Abstract
rRNA synthesis by RNA polymerase I requires both the promoter selectivity factor 1, which is composed of TATA binding protein (TBP) and three TBP-associated factors, and the activator upstream binding factor (UBF). Whereas there is strong evidence implicating a role for phosphorylation of UBF in the control of growth-induced increases in rRNA transcription, the mechanism of this effect is not known. Results of immunoprecipitation studies with TBP antibodies showed increased recovery of phosphorylated UBF from growth-stimulated smooth muscle cells. Moreover, using an immobilized protein-binding assay, we found that phosphorylation of UBF in vivo in response to stimulation with different growth factors or in vitro with smooth muscle cell nuclear extract increased its binding to TBP. Finally, we demonstrated that UBF–TBP binding depended on the C-terminal ‘acidic tail’ of UBF that was hyperphosphorylated at multiple serine sites after growth factor stimulation. Results of these studies suggest that phosphorylation of UBF and subsequent binding to TBP represent a key regulatory step in control of growth-induced increases in rRNA synthesis.
A critical component of the cellular response to growth factors is an increase in rRNA synthesis (1). Transcription of 18S and 28S rRNA genes by RNA polymerase I requires the cooperative binding of the multimeric protein complex termed selectivity factor I (SL1) and upstream binding factor (UBF) (2, 3). SL1 confers RNA polymerase I selectivity and is composed of TATA-binding protein (TBP) and three TBP-associated factors, (TAF)I110, TAFI63, and TAFI48 (4, 5). However, the rRNA promoter lacks a TATA box, and SL1 does not bind efficiently to the rRNA promoter by itself (4). Stable binding of SL1 requires UBF, a 94/97-kDa polypeptide that binds to the upstream control element of the rRNA promoter (5, 6). The precise mechanism of the UBF–SL1 interaction is not known, although recent in vitro studies have shown that UBF interacts directly with TBP (7, 8) and TAFI48 (9) but does not interact with TAFI63 or TAFI110. (9). A key unresolved issue is how growth factors modulate the activity of the rRNA transcriptional machinery.
We and others have demonstrated previously that phosphorylation of UBF is increased during cellular growth (10–14). Specifically, we have shown that angiotensin-II (A-II)-induced hypertrophy of vascular smooth muscle cells (SMC) was associated with increased rRNA content (15). Subsequently, we showed that A-II stimulated rRNA transcription and increased the serine phosphorylation and nucleolar localization of UBF (13). Similar responses have been shown in a wide variety of cell types in response to a wide plethora of growth factors (10–13). As such, growth-induced increases in phosphorylation of UBF and subsequent increases in rRNA transcription appear to represent a highly conserved required growth regulatory pathway. Despite clear evidence demonstrating that phosphorylation of UBF dramatically increases its activity in in vitro transcription reactions (10, 16), the mechanism by which the phosphorylation of UBF promotes rRNA transcription is not known. Given the recent observations that UBF bound specifically to the TBP component of SL1 (7, 8), we hypothesized that growth-induced hyperphosphorylation of UBF promotes its interaction with the TBP component of SL1 and consequently enhances rRNA transcription. The goal of the present studies was to determine whether the UBF–TBP interaction in cultured SMC was related to growth state and to determine whether the phosphorylation state of UBF plays a regulatory role in the binding between UBF and TBP.
METHODS
Culture of Vascular SMC.
Rat aortic vascular SMC were isolated, cultured, and grown to confluency as described previously (15). The growth medium was replaced with serum-free medium [(SFM), 1:1 DMEM/F12 media containing 5 × 10−7 M insulin/5 μg/ml transferrin/0.2 mM ascorbic acid/6.25 ng/ml selenium] for 5 days to reversibly growth-arrest cells in a balanced protein state, as shown previously (15). On the day of the experiment, the cells were treated for 1 h with either fetal bovine serum (FBS, 10%), 10−6 M A-II, platelet-derived growth factor BB (PDGF-BB, 10 ng/ml) or the PDGF-BB vehicle (2 mg/ml BSA in 10 mM acetic acid) or SFM and were harvested in lysis buffer (150 mM NaCl/50 mM Tris, pH 8.0/1% Nonidet P-40 with protease inhibitors) and total protein was assayed (Bio-Rad).
Immunoprecipitation Assays.
Quiescent and growth-stimulated SMC were harvested in lysis buffer (above), precleared, and incubated with a TBP antibody (Santa Cruz Biotechnology) for 1 h at 4°C. Proteins were precipitated with Protein-A agarose. Precipitates were collected by centrifugation and the pellet was washed three times with lysis buffer. Laemmeli sample buffer was added to the pellet and was boiled for 5 min. Samples were resolved by SDS/PAGE gel and were transferred to poly(vinylidene difluoride) membrane. The membrane was blocked in 5% milk and was probed with either TBP or UBF antibody.
In Vitro Phosphorylation of Recombinant UBF (rUBF)1.
rUBF1 was incubated with smooth muscle nuclear extract (0.2 μg) and 200 μM [γ−32P]ATP (250 cpm/pmol) in a 30-mM MgCl2 solution at 25°C for the times indicated. The reactions were terminated by the addition of Laemmeli sample buffer and by boiling for 5 min.
Far Western Analysis.
Samples were resolved on a 12% SDS/PAGE electrophoretically transferred (105 V for 1 h at 4°C) to a poly(vinylidene difluoride) membrane. The poly(vinylidene difluoride) membranes were incubated in a blocking solution (50 mM Tris⋅HCl/50 mM NaCl/1 mM EDTA/1 mM DTT/3% BSA, pH 7.9) for 1 h at 4°C. Human recombinant TBP (Santa Cruz Biotechnology, 200 ng in 10 ml blocking solution) was added and the membrane was gently agitated for 2 h at 4°C. The membrane was thoroughly washed (20 mM Tris⋅HCl/50 mM NaCl/1 mM EDTA/1 mM DTT/0.2% Tween-20, pH 7.9), and bound TBP was detected by immunoblotting with an affinity-purified α-hTBP antibody (a generous gift from Hyockman Kwon and Michael R. Green) and enhanced chemiluminescence detection was performed as recommended by the supplier (ECL, Amersham).
Two-Dimensional Phosphotryptic Peptide Maps.
UBF from 32P-labeled cultured SMC was resolved by SDS/PAGE and the portion of the gel containing UBF was excised and minced. The pieces of gel were dehydrated with the addition of 200 μl 100% methanol and were allowed to air dry. The gel pieces were rehydrated in 100 μl of 50 mM ammonium bicarbonate containing 2 μg 1-tosylamido-2-phenylethyl chloromethyl ketone-treated trypsin (1,000 units/mg, Sigma). Another 2 μg of trypsin was added 1 h later and the reaction was allowed to digest overnight at 37°C. The digest was washed three times with H2O and dried with the use of a speed-vacuum concentrator. The digest was washed once with 300 μl of pH 1.9 buffer [79 ml glacial acetic acid/25 ml formic acid (88%)/897 ml H20] before being resuspended in 5 μl of pH 1.9 buffer and counted by means of the Cerenkov method. Equal counts were spotted onto a TLC plate and separated electrophoretically in the first dimension for 30 min at 1,000 V. The plates were allowed to dry and then were placed in a chromatography tank containing isobutyric acid buffer (62.5% isobutyric acid/1.9% n-butanol/4.8% pyridine/2.9% acetic acid). The plates were resolved in the second dimension until the solvent front was 2 cm from the top of the TLC plate. The plates were allowed to dry before being covered with plastic wrap and were exposed to film for 2 days.
Preparation of rUBF1.
rUBF1 was prepared by infecting an Sf9 culture at 1.5 × 105 cells/ml with a UBF expression baculovirus (titer 2.5 × 105). Cells were incubated as a suspension in a spin flask at 27°C for 60–72 h. At the termination of the incubation, the cells were pelleted and resuspended in BG buffer (50 mM β-glycerolphosphate/1.5 mM EGTA/150 μM Na3VO4/1 mM DTT). Cells were lysed by sonication and loaded onto a heparin agarose Affi-Gel column (Bio-Rad). Proteins were eluted from the column by a linear salt gradient from 200 to 800 mM KCl and were collected in fractions. Fraction samples were resolved by SDS/PAGE and were analyzed by Western blotting with a UBF antibody. rUBF1 eluted at approximately 400 mM KCl. The rUBF1-containing fractions were pooled, dialyzed, and concentrated, yielding a final rUBF1 concentration of 120 μg/ml.
RESULTS AND DISCUSSION
As an initial test of our hypothesis that growth-induced hyperphosphorylation of UBF promotes its interaction with the TBP component of SL1, we performed immunoprecipitation assays with a TBP antibody to determine whether growth-induced increases in UBF phosphorylation were associated with increased UBF–TBP binding. Precipitations were performed in cultured SMC that were initially growth-arrested in a defined SFM (15) and then stimulated with either the hypertrophic agent A-II or the mitogens, PDGF-BB or FBS. Cells were growth-arrested in this defined SFM containing insulin, since we have shown previously that this maintains them in a state whereby they continue to transcribe rRNA and maintain neutral protein balance (15). In contrast, we found that use of alternate growth arrest protocols involving serum withdrawal resulted in nearly complete cessation of rRNA transcription, a state of negative protein balance, and ultimate cell death.
Cells were harvested at 1 h after growth stimulation, since we previously showed that this induces maximal phosphorylation of UBF and a concomitant increase in rRNA transcription without altering UBF expression (13). Results demonstrated marked increases in UBF coprecipitation with TBP in SMC stimulated with A-II (Fig. 1A, lane 1 vs. lane 2), PDGF-BB (Fig. 1A, lane 3 vs. lane 4), or FBS (Fig. 1A, lane 1 vs. lane 5). These differences were not because of variations in the amount of TBP present in the precipitates (Fig. 1B), nor were any changes in total TBP concentration detected in growth-stimulated SMC at this 1-h time point based on Western analysis of cell lysates (data not shown). Interestingly, TBP immunoprecipitations were performed also on SMC labeled with 32P-orthophosphoric acid for 12 h before growth stimulation. Results of these experiments showed a dramatic increase in the total amount of 32P-labeled UBF present in the TBP immunoprecipitate from the growth-stimulated cells. These increases closely paralleled increases in UBF content (Fig. 1A), indicating that the specific activity of 32P in TBP-associated UBF was unchanged in growth-stimulated vs. quiescent cells. Thse results suggest that growth stimulation was associated with increases in the stoichiometry of UBF phosphorylation, not phosphorylation of new sites.
Figure 1.
Western blot analysis showing increased levels of UBF in TBP immunoprecipitates from growth-stimulated vs. quiescent cells. (A) Western blot analysis of UBF levels in TBP immunoprecipitates from SMC growth arrested in a defined SFM and stimulated with A-II, PDGF-BB, and FBS, as described in Methods. Cells were harvested and immunoprecipitations were performed as described in Methods. The blots were probed with a UBF antibody (a generous gift from L. Rothblum, Weis Research Institute, Danville, PA). Blots were detected with enhanced chemiluminescence (ECL, Amersham) according to manufacturer’s directions. (B) The same TBP immunoprecipitates assayed in A were assayed for TBP by Western blotting with a TBP antibody (Santa Cruz Biotechnology) as described in Methods. Because of a greater recovery of TBP, the loadings in B were proportionally reduced for all samples by a factor of eight, as compared with the corresponding lane in A.
Although SMC express the two UBF isoforms equally, results showed substantially more UBF1 than UBF2 in the TBP immunoprecipitates. A previous study (7) showed approximately equal appearance of UBF1 and UBF2 in TBP immunoprecipitation assays by using slightly different assay conditions as well as a different TBP antibody. One possible explanation that might reconcile these apparent differences is that there is preferential binding of TBP to UBF1 that is complexed with UBF2, but UBF2 is dissociated under our assay conditions. Additional studies will be required to address this issue definitively. Nonetheless, our data provide strong evidence that after stimulation of SMC with either a hypertrophic agent (A-II) or hyperplastic agents (PDGF-BB and FBS), there is enhanced association of TBP and UBF in the absence of detectable changes in either TBP or UBF concentration.
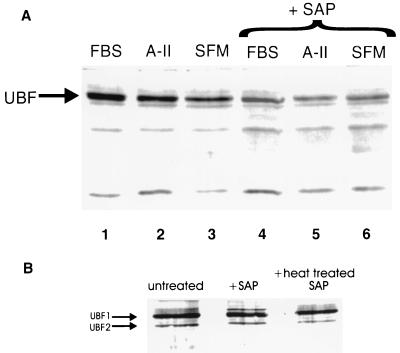
The preceding immunoprecipitation assays indicate that TBP–UBF interaction is enhanced under conditions in which UBF is hyperphosphorylated but provide no direct evidence that UBF phosphorylation regulates TBP binding. Previous studies by others (7, 8) provided evidence for direct interaction of UBF and TBP based on immunoprecipitation and in vitro immobilized protein-binding assays (Far Western analysis). However, these studies did not examine the effects of growth stimulation on this interaction nor did they examine whether the phosphorylation state of UBF influenced TBP interaction. We thus performed TBP Far Western analysis on nuclear lysates from FBS- or A-II-treated SMC (Fig. 2A) with or without pretreatment of lysates with shrimp alkaline phosphatase (SAP) to dephosphorylate UBF. Results demonstrated that TBP bound to several proteins within the nuclear extract, including strong binding to a protein doublet with molecular masses corresponding to UBF1 and UBF2 (Fig. 2A). TBP binding to UBF was increased in FBS- and A-II-stimulated cells, as compared with SFM (vehicle)-treated controls (Fig. 2A, lanes 1 and 2 vs. lane 3). This binding was substantially reduced by treatment with SAP Fig. 2A, lanes 1–3 vs. lanes 4–6). Control Western blot analysis confirmed that the principal TBP interactive protein observed in Fig. 2A was UBF and showed that phosphatase treatment substantially reduced UBF phosphorylation (data not shown) but did not alter UBF protein content (Fig. 2B).
Figure 2.
TBP Far Western blot analysis of SMC nuclear extracts showing enhanced TBP–UBF binding in growth-stimulated SMC that was reduced by treatment with SAP. (A) Far Western blot of TBP binding proteins in SMC nuclear extracts. Cells were growth-arrested in SFM and then stimulated with A-II or FBS as described in Methods. Nuclear extracts were prepared as described (23). Four μg of each extract were treated with either SAP (10 units/ml, United States Biochemical) or glycerol (vehicle) and were incubated for 1 h at 37°C before electrophoresis and Far Western blotting. Lanes 1–3 show blots of nuclear extracts from SMC treated with 10% FBS, A-II, or SFM vehicle, respectively. Lanes 4–6 show blots of nuclear extracts treated with SAP. The band corresponding to the UBF1/UBF2 doublet is indicated, although the two UBF isoforms are not well resolved under the conditions of these assays. (B) Control Western blot analyses with a UBF antibody confirmed the position of UBF and demonstrated that treatment with SAP had no effect on UBF protein content. Similar results to these were obtained when TBP Far Western analyses were performed by using immunoprecipitates of UBF derived from vehicle and growth factor-stimulated SMC (data not shown), as compared with the whole nuclear extracts as shown here.
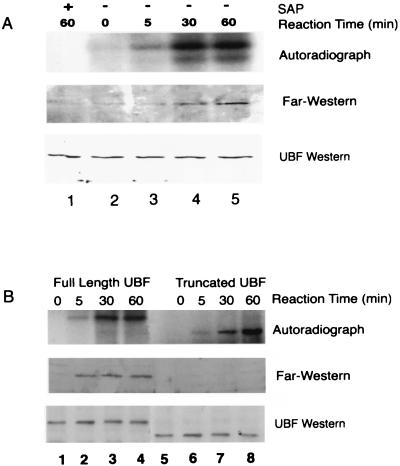
To test directly whether phosphorylation of UBF regulates its binding to TBP, we performed Far Western analyses with purified rat rUBF1 phosphorylated in vitro with nuclear extracts from A-II-treated SMC. Experiments with extracts from FBS- and PDGF-BB-treated cells yielded identical results (data not shown). Results from these experiments showed that time-dependent increases in rUBF1 phosphorylation (Fig. 3A Top) were associated with corresponding increases in TBP binding (Fig. 3A Middle). SAP treatment markedly decreased UBF phosphorylation and TBP binding (Fig. 3A, lanes 1 vs. 5) but did not reduce UBF protein content (Fig. 3A Bottom. These data are consistent with results of studies in cultured SMC shown in Fig. 1 and provide strong evidence that UBF phosphorylation enhances its binding to TBP. Although a time-dependent activation of TBP or other intermediary factors may also have contributed to the growth-induced increases in the UBF–TBP interactions seen in the immunoprecipitation experiments (Fig. 1), such changes could not have contributed to the phosphorylation-dependent UBF–TBP interaction seen in the in vitro analyses shown in Fig. 3A.
Figure 3.
Far Western analysis of rUBF1 phosphorylated in vitro showed phosphorylation-dependent UBF–TBP binding. (A) 32P-autoradiograph analysis (Top) of rUBF1 phosphorylated in vitro by SMC nuclear extract. The phosphorylation reaction was performed as described in Methods. SAP was added for the last 30 min of one of the 60-min time point samples. The membrane was dried and exposed to film for 18 h to detect 32P incorporation into rUBF1. Far Western analysis (Middle) of TBP binding to rUBF1 was performed as described in Methods. Control UBF Western blot (Bottom) showing equal amounts of rUBF in each of the experimental groups. (B) Autoradiographic analysis of 32P incorporation (Top) or Far Western blot analysis of TBP binding (Middle) by using truncated rUBF lacking the C-terminal acidic tail (amino acid residues 1–656, a generous gift from L. Rothblum, Weis Research Institute, Danville, PA) or full-length rUBF. (Bottom) A UBF Western blot showing equal amounts of rUBF in each of the experimental groups. Phosphorylation reactions and Far Western analyses were performed as described in Methods, with incubation times as indicated. Western blot analyses with UBF antibody showed that equivalent amounts of truncated and full-length rUBF1 were present in each lane (data not shown).
Several studies have shown that phosphorylation of the C-terminal ‘acidic tail’ of UBF is required for its transcriptional activity (5, 14, 16). To determine whether this region was also required for UBF–TBP interaction, we performed Far Western analysis using a deletion mutant of UBF lacking the acidic tail (amino acids 657–754 were deleted). Results demonstrated that although truncated UBF was phosphorylated by SMC nuclear extract (Fig. 3B Top, lanes 5–8), it did not bind to TBP (Fig. 3B Middle, lanes 5–8). The results of our Far Western studies indicate that the ‘acidic tail’ of UBF is required for TBP binding but does not prove that the regulatory phosphorylation sites actually lie within the acidic tail.
Results of our TBP immunoprecipitation studies on 32P-labeled SMC suggested that the enhanced TBP binding activity of UBF after growth stimulation was associated with increases in the stoichiometry of UBF phosphorylation or hyperphosphorylation as opposed to the phosphorylation of new sites. To test this directly, we carried out two-dimensional phosphotryptic peptide mapping studies of UBF phosphorylated in vivo (Fig. 4A). Results showed the presence of at least 11 labeled fragments that were resolvable under our assay conditions. Consistent with our immunoprecipitation results, we were unable to identify any qualitative differences in phosphotryptic peptide maps between quiescent and growth-stimulated cells. These phosphotryptic peptide maps indicate that A-II and FBS do not induce the phosphorylation of new sites. Rather, they appear to stimulate an increase in the stoichiometry of phosphorylation of existing sites.
Figure 4.
Two-dimensional phosphotryptic peptide maps of UBF phosphorylated in vivo showed that growth stimulation did not induce the phosphorylation of new sites. (A) Two-dimensional phosphotryptic peptide maps of phosphorylated UBF from quiescent and growth-stimulated SMC. Postconfluent growth-arrested SMC were switched to a low-phosphate SFM and labeled in vivo with 32P-orthophosphoric acid (0.5 mCi/ml, 6,000 Ci/mmol) for 8 h before being treated with A-II (10−6M), SFM, or 10% FBS for 1 h. UBF was immunoprecipitated from SMC nuclear extracts as described previously (13). The maps were generated as described in Methods. (B) Two-dimensional phosphotryptic peptide maps of full-length and truncated rUBF1 that was phosphorylated in vitro by SMC nuclear extract. One μg of either full-length rUBF1 or truncated rUBF1 was phosphorylated in vitro by A-II-treated SMC crude nuclear extract (0.2 μg) and 300 μM [γ −32P]ATP (250 cpm/pmol), 7.5 μM MgCl2. The reactions were terminated by the addition of SDS sample buffer. The maps were generated as described in Methods.
We also performed two-dimensional phosphotryptic mapping studies on the full-length vs. the C-terminal-truncated UBF that were phosphorylated in vitro. The maps of full-length rUBF1 phosphorylated in vitro were similar, but not identical, to those obtained with UBF phosphorylated in vivo (Fig. 4 A vs. B). For example, maps of rUBF1 phosphorylated in vitro showed nine of the eleven phosphopeptides seen with UBF phosphorylated in vivo (phosphopeptides numbers 7 and 10 were missing) as well as a unique phosphopeptide (designated number 12, Fig. 4B Left) not seen in UBF from cultured SMC. However, it appears that these differences are not necessary for TBP binding activity, since recombinant UBF phosphorylated in vitro bound to TBP (Fig. 3 A and B). Thus the candidate regulatory phosphorylation site or sites are contained within phosphotryptic peptides 1–6, 8, 9, or 11 (or some phosphopeptide not resolved in these assays). Unfortunately, we were unable to determine the specific region of UBF that gives rise to individual phosphotryptic fragments because of the complex pattern of tryptic fragments generated in these studies (17). Phosphotryptic maps of truncated rUBF1 (Fig. 4B Right) showed phosphorylation of only a subset of fragments (i.e., phosphopeptides labeled 3–6, 8, 9, and 11) observed with full-length UBF phosphorylated in vivo (Fig. 4A) or rUBF phosphorylated in vitro (Fig. 4B Left) indicating the presence of phosphorylation sites both inside and outside the acidic tail of UBF.
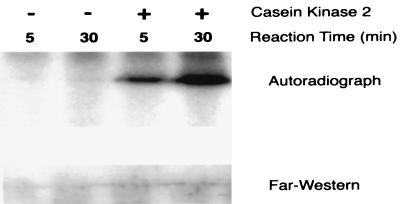
Although the kinase or kinases that phosphorylate UBF have not been identified, the acidic tail is rich with consensus casein kinase II (CKII) sites. Moreover, there is evidence showing that growth factors induce increases in CKII activity and nuclear translocation (18, 19). Voit et al. (16) presented evidence that consensus CKII phosphorylation sites within the acidic tail were involved in UBF transcriptional activity in vitro but that CKII alone was not sufficient for transcriptional activation. We thus tested the ability of CKII to phosphorylate UBF and activate TBP binding. Results demonstrated that although CKII stimulated marked increases in UBF phosphorylation in vitro (Fig. 5 Upper), it did not increase UBF–TBP binding (Fig. 5 Lower). In addition, two-dimensional phosphotryptic peptide maps of rUBF1 phosphorylated in vitro showed that CKII phosphorylated two of the eleven sites (i.e., phosphotryptic peptides 9 and 11) phosphorylated by SMC extract (data not shown). Thus it is possible that CKII may regulate UBF activity in concert with another kinase or other kinases. We also tested a variety of other kinases with consensus sites within UBF, including casein kinase I, Cdc2, and glycogen synthase kinase. Whereas these three enzymes each phosphorylated UBF, they showed incomplete phosphotryptic maps and also failed to enhance the TBP binding activity of rUBF (data not shown).
Figure 5.
Far Western analysis showed that CKII phosphorylated rUBF1 in vitro but did not increase UBF–TBP binding. Autoradiographic analysis of 32P incorporation (Upper) or Far Western blot rUBF1 (Lower) phosphorylated in vitro by CKII. Phosphorylation reactions and Far Western analysis were performed as described in Methods but with substitution of 20 ng of CKII (750 units/mg activity, Upstate Biotechnology, Lake Placid, NY) in place of the SMC nuclear extract.
Results of the present studies provide evidence that UBF–TBP interaction in cultured cells is increased markedly after stimulation with a variety of different growth factors. We also provide strong evidence based on in vitro Far Western assays that this association is regulated at least in part by the phosphorylation state of UBF. Moreover, in separate studies we have found that extracts of rat liver exhibit nearly identical activity as SMC in stimulating increased phosphorylation of UBF and its binding to TBP (A.J.K and G.K.O., unpublished results). As such, it appears that our results are not restricted to smooth muscle and may be more generally applicable to multiple cell types. The fact that phosphorylation of UBF has been implicated in the control of rRNA transcription in a wide variety of different cell types (3) and our observations that a variety of growth factors stimulated identical UBF phosphorylation patterns indicate that this may represent a general mechanism for the growth-factor regulation of rRNA transcription in many cell types. This model is consistent with previous studies showing the importance of cooperative protein–protein interactions between UBF and SL1 for targeting of SL1 to the rRNA promoter (4). Of interest, results of in vitro rRNA ‘order of addition’ transcription studies indicate that binding of UBF to the rRNA promoter elements is one of the earliest events in formation of the RNA polymerase I transcription initiation complex (20). However, UBF phosphorylation does not appear to regulate DNA binding (16, 21). Rather, our results suggest that phosphorylation of UBF regulates its ability to recruit TBP/SL1 to the rRNA promoter.
Several other groups (10, 16) have presented contrasting UBF tryptic phosphopeptide maps between growth-stimulated and quiescent cells suggesting that there are fundamentally different mechanisms involved in control of rRNA transcription in different growth conditions. However, those experiments involved growth arrest of cells in media lacking any serum or hormonal supplements under conditions in which cells were likely in a state of negative protein balance. Indeed, in at least one study (10), 72 h after serum withdrawal no immunoprecipitable UBF was detected. Our culture conditions involved a well-established method of growth arrest in a defined SFM containing 5 × 10−7 M insulin that maintained our SMC in a state in which they continue to transcribe rRNA and where there is no net loss or gain of cellular protein — a condition that better mimics normal cells in vivo (15). However, a potential caveat of our studies is that cultured SMC are known to produce a variety of autocrine growth factors that do not stimulate proliferative growth of the SMC but may have contributed to basal phosphorylation of UBF (22). Thus, it is highly likely that the differences in UBF phosphorylation patterns between quiescent and growth–stimulated cells in previous studies and ours are a function of differences in the culture conditions, not differences between cell types or the importance of UBF phosphorylation in the regulation of rRNA transcription.
In summary, our studies provide one potential mechanism to explain how growth factors regulate recruitment of TBP–TAF to the rRNA promoter. In addition, they show that growth stimulation, at least under the conditions of our studies, did not result in induction of new sites of UBF phosphorylation but rather a hyperphosphorylation or increased stoichiometry of sites phosphorylated in cells under basal conditions. Such a change perhaps should not be surprising, since one would not expect fundamentally different mechanisms to regulate rRNA transcription in quiescent vs. growing cells. Finally, our demonstration that phosphorylation of UBF activates its ability to bind TBP and that this can be measured in vitro now provides a critical (high-throughput) experimental assay system that will allow us to: (i) determine specific phosphoserine sites within UBF that mediate TBP binding; (ii) identify the kinase or kinases that phosphorylate UBF; and (iii) determine molecular mechanisms as to how phosphorylation of UBF alters its interaction with TBP and possibly other components of the RNA polymerase I transcription complex.
Acknowledgments
The authors gratefully acknowledge the expert technical assistance of Ms. Andrea Tanner and Ms. Diane Raines. This work was supported by National Institutes of Health grant P01 HL19242 (G.K.O.) and training grant 5T32-HL07284 (C.S.M., J.C.H., and A.J.K.), as well as by Fellowship grants from the Virginia Affiliate of the American Heart Association, Richmond, VA, to A.J.K. (VA-97-F-17), J.C.H. (VA-94-F-22), and C.S.M. (VA-95-F-18).
ABBREVIATIONS
- SL1
selectivity factor 1
- UBF
upstream binding factor
- TBP
TATA-binding protein
- A-II
angiotensin-II
- SMC
smooth muscle cells
- SFM
serum-free medium
- FBS
fetal bovine serum
- PDGF-BB
platelet-derived growth factor BB
- rUBF
recombinant UBF
- SAP
shrimp alkaline phosphatase
- CKII
casein kinase II
References
- 1.Baserga A. Exp Cell Res. 1998;151:1–5. doi: 10.1007/978-3-642-67986-5_1. [DOI] [PubMed] [Google Scholar]
- 2.Learned R M, Learned T K, Haltiner M M, Tjian R. Cell. 1986;45:847–857. doi: 10.1016/0092-8674(86)90559-3. [DOI] [PubMed] [Google Scholar]
- 3.Moss T, Stefanovsky V Y. Prog Nucleic Acid Res Mol Biol. 1995;50:25–66. doi: 10.1016/s0079-6603(08)60810-7. [DOI] [PubMed] [Google Scholar]
- 4.Bell S P, Learned R M, Jantzen H-M, Tjian R. Science. 1988;241:1192–1197. doi: 10.1126/science.3413483. [DOI] [PubMed] [Google Scholar]
- 5.Hempel W M, Cavanaugh A H, Hannan R, Taylor L, Rothblum L I. Mol Cell Biol. 1996;16:557–563. doi: 10.1128/mcb.16.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Putnam C D, Copenhaver G P, Denton M L, Pikaard C S. Mol Cell Biol. 1994;14:6476–6488. doi: 10.1128/mcb.14.10.6476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kwon H, Green M R. J Biol Chem. 1994;269:30140–30146. [PubMed] [Google Scholar]
- 8.Hempel W M, Cavanaugh A H, Hannan R D, Taylor L, Rothblum L I. Mol Cell Biol. 1996;16:557–563. doi: 10.1128/mcb.16.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beckman H, Chen J-L, O’Brien T, Tjian R. Science. 1995;270:1506–1509. doi: 10.1126/science.270.5241.1506. [DOI] [PubMed] [Google Scholar]
- 10.O’Mahony D J, Xie W, Smith S D, Singer H A, Rothblum L I. J Biol Chem. 1992;267:35–38. [PubMed] [Google Scholar]
- 11.Hannan R D, Lukyen J, Rothblum L I. J Biol Chem. 1996;271:3213–3220. doi: 10.1074/jbc.271.6.3213. [DOI] [PubMed] [Google Scholar]
- 12.Luyken J, Hannan R D, Cheung J Y, Rothblum L I. Circ Res. 1996;78:354–361. doi: 10.1161/01.res.78.3.354. [DOI] [PubMed] [Google Scholar]
- 13.Hershey J C, Hautmann M, Thompson M M, Rothblum L I, Haystead T A J, Owens G K. J Biol Chem. 1995;270:25096–25101. doi: 10.1074/jbc.270.42.25096. [DOI] [PubMed] [Google Scholar]
- 14.Voit R, Schnapp A, Kuhn A, Rosenbauer H, Hirschmann P, Stunnenberg H G, Grummt I. EMBO J. 1992;11:2211–2218. doi: 10.1002/j.1460-2075.1992.tb05280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geisterfer A A, Peach M J, Owens G K. Circ Res. 1988;62:749–756. doi: 10.1161/01.res.62.4.749. [DOI] [PubMed] [Google Scholar]
- 16.Voit R, Kuhn A, Sander E E, Grummt I. Nucleic Acids Res. 1995;23:2593–2599. doi: 10.1093/nar/23.14.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Genetics Computer Group. Program Manual for the Wisconsin Package 8. Madison, WI: Genetics Computer Group; 1994. [Google Scholar]
- 18.Klarlund J K, Czech M P. J Biol Chem. 1988;263:15872–15875. [PubMed] [Google Scholar]
- 19.Ackerman P, Osheroff N. J Biol Chem. 1989;264:11958–11965. [PubMed] [Google Scholar]
- 20.Schnapp A, Grummt I. J Biol Chem. 1991;266:24588–24595. [PubMed] [Google Scholar]
- 21.O’Mahony D J, Smith S D, Xie W, Rothblum L I. Nucleic Acids Res. 1992;20:1301–1308. doi: 10.1093/nar/20.6.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwartz S M, Heimark R L, Majesky M W. Physiol Rev. 1990;70:1177–1209. doi: 10.1152/physrev.1990.70.4.1177. [DOI] [PubMed] [Google Scholar]
- 23.Dignam J D, Lebovitz R M, Roeder R G. Nucleic Acids Res. 1983;11:1575–1583. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]