Abstract
Retinoids, synthetic and natural analogs of retinoic acid, exhibit potent growth inhibitory and cell differentiation activities that account for their beneficial effects in treating hyperproliferative diseases such as psoriasis, actinic keratosis, and certain neoplasias. Tazarotene is a synthetic retinoid that is used in the clinic for the treatment of psoriasis. To better understand the mechanism of retinoid action in the treatment of hyperproliferative diseases, we used a long-range differential display–PCR to isolate retinoid-responsive genes from primary human keratinocytes. We have identified a cDNA, tazarotene-induced gene 3 (TIG3; Retinoic Acid Receptor Responder 3) showing significant homology to the class II tumor suppressor gene, H-rev 107. Tazarotene treatment increases TIG3 expression in primary human keratinocytes and in vivo in psoriatic lesions. Increased TIG3 expression is correlated with decreased proliferation. TIG3 is expressed in a number of tissues, and expression is reduced in cancer cell lines and some primary tumors. In breast cancer cell lines, retinoid-dependent TIG3 induction is observed in lines that are growth suppressed by retinoids but not in nonresponsive lines. Transient over-expression of TIG3 in T47D or Chinese hamster ovary cells inhibits colony expansion. Finally, studies in 293 cells expressing TIG3 linked to an inducible promoter demonstrated decreased proliferation with increased TIG3 levels. These studies suggest that TIG3 may be a growth regulator that mediates some of the growth suppressive effects of retinoids.
Retinoic acid (RA) and its synthetic analogs are therapeutically effective in the treatment of hyperproliferative dermatological diseases, such as psoriasis and cutaneous neoplasias, and in certain cancers (1–6). The biological effects of retinoids are mediated through two families of nuclear receptors, retinoic acid receptor (RARα, -β, and -γ) and retinoid X receptor (receptors α, β, and γ) (6, 7). RARs and retinoid X receptors are ligand-dependent transcription factors that, as heterodimers, function by changing the expression of RA-responsive genes (5–8). Tazarotene is an RARβ/γ-selective synthetic retinoid that is used clinically for the treatment of psoriasis, a skin disorder characterized by epidermal hyperproliferation and inflammation (9). Despite the therapeutic efficacy of retinoids in various hyperproliferative dermatological diseases, the molecular basis of their action in skin is largely unknown. Various genes whose expression is induced in cell culture systems by retinoids have been described. Of these, cellular retinoic acid binding protein II and tazarotene-induced genes (TIG) 1 and 2 are the only markers whose expression is induced in a retinoid-dependent manner in skin (10–12). The increased levels of cellular retinoic acid binding protein II in psoriatic plaques indicate that it may not be a useful efficacy marker of retinoid action in psoriatic lesions. In addition, several lines of evidence suggests that cellular retinoic acid binding protein II may function to modulate retinoid action by sequestering RA away from the nucleus and by enhancing RA metabolism (13, 14). Although TIG1 and TIG2 are induced in vivo in a retinoid-dependent manner (11, 12), in the absence of any knowledge about their function, they may not be useful efficacy markers of retinoid action. To better understand the mechanism of antiproliferative action of retinoids in diseased skin, we searched for genes induced by the antipsoriatic RARβ/γ-selective retinoid, tazarotene. We identified a cDNA, TIG3, by differential display (DD)–PCR that appears to function as a class II tumor suppressor and whose induction may explain some of the growth inhibitory effects of retinoids.
METHODS
Plasmids and Constructs.
The 0.6-kilobase (kb) TIG3 cDNA fragment and the 5′ rapid amplification of cDNA ends–PCR fragment were cloned into the pCR II TA vector (Invitrogen). To generate pcTIG3, total RNA from tazarotene treated keratinocytes was reverse transcribed by using oligo(dT), and this cDNA was used as a template for PCR amplification by using Pfu DNA polymerase (Stratagene) and primers having the sequences 5′-TTGGATCCTGTGGCTGCTTCAGGCTGTTGC-3′ containing a BamHI restriction enzyme site and 5′-TCAAGCTTCCACCATGGCTTCGCCACACCAAGAGCCCA-3′ containing a HindIII restriction enzyme site and a Kozak consensus sequence (CCACC) to ensure efficient translation. The amplification product was double-digested and cloned into the same sites of the pcDNA3 vector (Invitrogen). To generate pIND-TIG3, the TIG3 coding sequence was amplified by using Pfu polymerase and the primers 5′-CCACCATGGCTTCGCCACACCAA-3′ and 5′-TGTGGCTGCTTCAGGCTGTTGC-3′ followed by cloning into the pCR II TA cloning vector [pTA-TIG3(full)]. The TIG3 coding sequence was excised from pTA-TIG3 with HindIII and XhoI and was cloned into the same sites of the pIND vector (Invitrogen). The pcTIG3(AS) antisense expression vector was generated by excising TIG3 from pTA-TIG3(full) with EcoRI, followed by cloning into pcDNA3 in the same site and sequencing to determine orientation.
Cell Cultures and Transfections.
Primary cultures of human foreskin keratinocytes were purchased commercially and cultured in keratinocyte growth medium (Clonetics, San Diego). Colony forming assays in T47D cells were carried out in six-well plates. Equal numbers of cells were transfected with 2.5 μg of plasmid by using Lipofectamine (Life Technologies, Gaithersburg, MD) and 48 hours later were switched to selection medium containing 600 μg/ml of G418. Colonies were allowed to form for 3 weeks, at which time those containing >50 cells were counted. Colonies subsequently were isolated and expanded to cell lines for growth assays. Chinese hamster ovary (CHO) cells were transfected with 15 μg of pcDNA3 or pcTIG3 by using calcium phosphate. After 48 hours, the cells were shifted to medium containing 550 μg/ml G418, and colonies were allowed to expand for 2 weeks. Cells were fixed, were stained with hematoxylin, and were counted. EcR-293 embryonic kidney cells stably expressing the ecdysone receptor and the pIND vectors were purchased from Invitrogen. The EcR-293 cells were transfected with pIND-TIG3 or pIND-LacZ by electroporation in a Bio-Rad Gene Pulser II with a Bio-Rad Capacitance Extender II using 330 V, 1,000 μF, and infinity ohms in a 0.4-cm cuvette. Cells were selected for 3 weeks in medium containing 400 μg/ml of G418 and 400 μg/ml of Zeocin (Invitrogen). Surviving colonies were picked and expanded into cell lines. For induction of TIG3 or LacZ, cells were cultured in selection medium containing the appropriate amount of muristerone for 24 hours. Induction of TIG3 was assayed by Western blot analysis. Induction of LacZ was assayed by using the β-galactosidase Staining Kit (Invitrogen). For all growth assays, the cell lines were plated in 96-well plates and were cultured for 1 week (in selection medium when needed) in the presence of various concentrations of vehicle, muristerone, or retinoid and then were assayed by using a BrdUrd ELISA kit (Boehringer Mannheim).
Retinoids.
Tazarotene (AGN 190168; ethyl 6-[2-(4, 4-dimethylthiochroman-6-yl)-ethynyl] nicotinate), a RARβ/γ selective retinoid, was synthesized in the Department of Chemistry, Retinoid Research, Allergan (Irvine, CA).
DD-PCR.
Keratinocytes either were mock-treated or treated with tazarotene (1 μM) for 3 days and were harvested for total RNA preparation. DD-PCR was carried out by using a modified DD-PCR technique utilizing Expand polymerase (Boehringer Mannheim) and a Hieroglyph kit (Genomyx, Foster City, CA). Amplification was performed by using [33P]dATP in 0.2-ml thin-walled tubes in a Perkin–Elmer 9600 thermal-cycler. Amplified fragments were separated on an HR-1000 4.5% long-range high-resolution polyacrylamide gel (Genomyx) in a genomyxLR DNA Sequencer for 16 hours and were dried and subjected to autoradiography. Differentially expressed fragments were excised from the gel and were reamplified, and [32P]dCTP-labeled probes were generated by nick translation for use in Northern blot analysis.
Northern Hybridization.
Multiple-tissue and cancer cell line Northern blots were purchased from Clonetech. Total RNA blots containing RNA isolated from resected tumors and adjacent healthy, nontumor tissue were purchased from Invitrogen. All other blots were generated by using total RNA (15 μg) electrophoresed on 1% agarose/1.1 M formaldehyde gels, were transferred to Nytran (Schleicher & Schuell) membrane, and were probed with labeled cDNA probes at 52°C for 2 hours in Quickhyb (Stratagene). Blots were washed to 62°C in 0.1% standard saline citrate/1% SDS for 15 minutes and were exposed on a phosphorimager or were autoradiographed.
PCR Amplification of TIG3 in Biopsy RNA Samples.
Patients with long-standing bilateral plaque psoriasis (n = 20) were treated twice daily with vehicle or 0.1% tazarotene gel in a clinical study for up to 8 weeks. Punch biopsies were taken from 18 patients after 2 weeks of treatment, and patients were assessed for their clinical response to the drug after 8 weeks. Total pooled RNA (100 ng) from 15 responders (≥40% decrease in total clinical score at day 56) was reverse transcribed with oligo(dT) primers and PCR amplified by using primers to the 3′ end of TIG3 (5′-GCGACAGCCTGAAGCAGC-3′ and 5′-TTATTGATCCTTCAGTCTTG-3′) or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) primers as an internal control. The amplified TIG3 product was 190 bp. At each cycle starting at cycle 20, 10 μl were removed and visualized by ethidium bromide staining on a 1% agarose gel. Real time PCR of reverse-transcribed total RNA from biopsies was performed in the presence of an internal labeled oligonucleotide with the sequence 5′-fluorescein phosphoramidite (FAM)-CCTCTGTTTCCCTCTCTCGCTGGCA-6-caboxytetramethylrhodamine, succinimidyl ester (TAMRA)-3′ and the external primers 5′-GGGCAGATGGCTGTTTATTGATC-3′ and 5′-CCCTGTCTCAGGCGTTCTCTAGA-3′. Loss of fluorescence quenching because of hydrolysis of the labeled oligonucleotide by Taq polymerase 5′-3′ endonuclease activity was quantitated continuously in an Applied Biosystems Prism 7700.
5′ Rapid Amplification of cDNA Ends–PCR Amplification.
Total RNA (1 μg) from tazarotene-treated keratinocytes was used in the first strand synthesis reaction using the TIG3 primer 5′-TTCACCTCTGCACTGTTGCTC-3′. The resulting cDNA then was C-tailed on its 3′ end and then was used as a template in a PCR amplification reaction that used a nested TIG3 primer (5′-AGTGCTCATAGCCAAGGC-3′) and an oligo(dG) anchor primer. A product of ≈0.2 kb in length was amplified, was cloned into pCR II TA vector, and was sequenced.
Antibody Production and Western Blot Analysis.
TIG3 was PCR amplified by using the primers 5′-ATCATATGGCTTCGCCACACCAAGAGCC-3′ and 5′-GAGGATCCTCAGGCTGTTGCTTTTTTTTGGTATC-3′, which contain NdeI and BamHI restriction sites. The TIG3 cDNA was cloned into pET28a and was transformed into BL21(DE3) bacteria. TIG3 production was induced with isopropyl-1-thio-β-d-galactopyranoside, 50 ml of bacterial suspension was treated with lysozyme, and the resulting solution was subjected to sonication and freeze/thaw followed by centrifugation. The pellet was extracted with 6 M urea and 3% SDS and was electrophoresed on a 16% polyacrylamide gel. The TIG3 band was excised and injected into rabbits. For Western blot analysis of TIG3 expression, cells were washed twice in PBS, were incubated in 10 mM Tris, 0.1 mM DTT, 1 mM phenylmethylsulfonyl fluoride, 5 μg/ml leupeptin, and 5 μg/ml aprotinin for 30 minutes, and the scraped cells were lysed by sonication. Equal amounts of protein were boiled in SDS sample buffer for 5 minutes and were separated on a 4–20% denaturing SDS/PAGE gel. Proteins were electroblotted onto Protran (Schleicher & Schuell) nitrocellulose membranes by using a semi-dry apparatus. The membranes were blocked for one hour in TBST (10 mM Tris/150 mM NaCl/0.05% Tween 20) containing 5% nonfat dried milk and were incubated with anti-TIG3 antibody diluted 1:1,000 in TBST containing 5% nonfat dried milk overnight at 4°C. The blots were washed in TBST, were incubated for 1 hour with anti-rabbit horseradish peroxidase coupled antibody diluted 1:2,000 in TBST containing 5% nonfat dried milk, were washed again in TBST, and were visualized by using the ECL Kit (Amersham).
RESULTS AND DISCUSSION
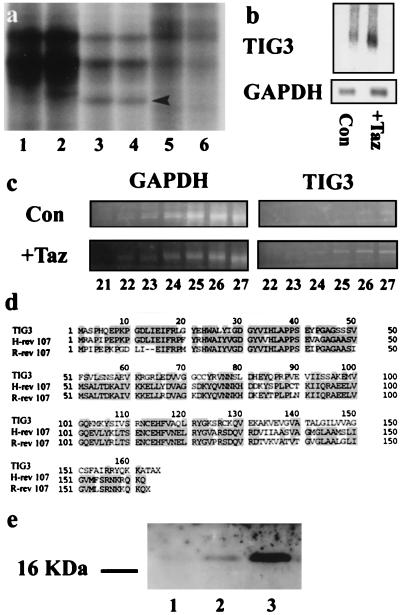
A long range DD-PCR strategy on mock and tazarotene-treated keratinocytes yielded a 600-bp fragment of a human complementary DNA, TIG3 (Retinoic Acid Receptor Responder 3) (Fig. 1a). TIG3 message was amplified from tazarotene-treated but not mock-treated keratinocytes or HeLa cells (Fig. 1a). The retinoid-dependent up-regulation of TIG3 was confirmed by Northern blot analysis in cultured keratinocytes (Fig. 1b). Northern blot analysis revealed a single TIG3 mRNA species migrating at 0.8 kb. Normalization to GAPDH revealed a >4-fold induction of the TIG3 message after retinoid treatment. To examine TIG3 regulation by tazarotene in vivo in psoriatic lesions, patients (n = 20) with long-standing bilateral lesions were treated topically with tazarotene (0.1% gel) or placebo (vehicle only) in a clinical study for up to 8 weeks. Punch biopsies taken from 15 patients after 2 weeks of treatment were pooled to prepare total RNA. In a reverse transcription–PCR reaction, a TIG3 signal was observed from 24 cycles of PCR amplification in retinoid-treated biopsies but does not appear until 26 cycles in control biopsies (Fig. 1c). This two-cycle difference corresponds to a 4-fold induction and subsequently was confirmed by using real-time quantitative PCR using Taq-Man technology in an Applied Biosystems 7700 PCR machine (data not shown).
Figure 1.
(a) Long-range DD-PCR of mock-treated keratinocyte RNA (lanes 1 and 2), tazarotene-treated keratinocyte RNA (lanes 3 and 4), and normal HeLa cell RNA as a control (lanes 5 and 6). The cDNA fragment identified as TIG3 (arrow) appears in lanes 3 and 4 but not in the other lanes. (b) Northern blot analysis of TIG3 expression in mock-treated (lane 1) or tazarotene-treated (lane 2) primary human foreskin keratinocytes. Cells were cultured in keratinocyte growth medium without serum until confluent, then were mock-treated or treated with 10−6 M tazarotene for 3 days. Cells were harvested, and 15 μg of total RNA was analyzed. The blot then was stripped and reprobed with GAPDH. (c) Semiquantitative reverse transcription–PCR analysis of week 2 biopsies from patients treated with topical tazarotene gel. The TIG3 message was detected initially at cycle 24 in treated biopsies but was not detected until cycle 26 in control untreated biopsies. (d) Multiple sequence alignment showing homology between TIG3, H-rev 107, and rat H-rev 107 (R-rev 107). (e) Western blot analysis of 15 μg of protein extract from mock-treated T47D cells (lane 1), T47D cells treated for 2 days with 1 μM tazarotene (lane 2), and ECR-293-TIG3 cells induced for 24 hours with 1 μM muristerone A (lane 3). Standards indicate the TIG3 protein runs at an apparent molecular mass of 18 kDa.
The partial mRNA sequence was extended by the rapid amplification of cDNA ends technique to obtain the full TIG3 message, which was 736 bp in length, contained an ORF from 30 to 523 bp, and coded for a putative protein product of 164 amino acids (GenBank accession no. AF060228). A search of the GenBank database revealed that TIG3 showed significant nucleotide and amino acid homology to the known class II tumor suppressor gene H-rev 107 (Fig. 1d). H-rev 107 encodes a protein of 18 kDa and was isolated by subtractive hybridization from a phenotypic revertant of H-ras transformed rat fibroblasts (15, 16). The predicted ORF of TIG3 was 57 and 52% identical to those of rat and human H-rev 107 proteins, respectively (Fig. 1d). The hydrophobicity plots of TIG3, human, and rat H-rev 107 proteins are almost identical and showed that the three shared an analogous carboxy-terminal hydrophobic domain, suggesting membrane association (data not shown). Rat H-rev 107 has, in fact, been shown to be a nuclear membrane-associated protein (15). TIG3 also contains three myristylation sites, which are conserved with human and rat H-rev 107 proteins. A polyclonal antibody was raised against bacterially expressed TIG3 protein, and this anti-TIG3 antibody reacted well with recombinant TIG3 protein (data not shown). Subsequent Western blot analysis showed that the endogenous TIG3 protein induced by retinoids in T47D cells and cloned TIG3 expressed in 293 HEK cells migrates at an apparent molecular weight of 18 (Fig. 1e).
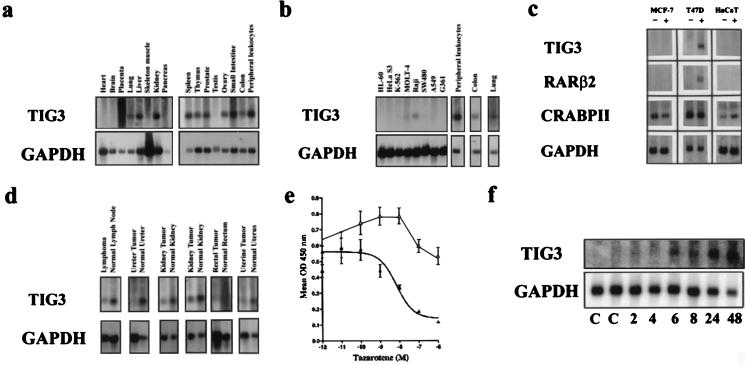
Multiple-tissue Northern blot analysis revealed that TIG3 was expressed in a variety of normal tissues (Fig. 2a) but not in several cancer cell lines corresponding to these tissues (Fig. 2 b and c). Significantly, decreased TIG3 expression also was observed in 6 of 25 primary tumors when compared with the adjacent normal tissue from which the tumor was obtained (Fig. 2d).
Figure 2.
(a) Multiple tissue Northern blot analysis revealed TIG3 expression in a variety of tissues, including lung, liver, kidney, spleen, thymus, prostate, ovary, small intestine, colon, and peripheral blood leukocytes. (b) TIG3 was not expressed in HL-60 peripheral blood acute promyelocytic leukemia cells, HeLa S3 cervical adenocarcinoma cells, K-562 chronic myelogenous leukemia cells, SW480 colon carcinoma, A549 lung carcinoma, or G361 melanoma cells. Low levels of expression were detected in MOLT-4 T-cell acute lymphoblastic leukemia cells and Raji B-cell Burkitt’s lymphoma cells. (c) TIG3 was not expressed in MCF-7 or T47D breast cancer cells, nor in HaCaT cells in the absence of tazarotene (−). TIG3 could be induced in T47D cells with the addition of 1 μM tazarotene (+) but was not induced in the MCF-7 or HaCaT cell lines. This in part may be because of a general lack of retinoid response in these cells as indicated by the lack of induction of RARβ2. (d) Expression of TIG3 was analyzed by Northern hybridization in biopsies from 25 primary human tumors of various tissues and was compared with expression levels in the adjacent normal tissues. Six tumors (lymphoma, ureter, two kidney, rectal, and uterine) showed significantly reduced or absent expression of TIG3 when GAPDH levels were used to control for variation in RNA levels. In no case was the expression of TIG3 increased in the tumor compared with normal tissue. (e) BrdUrd assay of T47D (closed triangle) and MCF-7 (open triangle) after 1 week of treatment with tazarotene shows that the growth of the T47D cell line but not the MCF-7 cell line is inhibited in a dose-responsive manner. (f) Time course induction of TIG3 message by tazarotene (1 μM) in T47D cells and analysis by Northern blot hybridization.
Tazarotene induced TIG3 expression in T47D but not in MCF-7 breast cancer cells (Fig. 2c). The proliferation of T47D cells was inhibited by tazarotene in a dose-dependent manner whereas the growth of MCF-7 cells was enhanced slightly at low concentrations and was relatively insensitive to tazarotene treatment at higher concentrations (Fig. 2e). Similarly, TIG3 was induced in a retinoid-dependent manner in retinoid-sensitive ZR75–1 and SKBR-3 but not in resistant MDA-MB-231 and MDA-MB-430 breast cancer cells (data not shown). To determine the time course of TIG3 induction, T47D cells were treated with vehicle or tazarotene (1 μM) for 2, 4, 6, 8, 24, or 48 hours, and total RNA was isolated. Northern blotting revealed that TIG3 message was induced 2- to 3-fold by 6 hours of treatment as quantitated by densitometry (Fig. 2f). These data suggest an association of retinoid induction of TIG3 and inhibition of breast cancer cell proliferation. It is also interesting to note that, although TIG3 is expressed at low levels and then is induced further by retinoids in keratinocytes (Fig. 1b), it neither is expressed nor can be induced in the HaCaT-transformed keratinocyte cell line (Fig. 2c).
To evaluate further the effect of TIG3 expression on cell proliferation, TIG3 cDNA was cloned into pcDNA3 (pcTIG3 in a sense and pcTIG3(AS) in an antisense orientation) and also was cloned into the ecdysone-inducible pIND vector (pIND-TIG3), all containing the neomycin resistance gene, and was transfected into T47D cells. The cells were allowed to proliferate for 3 weeks in G418 selection media, and the colonies were counted. Ectopic expression of pcTIG3 inhibited the proliferation of T47D by 60–70% in comparison to cells transfected with pcDNA3, antisense TIG3, or pIND-TIG3 in the absence of muristerone A (Fig. 3a). Similar experiments performed in CHO cells revealed that TIG3 expression also inhibited the proliferation of these cell cultures by 60–70% (Fig. 3 b and c). Isolation and expansion of pcTIG3 and pcDNA3 control T47D cell lines showed that the pcTIG3 lines grew at a significantly slower rate than the control neomycin resistant lines (Fig. 3d). Because these cell lines were difficult to propagate, we generated cell lines that express TIG3 under the control of the ecdysone receptor-inducible promoter. EcR-293 embryonic kidney cells stably expressing the ecdysone receptor and retinoid X receptor were transfected with the pIND-TIG3 or pIND-LacZ constructs, and cell lines were isolated. Of 12 pIND-LacZ cell lines isolated, 2 showed significant induction of β-galactosidase activity after induction with 1 μM muristerone A and were used as controls in subsequent growth experiments. Ten pIND-TIG3 cell lines were isolated, and levels of TIG3 expression after treatment with 1 μM muristerone A were assayed by Western blotting. Cell lines 4 and 8 showed the highest level of TIG3 expression (Fig. 3e). As shown in Fig. 3f, the growth rates of pIND-TIG3 lines 4 and 8 were reduced significantly after induction of TIG3 by muristerone A treatment as compared with the absence of muristerone A. The growth of pIND-LacZ-2-expressing cell line was not repressed significantly by muristerone A treatment. The data obtained in these lines clearly indicate that TIG3 expression is associated with growth repression.
Figure 3.
Colony formation in T47D (a) and CHO (b) cells containing the control expression vector (pcDNA3), the TIG3 expression vector (pcTIG3), the antisense TIG3 vector [pcTIG3(AS)], or the inducible vector in the absence of muristerone A (pIND-TIG3). Colony numbers represent the number of colonies per well (T47D) or per 50-cm2 dish (CHO) after 3 weeks in G418 selection medium. TIG3 expression significantly inhibited the colony formation in both cell types by 60–70%. Experiments were done in triplicate and were repeated at least two times. (c) Representative plates from the CHO cell colony formation experiments stained with hematoxylin. (d1 and d2) Several T47D cell lines from the control pcDNA3 and the pcTIG3 plates were isolated and expanded into cell lines, and their growth rates were assayed by BrdUrd incorporation. The pcTIG3-containing cell lines grew at a significantly slower rate than the control pcDNA3-containing cells. (e) Western blot analysis showing TIG3 expression in the 10 isolated EcR-293-TIG3 cell lines after 24 hours induction with 1 μM muristerone A. Cell lines 2, 3, 4, 8, and 10 showed some level of TIG3 expression, with 4 and 8 exhibiting the highest levels. (f) The growth rates of clones 4 and 8 were inhibited when cultured for 1 week in the presence of 1 μM muristerone A (+) when compared with those cells cultured in the vehicle alone (−). The pIND-LacZ cell line 2 was not inhibited significantly by 1 μM muristerone A.
Two regions of TIG3 cDNA also showed homology to one end of human chromosome 11 cosmid 187d6, localized to 11q23 (GenBank accession no. U73641). These two regions correspond to exons 1 (38 bp) and 2 (118 bp) of TIG3 and are separated by an intron of 2,680 bp. The cosmid sequence also contains 300 bp of intron 2. This cosmid contained 32 kb of promoter sequence for TIG3, in which we have identified at least six putative RA response elements to which RARs and retinoid X receptors might bind: a DR2 (direct repeat of 5′-PuG(G/T)TCA-3′ separated by two nucleotides) and a DR5 motif (same consensus sequence separated by five nucleotides) within 5 kb of exon 1, a DR5 motif within intron 1, a DR2 motif within intron 2, and distant DR2 and DR5 motifs at −10 and −15 kb, respectively (Fig. 4).
Figure 4.
Schematic representation of the genomic organization of the first two exons of TIG3 and the location of the putative RA response elements in the TIG3 promoter as determined by homology with human chromosome 11 cosmid 187d6, localized to 11q23.
Our results suggest that TIG3 is a retinoid-inducible anti-proliferative/class II tumor suppressor gene. Class II tumor suppressors are genes that are functionally intact but are expressed at an abnormally low level in cell lines and/or tumors. This low level of expression permits cell proliferation. TIG3 displays significant nucleotide and amino acid homology to the known class II tumor suppressor H-rev 107 and identifies this as a family of putative tumor suppressors. H-rev 107 is an 18-kDa protein expressed in normal rat fibroblasts, is down-regulated after ras-transformation, is reexpressed in ras revertants, and is expressed highly in ras-transformation resistant fibroblasts (15). H-rev 107 also is expressed in a number of normal tissues but not in various rat tumor cell lines and experimental tumors (15, 16). Further, over-expression of H-rev 107 in H-ras-transformed hepatoma cells or fibroblasts resulted in a 75% reduction of colony formation in vitro and an attenuation of tumor formation in vivo (16). TIG3 is induced in a retinoid dependent manner in vivo in psoriatic lesions in which tazarotene exerts therapeutic antiproliferative effects. TIG3 is expressed in several normal tissues but not in the corresponding cancer cell lines and some primary tumors. Further, TIG3 is inducible in retinoid-sensitive but not in resistant breast cancer cells. Finally, over-expression of TIG3 in several mammalian cell lines inhibits their growth. Of interest, TIG3 is localized on the 11q23 chromosomal region, which is thought to contain at least three distinct tumor suppressor genes and is translocated in some leukemia, lymphoma, and rhabdomyosarcoma as well as breast, lung, ovarian, and cervical carcinomas (17–21).
Identification of TIG3 and H-rev 107 as true tumor suppressor genes requires the identification of disabling mutations in the regulatory or coding sequences or loss of heterozygosity in patient tumor biopsies. The determination of the exact biological functions of TIG3 and H-rev 107 and an understanding of how they may interact with the signaling pathways in cells should be important in evaluating the roles these genes play in the proliferation of cells. In addition, analysis of the TIG3 promoter region may aid in the development of novel retinoids or other agents that can induce TIG3 levels and thereby exert antiproliferative effects. Retinoids exhibit anti-AP1 (22) and anti-NF-IL6 (23) activities, and the suppression of proliferative genes regulated by such transcription factors is likely to be important in the antiproliferative activities of retinoids. However, it is also possible that induction of antiproliferative genes such as p21 (24) and TIG3 also may play a significant role in the growth suppressive effects of retinoids.
Acknowledgments
We thank Armin Kasravi for his assistance with figure preparation. We also thank Drs. Scott Thacher and Larry Wheeler for critically reading this manuscript.
ABBREVIATIONS
- RA
retinoic acid
- RAR
RA receptor
- TIG
tazarotene-induced gene
- DD
differential display
- kb
kilobase
- CHO
Chinese hamster ovary
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF060228).
References
- 1.Lotan R. Cancer Res. 1994;54:1987s–1990s. [PubMed] [Google Scholar]
- 2.Lotan R. FASEB J. 1996;10:1031–1039. doi: 10.1096/fasebj.10.9.8801164. [DOI] [PubMed] [Google Scholar]
- 3.Hong W K, Itri L M. In: The Retinoids: Biology, Chemistry, and Medicine. Sporn M B, Roberts A B, Goodman D S, editors. New York: Raven; 1994. pp. 597–630. [Google Scholar]
- 4.Peck G L, DiGiovanna J J. In: The Retinoids: Biology, Chemistry & Medicine. Sporn M B, Roberts A B, Goodman D S, editors. New York: Raven; 1994. pp. 631–658. [Google Scholar]
- 5.Boehm M F, Heyman R A, Patel S, Stein R B, Nagpal S. Exp Opin Invest Drugs. 1995;4:593–612. [Google Scholar]
- 6.Nagpal S, Chandraratna R A S. Curr Pharmaceutical Design. 1996;2:295–316. doi: 10.2174/1381612003400146. [DOI] [PubMed] [Google Scholar]
- 7.Chambon P. Semin Cell Biol. 1994;5:115–125. doi: 10.1006/scel.1994.1015. [DOI] [PubMed] [Google Scholar]
- 8.Nagpal S, Friant S, Nakshatri H, Chambon P. EMBO J. 1993;12:2349–2360. doi: 10.1002/j.1460-2075.1993.tb05889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weinstein G D, Krueger G G, Lowe N J, Duvic M, Friedman D J, Jegasothy B V, Jorizzo J L, Shmunes E, Tschen E H, Lew-Kaya D A, et al. J Am Acad Dermatol. 1997;37:85–92. doi: 10.1016/s0190-9622(97)70216-0. [DOI] [PubMed] [Google Scholar]
- 10.Elder J T, Cromie M A, Griffiths C E M, Chambon P, Voorhees J J. J Invest Dermatol. 1993;100:356–359. doi: 10.1111/1523-1747.ep12471816. [DOI] [PubMed] [Google Scholar]
- 11.Nagpal S, Patel S, Asano A T, Duvic M, Chandraratna R A S. J Invest Dermatol. 1996;106:269–274. doi: 10.1111/1523-1747.ep12340668. [DOI] [PubMed] [Google Scholar]
- 12.Nagpal S, Patel S, Jacobe H, DiSepio D, Ghosn C, Malhotra M, Teng M, Duvic M, Chandraratna R A S. J Invest Dermatol. 1997;109:91–95. doi: 10.1111/1523-1747.ep12276660. [DOI] [PubMed] [Google Scholar]
- 13.Boylan J F, Gudas L J. J Cell Biol. 1991;112:965–979. doi: 10.1083/jcb.112.5.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fiorella P D, Napoli J L. J Biol Chem. 1991;266:16572–16579. [PubMed] [Google Scholar]
- 15.Hajnal A, Klemenz R, Schafer R. Oncogene. 1994;9:479–490. [PubMed] [Google Scholar]
- 16.Sers C, Emmenegger U, Husmann K, Bucher K, Andres A-C, Schafer R. J Cell Biol. 1997;136:935–944. doi: 10.1083/jcb.136.4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foulkes W D, Campbell I G, Stamp G W H, Trowsdale J. Br J Cancer. 1993;67:268–273. doi: 10.1038/bjc.1993.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hampton G M, Penny L A, Baergen R N, Larson A, Brewer C, Liao S, Busby-Earle R M, Williams A W, Steel C M, Bird C C, et al. Proc Natl Acad Sci USA. 1994;91:6953–6957. doi: 10.1073/pnas.91.15.6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hampton G M, Mannermaa A, Winquist R, Alavaikko M, Blanco G, Taskinen P J, Kiviniemi H, Newsham I, Cavenee W K, Evans G A. Cancer Res. 1994;54:4586–4589. [PubMed] [Google Scholar]
- 20.Rasio D, Negrini M, Maneti G, Dragani T, Croce C M. Cancer Res. 1995;55:3988–3991. [PubMed] [Google Scholar]
- 21.Tomlinson I P H, Gammack A J, Stickland J E, Mann G J, MacKie R M, Kefford R F, McGee J O. Genes Chromosomes Cancer. 1993;7:169–172. doi: 10.1002/gcc.2870070310. [DOI] [PubMed] [Google Scholar]
- 22.Nicholson R C, Mader S, Nagpal S, Leid M, Rochette-Egly C, Chambon P. EMBO J. 1990;9:4443–4454. doi: 10.1002/j.1460-2075.1990.tb07895.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DiSepio D, Malhotra M, Chandraratna R A S, Nagpal S. J Biol Chem. 1997;272:25555–25559. doi: 10.1074/jbc.272.41.25555. [DOI] [PubMed] [Google Scholar]
- 24.Liu M, Iavarone A, Freedman L P. J Biol Chem. 1996;271:31723–31728. doi: 10.1074/jbc.271.49.31723. [DOI] [PubMed] [Google Scholar]