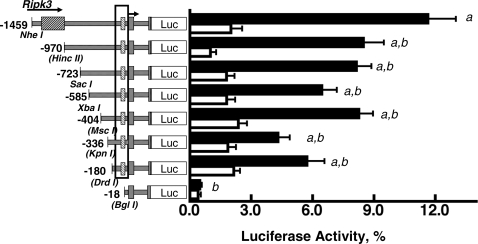
Figure 2.
Effects of 5′ deletions on Adcy4 promoter activity in Y1 and mutant 10r-6 cells. Y1 (black bars) and mutant 10r-6 cells (white bars) were transfected with luciferase expression vectors (0.41 pmol/dish) containing exon I, intron I, and part of exon II up to the initiator ATG and varying lengths of DNA flanking the transcription start site (arrow). The last exon of the Ripk3 gene is indicated, as is the sequence between −135 and −19 that is conserved among species (boxed). The restriction sites used to progressively truncate the 5′ end of the Adcy4 promoter also are shown; the sites destroyed by blunt end ligation are indicated with brackets. Results are expressed as mean percentages of the activity obtained with the pGL3 control vector (i.e. a luciferase reporter gene under control of the Simian virus 40 core promoter and enhancer) ± sem. Depending on the plasmid, data were compiled from eight to 20 independent experiments in Y1 cells and from four to 11 independent experiments for 10r-6 cells. Statistically significant differences, determined by ANOVA followed by the Newman-Keuls multiple comparison test, are: a, different from p-18Adcy4Luc, P < 0.001; b, different from p-1459Adcy4Luc, P < 0.001. All of the constructs, except p-336Adcy4Luc and p-18Adcy4Luc, had activities that were significantly higher (P < 0.05) in parent Y1 cells than in mutant 10r-6 cells.