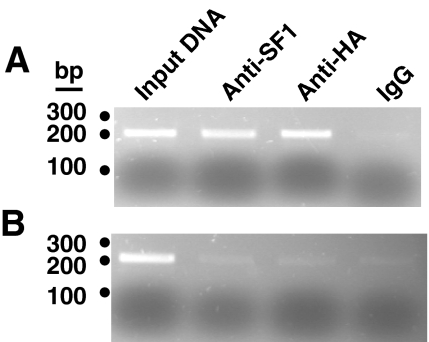
The interaction of SF1 with the proximal promoter region of Adcy4 also was evaluated by chromatin immunoprecipitation assay using cross-linked, sonicated nuclear extracts of Y1 cells expressing a myc- and hemagglutinin-tagged SF1. Immunoprecipitation with an anti-SF1 or antihemagglutinin antibody precipitated chromosomal DNA that could be amplified by primers directed to proximal promoter region of Adcy4 (from −199 to +4) but not by primers targeted to intron 4 of the gene (i.e. from +2548 to +2744). In contrast, immunoprecipitates obtained with a control IgG did not produce amplified fragments using either of the Adcy4 primer pairs (Fig. 5). These results indicate that SF1 interacts with the proximal promoter region of Adcy4 in situ and together with the EMSA data suggest that Sp1, Sp3 and SF1 may be responsible for the promoter regulatory activity associated with the conserved region of the Adcy4 promoter from −135 to −19.
Figure 5.
Interaction of SF1 with the proximal promoter region of Adcy4 in situ. Cross-linked nuclear extracts of Y1 cells expressing a myc- and hemagglutinin-tagged SF1 were sonicated, immunoprecipitated, and assayed for Adcy4 DNA by PCR as described in Materials and Methods. Where indicated, samples were immunoprecipitated with an SF1 antibody (anti-SF1; Upstate BioTechnology), a hemagglutinin epitope antibody (anti-HA; Santa Cruz Biotechnology Inc., Santa Cruz, CA), or a control IgG. A sample containing 1% of the sonicated DNA used for immunoprecipitation (input DNA) was included as a positive control for Adcy4. Immunoprecipitated DNA was amplified using primer pairs that spanned the proximal promoter region of Adcy4 from −199 to +4 (A) or primer pairs that spanned a region from +2548 to +2744 downstream of the transcription start site and within intron 4 of Adcy4 (B). Amplified fragments were sized using a 100-bp DNA ladder (Fermentas Canada, Burlington, Ontario, Canada).
Contributions of the Sp1 and SF1 binding sites to Adcy4 promoter activity
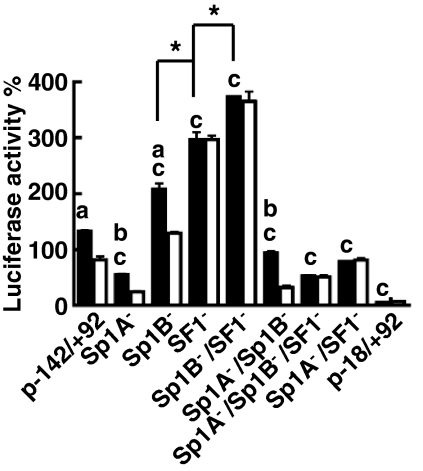
To test the contributions of the Sp1 and SF1 sites to Adcy4 promoter activity, a series of luciferase reporter gene plasmids were constructed with mutations that disrupted the Sp1A-, Sp1B-, and SF1-binding sites and tested for activity after transfection into Y1 and mutant 10r-6 cells (Fig. 6). These mutations were evaluated in the context of the Adcy4 promoter-regulatory sequences from −142 to +92 to obviate the inhibitory influences of the first intron (see Fig. 3). The −142/+92 construct had 25 times the activity of the truncated −18/+92 reporter construct in parent Y1 cells. Mutating the Sp1B site, the SF1 site, or both the Sp1B and SF1 sites further increased reporter gene activity 1.6-, 2.2-, and 2.8-fold respectively (P < 0.001; Fig. 6). Mutating the Sp1A site, either alone or in combination with the Sp1B and/or SF1 sites decreased reporter gene activity relative to the activity of p-142/+92Adcy4Luc (P < 0.001; Fig. 6).
Figure 6.
Contributions of the Sp1 and SF1 binding sites to Adcy4 promoter activity. Y1 (black bars) and mutant 10r-6 (white bars) cells were transfected with luciferase expression vectors (0.41 pmol/dish) driven by exon I of Adcy4 plus 142 bp of 5′flanking DNA (p-142/+92) or p-142/+92 constructs with mutations in the Sp1A (Sp1A−), Sp1B (Sp1B−), and/or SF1 (SF1−) sites as indicated. The activities of these constructs were compared with that of a truncated promoter extending only18 bp upstream of the Adcy4 transcription start site (p-18/+92). Results compiled from three independent experiments are expressed as described in the legend to Fig. 2. Bars (a and b), values that are significantly different in Y1 and mutant 10r-6 cells at P < 0.001 and < 0.05, respectively; bars (c), values obtained in Y1 cells that are significantly different from the value obtained using the wild-type −142/+92 construct. Bars (*), significant differences in activities in Y1 cells between the indicated plasmids (P < 0.001).
In the absence of the inhibitory influence of intron I, the p-142/+92Adcy4Luc reporter gene also had appreciable activity in the 10r-6 mutant (∼12 times the activity of the truncated p-18/+92Adcy4Luc) that was nevertheless significantly lower than the activity observed in parent Y1 cells (P < 0.001; Fig. 6). All of the constructs with an intact SF1-binding site had activities that were lower in the 10r-6 mutant cells than in parent Y1 cells, whereas all the constructs with the SF1 site disrupted had activities that were equivalent in the mutant and parent cell lines (Fig. 6). These latter observations indicate that SF1 contributes to the impaired activity of the Adcy4 promoter and consequently to the Adcy4 deficiency in the mutant cell line.
Suppressor effects of SF1 on Adcy4 promoter activity in Y1 cells
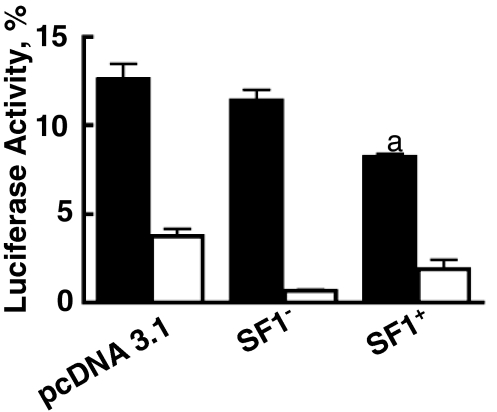
Inasmuch as the data presented above suggested a suppressor-like function of SF1 on the Adcy4 promoter, we evaluated the effects of SF1 on the activity of the Adcy4 promoter in parent Y1 adrenal cells. Cotransfection experiments with an antisense SF1 expression plasmid or the empty expression plasmid pcDNA3.1 served as controls. As shown in Fig. 7, an epitope-tagged SF1 expression vector suppressed the activity of p-970Adcy4Luc by 35% (P < 0.001), whereas the empty vector and the antisense epitope-tagged SF1 control were without effect. Given that 50% or less of these cells express both genes when cotransfected on two different plasmids (27), the 35% reduction in activity seen here (Fig. 7) implies a substantial suppressive effect of SF1 on the activity of the Adcy4 promoter. In contrast, the effect of the epitope-tagged SF1 expression vector on the activity of the truncated reporter gene, p-18Adcy4Luc, was not significantly different from that of the antisense epitope-tagged SF1 vector, although both vectors reduced the activity of the p-18Adcy4Luc reporter plasmid, compared with the pcDNA3.1 control (Fig. 7).
Figure 7.
Effect of an epitope-tagged SF1 on Adcy4 promoter activity in Y1 cells. Y1 cells were transfected with the luciferase reporter plasmids p-970Adcy4Luc (black bars) and p-18Adcy4Luc (white bars) together with equimolar amounts of a His-tagged SF1 expression vector (SF1+), a His-tagged SF1 antisense control (SF1−) or the empty expression plasmid, pcDNA3.1. The luciferase reporter gene constructs and the methods of data analysis are as indicated in Fig. 2. The activity of His-tagged SF1 was compiled from six independent experiments, whereas the activities of the two control plasmids were each compiled from three independent experiments. The activities of p-18Adcy4Luc were significantly different from those of p-970Adcy4Luc (P < 0.001) under all experimental conditions. The effect of His-tagged SF1 on p-970Adcy4Luc (a) was significantly different (P < 0.001) from that of pcDNA3.1 or the antisense His-tagged SF1.
Effects of SF1 knockdown on the expression of endogenous Adcy4
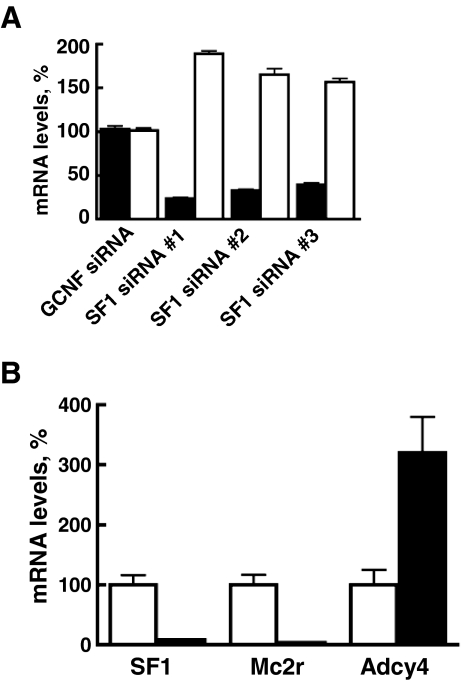
To test the contribution of SF1 to the Adcy4-deficient phenotype of the 10r-6 mutant, cells were transfected with plasmids encoding siRNAs targeted to three different parts of the SF1 transcript. Transformants were isolated from each transfection by selective growth in G418 and clonal isolates at first passage were assayed for SF1 and Adcy4 transcripts by quantitative RT-PCR. In cells transfected with the SF1 siRNAs, SF1 transcripts were reduced by 68% (range from 60 to 76%). In the same transformants, Adcy4 transcripts increased by 178% (range from 157 to 189%). This increase in Adcy4 transcripts after treatment of the mutant clone with SF1 siRNA is deemed to represent a partial recovery of Adcy4 expression because Adcy4 levels in parent and mutant cells differ by more than 4-fold (14). In contrast to the SF1 siRNA vectors, a siRNA plasmid targeted to an unrelated nuclear receptor, i.e. GCNF (Nr6a1), did not affect the levels of SF1 and Adcy4 transcripts in 10r-6 cells, although GCNF transcripts were reduced approximately 50% (Fig. 8A). Similar outcomes were obtained when SF1 was knocked down in Y1 cells after transfection with SF1 siRNA (Fig. 8B). In four different isolates recovered after transfection with SF1 siRNA, SF1 transcripts were reduced by 92% as were Mc2r transcripts that are expressed in an SF1-dependent manner (17,24,28). In contrast, Adcy4 transcripts were increased 3.2-fold, supporting a suppressor-like function of SF1 on Adcy4 expression.
Figure 8.
Effects of SF1 knockdown on Adcy4 mRNA levels. Mutant 10r-6 and parent Y1 cells were transfected with siRNAs targeted to specific regions of the SF1 transcript. Stable transformants were recovered and transcripts were assayed by quantitative RT-PCR. Where indicated, SF1 cDNA was amplified using TGGACTATTCGTACGACGAGG and GACTGTGCGCTTGAAGAAGC; Adcy4 cDNA was amplified using TGAGGAGGAAGACGAGAAGG and TGTCAAGGTGGCTTAGGTGG; Mc2r transcripts were amplified using CAGTGCTCACCTTCACATCG and AAGGATGGTTAGTGTCATGGC as forward and reverse primers, respectively. Results are expressed as the mean percentage of transcript levels obtained in untransfected cells ± sem. A, Mutant 10r-6 cells were transfected with siRNAs targeted to nucleotides 151/171 (SF1 siRNA no. 1), 565/585 (SF1 siRNA no. 2), and 1375/1395 (SF1 siRNA no. 3) of SF1 and results were compiled from three separate experiments with each transformant. Cells transfected with siRNA targeted to the nuclear receptor, germ cell nuclear factor (GCNF) (Nr6a1) served as a negative control. The levels of SF1 (black bars) and Adcy4 (white bars) observed in 10r-6 cells expressing SF1 siRNA were all significantly different from the corresponding levels obtained in 10r-6 cells expressing GCNF siRNA (determined by ANOVA followed by the Newman-Keuls multiple comparison test). B, Y1 cells were transfected with SF1 siRNA no. 1, and results were compiled from four independently isolated SF1 siRNA expressing clones. The levels of SF1, Mc2r, and Adcy4 observed in cells expressing SF1 siRNA (black bars) were all significantly different from the levels obtained in control samples (white bars) as determined by ANOVA followed by the Bonferroni multiple comparison test after log transformation of the data to reduce sample variances (53).
Discussion
The results from primer extension of Y1 adrenal cell and B57Bl/6 mouse liver RNAs and the examination of selected full-length RIKEN clones suggest that the mouse Adcy4 has multiple transcription start sites that are clustered within a small contiguous region of approximately 56 bp and that are used in a tissue-selective manner (Fig. 1B). In silico analysis of the 5′ ends of Adcy4 transcripts using the mouse CAGE (cap analysis gene expression) database (29) also indicated that Adcy4 has multiple transcription start sites clustered within 50 bp of the major site used by Y1 cells and mouse liver (data not shown). This spread of transcription start sites is typical of promoters without classical core elements such as TATA boxes and initiator-like elements and is often associated with ubiquitously expressed genes (30). The proximal portion of the Adcy4 5′-flanking DNA is GC-rich and contains several Sp1 binding sites; one or more of these may facilitate positioning of the transcription preinitiation complex as reported for certain other genes (31). Several lines of evidence support the importance of these Sp1 sites for Adcy4 expression. These include the highly conserved nature of these sites in similar parts of the Adcy4 promoter in several mammalian species (Fig. 1C) and the loss of Adcy4 promoter activity upon deletion (Figs. 2 and 3) or mutation (Fig. 6) of these sites. In the mouse, the Sp1A site appears to have a positive effect on Adcy4 promoter activity, whereas the Sp1B site appears to have an inhibitory influence. Both positive and negative effects of Sp1 sites on gene transcription have been reported previously and appear to be governed by a number of variables, including the relative abundance of Sp1 and the long and short (inhibitory) forms of Sp3; the posttranslational modifications of Sp1 and Sp3; the levels of Sp1- and Sp3-interacting proteins; and their residence times on Sp1 higher-order complexes (32). The Y1 and mutant adrenal cell lines express Sp1 and both the long and short forms of Sp3, as determined by Western blot analysis (data not shown), and the Sp1A and Sp1B sites in the Adcy4 promoter bind both Sp1 and Sp3 in EMSA using naked DNA (Fig. 4B); however, in the context of chromatin, the relative selectivity of Sp1 and the different Sp3 isoforms for these sites may be different (33).
Intriguingly, SF1 appears to have a negative regulatory effect on mouse Adcy4 expression through a site that overlaps with the Sp1B site in the proximal region of the promoter. Whereas the SF1 and Sp1B sites overlap in the flanking regions of mouse and rat Adcy4, these sites are not conserved in the corresponding regions in primates or dog (Fig. 1C); however, there is an SF1-like site at −134 bp in primates that may have a corresponding effect on Adcy4 expression in these species. The SF1 site in Adcy4 is most like the SF1 binding sites in the promoters of the horse and rat LHB (TGGCCTTG) (34,35) and the human MC2R (CCAAGTCC on the complementary strand) (36); however, it differs from each by a single nucleotide. Because slight variations in the sequence of DNA binding elements can have profound allosteric effects on transcription factor activity (37,38,39), this variant binding sequence may contribute to the suppressor activity of SF1 at the Adcy4 promoter. Alternatively, other contextual features of this SF1 site may be responsible for its inhibitory effect. Our data suggest that SF1 does not act cooperatively with Sp1 or Sp3 at this overlapping site in the mouse; rather, SF1 appears to act competitively with Sp1 and Sp3 forming an independent complex. In support of this suggestion, we find that Sp1 and Sp3 competitor oligonucleotides and Sp1 and Sp3 antibodies all fail to displace SF1 bound to this site; conversely, SF1 competitor oligonucleotides and antibodies fail to displace those complexes ascribed to Sp1 or Sp3. In addition, displacement of Sp1 and Sp3 from this site by competitor oligonucleotides or mutation of the Sp1 binding sequence increases the amount of SF1 bound (Figs. 4, A and B).
Our findings that disruption of the SF1 site by mutation abolishes the differential activity of Adcy4 promoter constructs in Y1 and mutant adrenal cells (Fig. 6), that cotransfection of SF1 with an Adcy4 promoter construct suppresses Adcy4 promoter activity in Y1 cells (Fig. 7), and that knockdown of SF1 using specific siRNAs increases Adcy4 transcript accumulation in the Adcy4-deficient mutant as well as in parental Y1 cells (Fig. 8) support the conclusion that SF1 is an important negative regulator of Adcy4 expression in SF1-expressing cells. We cannot tell, however, whether the Adcy4 deficiency in the 10r-6 mutant results from the overexpression of SF1 or from its impaired function because either scenario could enhance the suppressive effect of SF1.
SF1 is expressed in a tissue-selective manner to regulate the expression of genes associated with steroidogenesis and reproduction (40,41). Although SF1 has been shown to interact with transcriptional corepressors (reviewed in Ref. 41) and to interact with inhibitory ligands (42), SF1 is generally regarded as an activator of gene expression at its target genes (40,41). Our finding that SF1 acts as a repressor at the Adcy4 promoter is consistent with the behavior of other nuclear receptors that exhibit gene-specific repressor and activating functions (e.g. the glucocorticoid receptor and the thyroid hormone receptor) (43). The suppression of Adcy4 expression by SF1 may explain why Adcy4 is the least abundant of the five Adcy isoforms expressed in the Y1 cell line (14). Inasmuch as Adcy4 is regulated by the β/γ subunit complex of guanyl nucleotide regulatory proteins (44), whereas the other isoforms expressed in Y1 cells are affected by calcium (14), the net effect of SF1 would be to decrease the influence of the β/γ subunit complex of guanyl nucleotide regulatory proteins on cAMP signaling and increase the influence of calcium.
The extent to which SF1 mediates gene repression globally has not been evaluated extensively. In H295R adrenal tumor cells (45), tetracycline-induced overexpression of SF1 resulted in both up-regulation and down-regulation of distinct sets of transcripts that accompanied an increase in cell proliferation and decreases in apoptosis and glucocorticoid biosynthesis. Whether the changes in transcript accumulation seen upon overexpression of SF1 reflect inductive and suppressive effects of SF1 on gene expression or are secondary to effects on growth, apoptosis, or steroidogenesis is uncertain. Future studies that correlate genome-wide SF1 binding with genome-wide expression data will be required to determine the global influences of SF1 on gene repression. It is tempting to speculate that gene repression by SF1 might contribute to the functional zonation of the adrenal cortex. As inferred from transfection experiments, SF1 suppresses the effects of NURR1, NGFIB, and COUP-TF on CYP11B2 expression (46,47) and the effects of NGFIB on HSD3B2 expression (48) in H295R adrenocortical tumor cells. These suppressive effects might influence the relative expression of these genes in the three zones of the adrenal cortex and consequently impact on the zone-specific synthesis of aldosterone, cortisol and dehydroepiandrosterone.
Finally, the first intron of Adcy4 also seems to contribute independently to the low levels of Adcy4 relative to the other Adcy isoforms expressed in these adrenal cell lines (14) through its inhibitory effect on the reporter gene (Fig. 3). We have not explored the mechanism of this inhibitory effect; however, negative influences of introns on gene expression (49) and mRNA export from the nucleus have been reported previously (50), and one of these mechanisms may be operative here.
Acknowledgments
We thank Ms. Shara Hong for technical assistance and Dr. Ken Morohashi (National Institute for Basic Biology, Aichi, Japan) for the rabbit anti-SF1 antiserum.
Footnotes
This work was supported by Research Grant MOP-64325 (to B.P.S.) from the Canadian Institutes of Health Research, Research Grant R01-DK62027 from the National Institutes of Health (to G.D.H.), and a Canada Graduate Scholarship from the Natural Sciences and Engineering Research Council of Canada and a University of Toronto Open Studentship Award (to X.R.).
Disclosure Statement: X.R., J.T. and J.O.S. have nothing to disclose. G.D.H. receives consulting fees from HRA Pharma and B.P.S. receives royalties from McGraw-Hill Inc. for his contributions as an author of book, Goodman and Gilman’s The Pharmacological Basis of Therapeutics (11th ed. New York: McGraw-Hill, Inc.).
First Published Online April 3, 2008
Abbreviations: Adcy, Adenylyl cyclase; GCNF, germ cell nuclear factor; Mc2r, ACTH receptor; SF1, steroidogenic factor 1; siRNA, small interfering RNA; Sp, specificity protein.
References
- Hanoune J, Defer N 2001 Regulation and role of adenylyl cyclase isoforms. Annu Rev Pharmacol Toxicol 41:145–174 [DOI] [PubMed] [Google Scholar]
- Ludwig MG, Seuwen K 2002 Characterization of the human adenylyl cyclase gene family: cDNA, gene structure, and tissue distribution of the nine isoforms. J Recept Signal Transduct Res 22:79–110 [DOI] [PubMed] [Google Scholar]
- Wong ST, Trinh K, Hacker B, Chan GC, Lowe G, Gaggar A, Xia Z, Gold GH, Storm DR 2000 Disruption of the type III adenylyl cyclase gene leads to peripheral and behavioral anosmia in transgenic mice. Neuron 27:487–497 [DOI] [PubMed] [Google Scholar]
- Wang MM, Tsai RY, Schrader KA, Reed RR 1993 Genes encoding components of the olfactory signal transduction cascade contain a DNA binding site that may direct neuronal expression. Mol Cell Biol 13:5805–5813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong ST, Athos J, Figueroa XA, Pineda VV, Schaefer ML, Chavkin CC, Muglia LJ, Storm DR 1999 Calcium-stimulated adenylyl cyclase activity is critical for hippocampus-dependent long-term memory and late phase LTP. Neuron 23:787–798 [DOI] [PubMed] [Google Scholar]
- Chan GC, Lernmark U, Xia Z, Storm DR 2001 DNA elements of the type 1 adenylyl cyclase gene locus enhance reporter gene expression in neurons and pinealocytes. Eur J Neurosci 13:2054–2066 [DOI] [PubMed] [Google Scholar]
- Schaefer ML, Wong ST, Wozniak DF, Muglia LM, Liauw JA, Zhuo M, Nardi A, Hartman RE, Vogt SK, Luedke CE, Storm DR, Muglia LJ 2000 Altered stress-induced anxiety in adenylyl cyclase type VIII-deficient mice. J Neurosci 20:4809–4820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson EL, Jacobson KL, Singh JC, Idzerda R, Ott SM, DiJulio DH, Wong ST, Storm DR 2000 The type 8 adenylyl cyclase is critical for Ca2+ stimulation of cAMP accumulation in mouse parotid acini. J Biol Chem 275:14691–14699 [DOI] [PubMed] [Google Scholar]
- Chao JR, Ni YG, Bolanos CA, Rahman Z, DiLeone RJ, Nestler EJ 2002 Characterization of the mouse adenylyl cyclase type VIII gene promoter: regulation by cAMP and CREB. Eur J Neurosci 16:1284–1294 [DOI] [PubMed] [Google Scholar]
- Schimmer BP 1980 Cyclic nucleotides in hormonal regulation of adrenocortical function. Adv Cyclic Nucleotide Res 13:181–214 [PubMed] [Google Scholar]
- Sewer MB, Waterman MR 2003 ACTH modulation of transcription factors responsible for steroid hydroxylase gene expression in the adrenal cortex. Microsc Res Tech 61:300–307 [DOI] [PubMed] [Google Scholar]
- Shen T, Suzuki Y, Poyard M, Best-Belpomme M, Defer N, Hanoune J 1997 Localization and differential expression of adenylyl cyclase messenger ribonucleic acids in rat adrenal gland determined by in situ hybridization. Endocrinology 138:4591–4598 [DOI] [PubMed] [Google Scholar]
- Cote M, Guillon G, Payet MD, Gallo-Payet N 2001 Expression and regulation of adenylyl cyclase isoforms in the human adrenal gland. J Clin Endocrinol Metab 86:4495–4503 [DOI] [PubMed] [Google Scholar]
- Al-Hakim A, Rui X, Tsao J, Albert PR, Schimmer BP 2004 Forskolin-resistant Y1 adrenal cell mutants are deficient in adenylyl cyclase type 4. Mol Cell Endocrinol 214:155–165 [DOI] [PubMed] [Google Scholar]
- Rui X, Al-Hakim A, Tsao J, Albert PR, Schimmer BP 2004 Expression of adenylyl cyclase-4 (AC-4) in Y1 and forskolin-resistant adrenal cells. Mol Cell Endocrinol 215:101–108 [DOI] [PubMed] [Google Scholar]
- Frigeri C, Schimmer BP 2000 The activation function of steroidogenic factor-1 is impaired in ACTH-resistant Y1 mutants. Endocr Res 26:1005–1009 [DOI] [PubMed] [Google Scholar]
- Frigeri C, Tsao J, Czerwinski W, Schimmer BP 2000 Impaired steroidogenic factor 1 (NR5A1) activity in mutant Y1 mouse adrenocortical tumor cells. Mol Endocrinol 14:535–544 [DOI] [PubMed] [Google Scholar]
- Frigeri C, Tsao J, Cordova M, Schimmer BP 2002 A polymorphic form of steroidogenic factor-1 is associated with ACTH resistance in Y1 mouse adrenocortical tumor cell mutants. Endocrinology 143:4031–4037 [DOI] [PubMed] [Google Scholar]
- Schimmer BP, Cordova M, Tsao J, Frigeri C 2003 A polymorphic form of steroidogenic factor 1 associated with ACTH receptor deficiency in mouse adrenal cell mutants. In: Barsh G, Cone R, Van der Ploeg LHT, eds. The melanocortin system. New York: New York Academy of Sciences; 147–153 [DOI] [PubMed] [Google Scholar]
- Schimmer BP, Cordova M, Tsao J, Frigeri C 2002 SF1 polymorphisms in the mouse and steroidogenic potential. Endocr Res 28:519–525 [DOI] [PubMed] [Google Scholar]
- Rainey WE, Saner K, Schimmer BP 2004 Adrenocortical cell lines. Mol Cell Endocrinol 228:23–28 [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K 2007 Current protocols in molecular biology. New York: John Wiley, Sons [Google Scholar]
- Chirgwin JM, Przybyla AE, MacDonald RJ, Rutter WJ 1979 Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry 18:5294–5299 [DOI] [PubMed] [Google Scholar]
- Winnay JN, Hammer GD 2006 Adrenocorticotropic-mediated signaling cascades coordinate a cyclic pattern of steroidogenic factor-1-dependent transcriptional activation. Mol Endocrinol 20:147–166 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ 1990 Basic local alignment search tool. J Mol Biol 215:403–410 [DOI] [PubMed] [Google Scholar]
- Schafer G, Cramer T, Suske G, Kemmner W, Wiedenmann B, Hocker M 2003 Oxidative stress regulates vascular endothelial growth factor-A gene transcription through Sp1- and Sp3-dependent activation of two proximal GC-rich promoter elements. J Biol Chem 278:8190–8198 [DOI] [PubMed] [Google Scholar]
- Schimmer BP, Kwan WK, Tsao J, Qiu R 1995 Adrenocorticotropin-resistant mutants of the Y1 adrenal cell line fail to express the adrenocorticotropin receptor. J Cell Physiol 163:164–171 [DOI] [PubMed] [Google Scholar]
- Cammas FM, Pullinger GD, Barker S, Clark AJL 1997 The mouse adrenocorticotropin receptor gene: cloning and characterization of its promoter and evidence for a role for the orphan nuclear receptor steroidogenic factor 1. Mol Endocrinol 11:867–876 [DOI] [PubMed] [Google Scholar]
- Shiraki T, Kondo S, Katayama S, Waki K, Kasukawa T, Kawaji H, Kodzius R, Watahiki A, Nakamura M, Arakawa T, Fukuda S, Sasaki D, Podhajska A, Harbers M, Kawai J, Carninci P, Hayashizaki Y 2003 Cap analysis gene expression for high-throughput analysis of transcriptional starting point and identification of promoter usage. Proc Natl Acad Sci USA 100:15776–15781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandelin A, Carninci P, Lenhard B, Ponjavic J, Hayashizaki Y, Hume DA 2007 Mammalian RNA polymerase II core promoters: insights from genome-wide studies. Nat Rev Genet 8:424–436 [DOI] [PubMed] [Google Scholar]
- Smale ST, Kadonaga JT 2003 The RNA polymerase II core promoter. Annu Rev Biochem 72:449–479 [DOI] [PubMed] [Google Scholar]
- Li L, He S, Sun JM, Davie JR 2004 Gene regulation by Sp1 and Sp3. Biochem Cell Biol 82:460–471 [DOI] [PubMed] [Google Scholar]
- Li B, Adams CC, Workman JL 1994 Nucleosome binding by the constitutive transcription factor Sp1. J Biol Chem 269:7756–7763 [PubMed] [Google Scholar]
- Kaiser UB, Halvorson LM, Chen MT 2000 Sp1, steroidogenic factor 1 (SF-1), and early growth response protein 1 (egr-1) binding sites form a tripartite gonadotropin-releasing hormone response element in the rat luteinizing hormone-β gene promoter: an integral role for SF-1. Mol Endocrinol 14:1235–1245 [DOI] [PubMed] [Google Scholar]
- Wolfe MW 1999 The equine luteinizing hormone β-subunit promoter contains two functional steroidogenic factor-1 response elements. Mol Endocrinol 13:1497–1510 [DOI] [PubMed] [Google Scholar]
- Marchal R, Naville D, Durand P, Begeot M, Penhoat A 1998 A steroidogenic factor-1 binding element is essential for basal human ACTH receptor gene transcription. Biochem Biophys Res Commun 247:28–32 [DOI] [PubMed] [Google Scholar]
- Hall JM, Korach KS 2002 Analysis of the molecular mechanisms of human estrogen receptors α and β reveals differential specificity in target promoter regulation by xenoestrogens. J Biol Chem 277:44455–44461 [DOI] [PubMed] [Google Scholar]
- Hall JM, McDonnell DP, Korach KS 2002 Allosteric regulation of estrogen receptor structure, function, and coactivator recruitment by different estrogen response elements. Mol Endocrinol 16:469–486 [DOI] [PubMed] [Google Scholar]
- Lefstin JA, Yamamoto KR 1998 Allosteric effects of DNA on transcriptional regulators. Nature 392:885–888 [DOI] [PubMed] [Google Scholar]
- Hammer GD, Parker KL, Schimmer BP 2005 Transcriptional regulation of adrenocortical development. Endocrinology 146:1018–1024 [DOI] [PubMed] [Google Scholar]
- Parker KL, Rice DA, Lala DS, Ikeda Y, Luo X, Wong M, Bakke M, Zhao L, Frigeri C, Hanley NA, Stallings N, Schimmer BP 2002 Steroidogenic factor 1: an essential mediator of endocrine development. Recent Prog Horm Res 57:19–36 [DOI] [PubMed] [Google Scholar]
- Urs AN, Dammer E, Sewer MB 2006 Sphingosine regulates the transcription of CYP17 by binding to steroidogenic factor-1. Endocrinology 147:5249–5258 [DOI] [PubMed] [Google Scholar]
- Aranda A, Pascual A 2001 Nuclear hormone receptors and gene expression. Physiol Rev 81:1269–1304 [DOI] [PubMed] [Google Scholar]
- Gao BN, Gilman AG 1991 Cloning and expression of a widely distributed (type IV) adenylyl cyclase. Proc Natl Acad Sci USA 88:10178–10182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doghman M, Karpova T, Rodrigues GA, Arhatte M, De Moura J, Cavalli LR, Virolle V, Barbry P, Zambetti GP, Figueiredo BC, Heckert LL, Lalli E 2007 Increased steroidogenic factor-1 dosage triggers adrenocortical cell proliferation and cancer. Mol Endocrinol 21:2968–2987 [DOI] [PubMed] [Google Scholar]
- Bassett MH, Suzuki T, Sasano H, White PC, Rainey WE 2004 The orphan nuclear receptors NURR1 and NGFIB regulate adrenal aldosterone production. Mol Endocrinol 18:279–290 [DOI] [PubMed] [Google Scholar]
- Shibata H, Kobayashi S, Kurihara I, Suda N, Yokota K, Murai A, Ikeda Y, Saito I, Rainey WE, Saruta T 2004 COUP-TF and transcriptional co-regulators in adrenal steroidogenesis. Endocr Res 30:795–801 [DOI] [PubMed] [Google Scholar]
- Bassett MH, Suzuki T, Sasano H, De Vries CJ, Jimenez PT, Carr BR, Rainey WE 2004 The orphan nuclear receptor NGFIB regulates transcription of 3β-hydroxysteroid dehydrogenase. implications for the control of adrenal functional zonation. J Biol Chem 279:37622–37630 [DOI] [PubMed] [Google Scholar]
- Maston GA, Evans SK, Green MR 2006 Transcriptional regulatory elements in the human genome. Annu Rev Genomics Hum Genet 7:29–59 [DOI] [PubMed] [Google Scholar]
- Luo MJ, Reed R 1999 Splicing is required for rapid and efficient mRNA export in metazoans. Proc Natl Acad Sci USA 96:14937–14942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüppel R, Dietze P, Lehnberg W, Frech K, Wingender E 1994 TRANSFAC retrieval program: a network model database of eukaryotic transcription regulating sequences and proteins. J Comput Biol 1:191–198 [DOI] [PubMed] [Google Scholar]
- Grillo G, Licciulli F, Liuni S, Sbisa E, Pesole G 2003 PatSearch: a program for the detection of patterns and structural motifs in nucleotide sequences. Nucleic Acids Res 31:3608–3612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bland JM, Altman DG 1996 Transformations, means, and confidence intervals. BMJ 312:1079 [DOI] [PMC free article] [PubMed] [Google Scholar]