Abstract
The neutrophil serine protease proteinase 3 (PR3) is a main autoantigen in anti-neutrophil cytoplasmic antibody-associated vasculitis. PR3 surface presentation on neutrophilic granulocytes, the main effector cells, is pathogenically important. PR3 is presented by the NB1 (CD177) glycoprotein, but how the presentation develops during neutrophil differentiation is not known. An N-terminally unprocessed PR3 (proPR3) is produced early during neutrophil development and promotes myeloid cell differentiation. We therefore investigated if PR3 presentation depended on NB1 during neutrophil differentiation and if PR3 and proPR3 could both be presented by NB1. In contrast to mature neutrophils, differentiating neutrophils showed an early NB1-independent PR3 surface display that was recognized by only two of four monoclonal anti-PR3 antibodies and occurred in parallel with proPR3, but not PR3 secretion, suggesting that the NB1-independent surface PR3 was proPR3. PR3 gene expression preceeded NB1. When the NB1 receptor was detected on the surface, a mode of PR3 surface display similar to mature neutrophils developed together with the degranulation system. Ectopic expression studies showed that NB1 was a sufficient receptor for PR3 but not proPR3. ProPR3 display on the plasma membrane may influence the bone marrow microenvironment. NB1-mediated PR3 presentation depended on PR3 N-terminal processing implicating the PR3–N-terminus as NB1-binding site.
Keywords: ANCA vasculitis, NB1, neutrophil differentiation, proteinase 3
Introduction
Anti-neutrophil cytoplasmic antibody (ANCA)-associated vasculitides are characterized by small vessel inflammation, often presenting with life-threatening pulmonary and renal manifestations including haemorrhage and necrotizing crescentic glomerulonephritis with acute renal failure [1–3]. Neutrophilic granulocytes (neutrophils) are key effector cells of the disease [4]. ANCA can induce neutrophils to produce superoxide and alter their apoptosis [5–7]. Proteinase 3 (PR3) and myeloperoxidase (MPO) are the main ANCA autoantigens [8,9]. PR3 has the unique feature to be displayed on a percentage of neutrophils in most individuals [10–13]. A high percentage of membrane PR3-positive neutrophils is a risk factor for both onset and worse outcome of the disease [12,13]. The PR3 displaying subpopulation is identical to the subpopulation that carries the NB1 glycoprotein, a glycophosphatidylinositol-linked membrane receptor [14,15]. We have provided evidence recently that NB1 acts as a PR3 receptor on the neutrophil plasma membrane.
Neutrophil differentiation is a complex process of sequential gene expression and protein packing into granule subtypes [16]. PR3 mRNA expression occurs early during myeloid differentiation [17–19]. From gene array data, PR3 and NB1 appear to be expressed sequentially [20]. It is currently not clear what determines a neutrophil progenitor to express NB1 mRNA [21]. Parallel studies concerning PR3 and NB1 protein expression and granule localization are lacking. It has also not been reported when NB1 and PR3 surface presentation develop. While others and we have shown that PR3 and NB1 can associate on the cell surface, it is unclear at present if they already associate in the cell [14,22]. This would have implications for possible future therapeutic interventions aiming at inhibition of PR3 surface display.
The NB1–PR3 binding mechanism has not yet been elucidated. Exogenous purified neutrophil PR3 can bind ectopically expressed NB1; however, there are currently no published data addressing the question if other specific myeloid molecules are necessary for trafficking of PR3 to the surface and how PR3 processing affects binding to NB1. PR3 mRNA includes regions for an N-terminal signal peptide and an N-terminal dipeptide [23,24]. While the signal peptide is removed immediately, a form containing the N-terminal dipeptide (proPR3) is secreted early during myeloid differentiation [18,25,26]. Dipeptide removal is required for PR3 proteolytic activity [23,27–30]. The dipeptide is not removed when PR3 is expressed ectopically in HEK293 epithelial cells [29,31]. ProPR3 has differentiating and anti-proliferative effects on myeloid progenitors [26,32].
To characterize the development of NB1–PR3 binding, we studied PR3 membrane presentation during neutrophil differentiation and found that an additional mode of PR3, most probably proPR3, presentation existed that was independent of NB1. NB1 was sufficient for PR3 surface presentation after ectopic co-expression non-myeloid cells. NB1-mediated surface display depended on PR3 N-terminal processing.
Methods
Subjects, neutrophil and stem cell isolation and culture
Neutrophils were obtained from normal volunteers after informed consent was obtained, according to Internal Review Board (IRB) requirements. Umbilical cord blood was obtained from normal term deliveries at this hospital, also after informed IRB-designated consent. Neutrophils were isolated from heparinized whole blood by red blood cell sedimentation with dextran 1%, followed by Ficoll-Hypaque density gradient centrifugation and hypotonic erythrocyte lysis, as described previously [33]. Cell viability was detected in every cell preparation by trypan blue exclusion and exceeded 99%. The neutrophil percentage in the suspension was > 95% by Wright–Giemsa staining. For isolation of CD34+ progenitors, procedures were as described [12,14]. Mononuclear cells from heparin anti-coagulated cord blood were obtained by centrifugation over a LSM1077 (PAA, Pasching, Austria) gradient (800 g, 30 min). Cells were washed and stained with direct CD34+ progenitor isolation kit (Miltenyi, Bergisch-Gladbach, Germany) and sorted according to the manufacturer's instructions. Cells were cultivated in stem span serum-free medium (Stem Cell Technologies, Vancouver, Canada) supplemented with penicillin/streptomycin, 100 ng/ml stem cell factor, 20 ng/ml TPO and 50 ng/ml FLT3-L (Peprotech, London, UK) for expansion. Neutrophil differentiation was in RPMI-1640 with 10% fetal calf serum (FCS) and 10 ng/ml granulocyte–colony-stimulating factor (G-CSF) (Peprotech).
Nitrogen bomb cavitation and Percoll density gradient
Neutrophil disruption and two-layer density gradient centrifugation were essentially as described [34]. The three granule bands and cytosolic supernatant were aspirated and remaining Percoll removed by centrifugation at 100 000 g for 2 h. In the resulting fractions, MPO was assessed as a marker of primary granules and activity determined by MPO activity assay with the substrate 2,2′-azino-bis(3-ethylbenzthiazoline-6-sulphonic acid) and alkaline phosphatase (AP) served as a marker of plasma membranes measured the substrate p-nitrophenylphosphate (both Sigma-Aldrich, St Louis, MO, USA), according to the manufacturer's instructions. Optical densities (OD) were measured in a Versa Max tunable microplate reader (Molecular Devices, Sunnyvale, CA, USA). Enzymatic activities in each fraction were determined immediately after density gradient centrifugation. The results expressed as a percentage of total enzymatic activity of the preparation were as follows: MPO; primary granules: 63% ± 7, secondary/tertiary granules: 29% ± 5, secretory vesicles and plasma membranes 7% ± 3, cytosol 2% ± 1 and AP; primary granules: 4% ± 4, secondary/tertiary granules: 8% ± 5 secretory vesicles and plasma membranes: 87% ± 7, cytosol: 1% ± 1. Protein content of each fraction was determined by Coomassie protein assay (Pierce, Rockford, IL, USA). We wish to thank Professor N. Borregaard for help with gradient centrifugation.
Reverse transcription–polymerase chain reaction
Total RNA were isolated with Trizol™ according to the manufacturer's instructions (Invitrogen, Karlsruhe, Germany) and treated with DNAse (Promega, Mannheim, Germany). cDNA was transcribed with Superscript II following the manufacturers protocol (Invitrogen). Quantitative Reverse transcription–polymerase chain reaction (RT–PCR) (qPCR) was performed using TaqMan technology (Applied Biosystems, Darmstadt, Germany). The following oligonucleotides were used for human NB1 (hCD177): forward primer 5′-TTGATGCTCATTGAGAGCGG-3′, reverse primer 5′-GCCTCCGTGCAGCCCT-3′, and the probe Fam 5′-CCCCAAGTGAGCCTGGTGCTCTCC-3′ Tamra. For PR3, we used human PR3 (hPR3) 465F 5′- TGTCACCGTGGTCACCTTCTT-3′, hPR3611R 5′-CCCCAGATCACGAAGGAGTCTAT-3′ and the probe hPR3504 Fam 5′-TTGCACTTTCGTCCCTCGCCG-3′ Tamra (Biotez, Berlin, Germany). RT–PCR and quantification were performed using an Applied Biosystems 7700 Sequence detector and qPCR Mastermix Plus (Eurogentec, Köln, Germany). Each sample was measured in triplicate, and expression levels were normalized to 18S housekeeper expression. For RNA quantification, the fluorescence signal was measured at each PCR cycle, and the increase in the fluorescence normalized reporter signal (RN) was documented in an amplification plot. Using non-template controls, the threshold was set in the log phase to subtract unspecific fluorescence signals.
Antibodies
PR3 was detected using monoclonal antibodies (mAbs) 12·8 (CLB, Amsterdam, the Netherlands), 4A5 and 6A6 (Wieslab, Lund, Sweden), and MCPR3-2 (Dianova, Hamburg, Germany), the NB1 antibody was clone MEM166 (BD Pharmingen and Biolegend, San Diego, CA, USA) and secondary fluorescein isothiocyanate (FITC) and phycoerythrin-conjugated F(ab)-fragments of goat anti-mouse immunoglobulin G (IgG) were from Dako (Hamburg, Germany), goat anti-mouse Alexa fluor 647 was from Invitrogen, anti-CD11b and CD66b were from Immunotech (Krefeld, Germany) and CD35 from Cymbus Biotech (Hants, UK). Horseradish peroxidase (HRP)-labelled goat anti-mouse IgG was from Santa Cruz Biotechnology (Santa Cruz, CA, USA) and HRP-labelled donkey anti-rabbit Ig (IgG) was from GE Healthcare (Munich, Germany). Polyclonal rabbit anti-PR3 was a gift from Professor R. Falk, Chapel Hill, NC, USA.
Flow cytometry
Flow cytometry was used as described previously to evaluate the membrane protein expression [33]. If indicated, cells were stimulated with 2 ng/ml tumour necrosis factor-α (Genzyme, Rüsselsheim, Germany) or N-formyl-Met-Leu-Phe (Sigma, Deisenhofen, Germany) for 20 min at 37°C. Incubation with neutrophil PR3 (The Binding Site, Birmingham, UK) was at 1 μg/ml on ice for 2 h, followed by washing twice. Samples were incubated on ice for antibody binding, washed and counted using a FACScan (Becton Dickinson, Heidelberg, Germany); 10 000 events per sample were collected and analysed with CellQuest Pro software (Becton Dickinson).
Confocal microscopy
Live cells were analysed immediately after staining using a Zeiss LSM 510 Meta mounted on an Axiovert 200 M using a 63× phase-contrast plan-apochromat oil objective (numerical aperture 1·4) at room temperature. Acquisition settings for all images were UV/488/543/633, and specific parameters for the fluorophores were FITC excitation at 488 nm light, detected with a 500–530 bandpass filter; Alexa 647 was excited at 633 nm and detected with a 650 nm longpass filter.
Cell lysis, sodium dodecyl sulphate-polyacrylamide gel electrophoresis, Western blot
Cells were incubated on ice in lysis buffer (20 mM Tris HCL pH 8·8, 138 mM NaCl, 10% glycerol, 2 mM ethylenediamine tetraacetic acid, 1% Triton X 100, 1% NP40 supplemented with proteinase inhibitors (10 mg/ml quercetin, 10 mg/ml leupeptin, 0·1 mM aprotinin, 5 mM iodoacetamid), 0·2 mM Na3VO4, 20 mM NaF and 1 mM phenylmethylsulphonyl fluoride) for 10 min, insoluble material was pelleted and samples were boiled without beta-mercaptoethanol and run in 10% sodium dodecyl sulphate-polyaerylamide gel electrophoresis gels. Protein was transferred to polyvinylidene difluoride-Western blot membranes (Roche, Mannheim, Germany), and detected by enhanced chemiluminescence Western blot reagent (Pierce, Rockford, IL, USA) on hyperfilm (GE Healthcare). OD were determined using arbitrary units with NIH image 1·61.
Vectors and cell line transfection
PR3 and NB1 plasmids have been described [29]. We wish to thank Professor U. Specks for the PR3 expression plasmids. pcDNA4 beta-galactosidase (Invitrogen) was used for control transfection. All plasmids were sequenced using an ABI PRISM 377 DNA sequencer (Applied Biosystems, Darmstadt, Germany) prior to use. Plasmids were amplified in Escherichia coli One Shot Top 10 (Invitrogen) and purified by endotoxin-free Qiagen Maxi-Prep kit (Qiagen, Hilden, Germany). HEK293 cells were cultured in Dulbecco's modified Eagle's medium high glucose supplemented with 10% FCS, l-glutamine and penicillin/streptomycin (Biochrom, Berlin, Germany), transfected with FugeneHD transfection kit (Roche, Indianapolis, IN, USA) and stained for flow cytometry 48–72 h after transfection. Transfection efficiency was > 80%.
Statistics
Statistics were calculated using StatView 4·5 (Abacus Concepts). Correlations are given with 95% confidence interval. Values are given with standard error of the mean. For comparison of two groups, a two-sided t-test was applied. P-values < 0·05 were considered significant.
Results
Proteinase 3 surface display on differentiating neutrophils
When exactly during neutrophil differentiation PR3 is brought to the neutrophil surface has not yet been reported. Live cell flow cytometry (Fig. 1a) of stem cells during neutrophilic differentiation showed that PR3 surface display occurred within 24 h of G-CSF exposure. This was found using two mAbs (MCPR3-2 and 4A5) that recognize two different PR3-epitopes [35]. However, two other mAbs (12·8 and 6A6), that also recognize neutrophil PR3, proPR3 and recombinant forms [31] and detect surface PR3 reliably on neutrophils, did not detect this surface PR3 form. The results were confirmed by confocal microscopy (Fig. 1b). In contrast to results with neutrophils from peripheral blood, we observed PR3 surface display on NB1 negative cells. Time–courses and percentages of membrane-positive cells during neutrophil differentiation were monitored by flow cytometry (Fig. 1c). NB1 was found on the cell surface from day 2 of G-CSF treatment and the percentage of NB1-positive cells increased up to day 7 of differentiation. The final percentage of NB1 positive cells after in vitro differentiations (38% ± 9 compared with 5% for isotype control) was very similar for different donors, with a wide variety of membrane positive cells in the peripheral blood. An exception were NB1-negative donors who remained negative during in vitro differentiation (n = 2). A PR3 surface presentation that was detected by all four mAbs developed after NB1 was present on the cell membrane and at day 7 was similar to NB1 (27% ± 8). These data suggest an NB1-independent mechanism of PR3 surface display early during neutrophil differentiation, whereas an NB1-dependent mechanism is involved at later time-points.
Fig. 1.
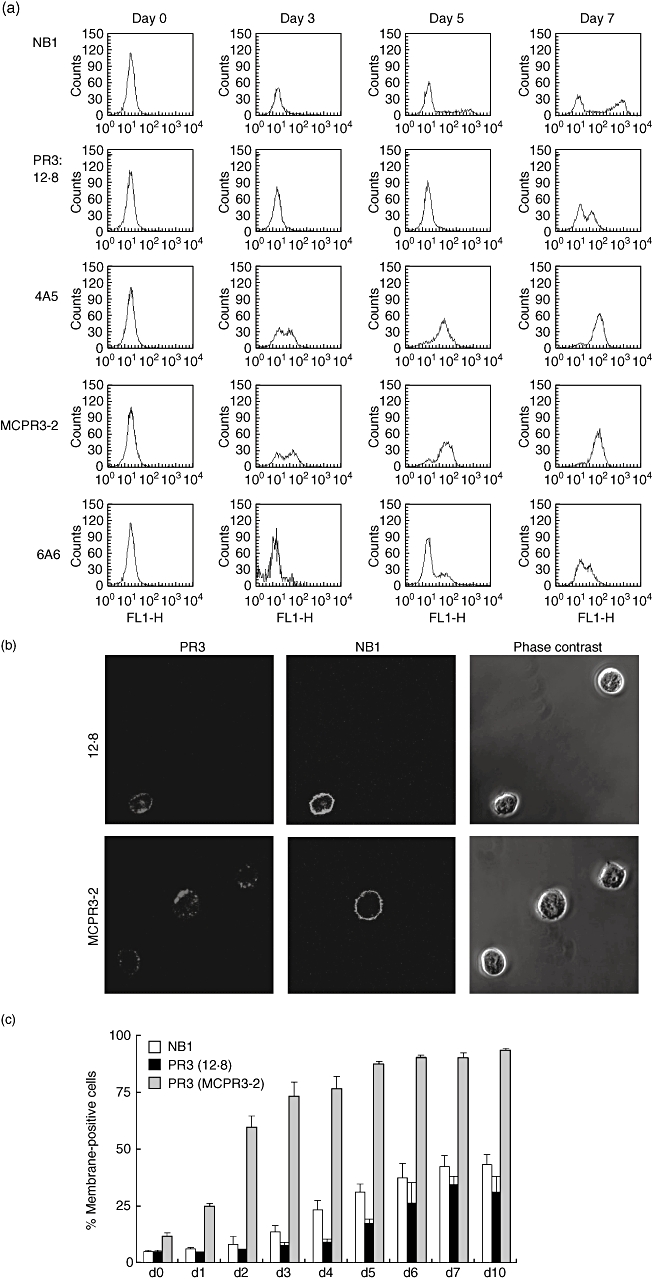
NB1 and proteinase 3 (PR3) surface display during neutrophilic differentiation in vitro. (a) Typical example of NB1 and PR3 surface presentation on live cells using 12·8, 4A5, 6A6 and MCPR3-2 monoclonal anti-PR3 antibodies that give two different patterns of membrane PR3 development. Confocal microscopy (b) demonstrates that the MCPR3-3 anti-PR3 antibody recognizes surface PR3 on developing neutrophils without NB1. (c) Quantification of NB1 and PR3 surface display development. Percentages of membrane-positive cells for either NB1 or PR3 cells are shown with the isotype set to 5% positive cells; n = 4 independent experiments per time-point during 10 days of differentiation.
Parallel PR3 and NB1 surface presentation on adult and neonatal neutrophils
To confirm the earlier finding of identical subsets of mature neutrophils displaying PR3 and NB1 on their membrane, we now tested all four mAbs in neutrophils from peripheral blood (Fig. 2). All PR3 antibodies gave identical results for adult neutrophils. In addition, we tested membrane presentation of both proteins in neonatal neutrophils that are distinct from adult in several surface markers and functional assays [36]. Again, we found very similar membrane PR3 and NB1 percentages, with all antibodies confirming and extending our previous results to neonatal cells.
Fig. 2.
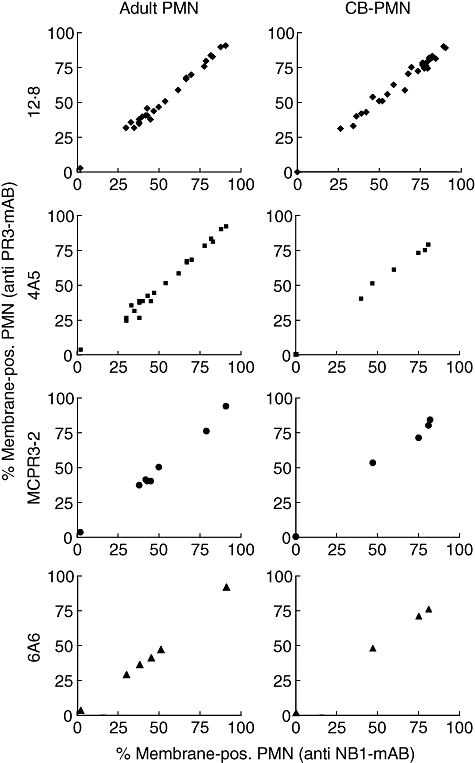
NB1 and proteinase 3 (PR3) surface display on neutrophils from peripheral blood. The correlation coefficient between percentage of PR3 and NB1-positive neutrophils was R = 0·99 in all cases, independent of the antibodies used (adult neutrophils [polymorphoneutrophil (PMN)]: n = 26 for 12·8, n = 22 for 4A5, n = 8 for MCPR3-2, n = 6 for 6A6). Neonatal, cord blood neutrophils (CB-PMN): n = 34 for 12·8, n = 8 for 4A5, n = 4 for MCPR3-2, n = 5 for 6A6).
Proteinase 3 and NB1-mRNA are expressed sequentially during neutrophil differentiation
To validate the neutrophil differentiation model from umbilical cord CD34+ haematopoietic stem cells for the longitudinal study of PR3 and NB1, we assessed mRNA levels during the differentiation phase. We observed a high level of PR3 mRNA expression that was down-regulated during differentiation with G-CSF, whereas NB1 expression was up-regulated in response to G-CSF and persisted during the time of in vitro culture (Fig. 3a). This is in accord with gene chip data in myeloid precursors from bone marrow [20]. Neutrophil proteins are sorted into different granule subtypes depending on their expression time-point [37]. Therefore, protein granule distribution in mature neutrophils should confirm their expression time. We assessed NB1 and PR3 specifically in granules from peripheral blood neutrophils after separation by Percoll density gradient centrifugation (Fig. 3b). Purity of fractions was confirmed by content of MPO and AP. In accordance with previous reports, we found that PR3 was concentrated in primary, NB1 in secondary and tertiary granules [38,39], supporting the conclusion that the sequence of gene expression in our differentiation model was correct.
Fig. 3.
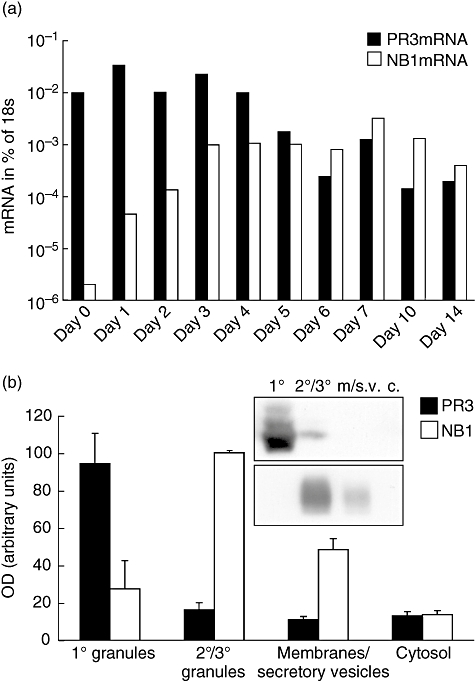
Proteinase 3 (PR3) and NB1 gene expression during neutrophil differentiation. (a) PR3-mRNA showed a gradual decline during in vitro differentiation, NB1 mRNA was up-regulated after addition of granulocyte–colony-stimulating factor and remained elevated during the observation period. Values are shown as percentage of housekeeper (18s). One typical of four independent experiments is shown. (b) PR3 and NB1 subcellular distribution in granules of peripheral blood neutrophils. Subcellular distribution is an indicator for the time of expression during development. PR3 was stored mainly in primary granules (OD = optical density: primary granules (1°): 94 ± 16, secondary/tertiary granules (2°/3°): 16 ± 4, secretory vesicles and plasma membranes (m./s.v) 11 ± 2, cytosol (c) 13 ± 2), NB1 in secondary and tertiary granules (OD: primary granules: 23 ± 15, secondary/tertiary granules: 100 ± 0, secretory vesicles and plasma membranes 28 ± 6, cytosol 14 ± 2). Immunoblot of a typical experiment and densitometric analysis from three (NB1) or four (PR3) independent experiments are shown.
Proteinase 3 and NB1 protein expression and translocation during neutrophilic differentiation
Protein expression was assessed by immunoblot analysis in cell lysates and supernatants of differentiating neutrophilic progenitors (Fig. 4). In the cell lysates, a low amount of high molecular weight PR3 was observed at the starting point of differentiation. After a few days of G-CSF exposure, mature PR3 appeared and high molecular weight forms disappeared. Accordingly, no high molecular weight PR3 was detected in granules from peripheral blood neutrophils (Fig. 3b). NB1 protein was first detected in cell lysates on day 2. In the cell culture supernatant, a high molecular weight PR3 (35 kDa), consistent with proPR3, was detected from day 2, similar to that described in another model of in vitro differentiation [28]. The 29 kDa PR3, together with NB1, appeared in the supernatant at later stages of differentiation (days 7–14). These data show that NB1 protein expression and cell surface display develop in parallel during neutrophilic differentiation and appear to be independent of detectable amounts of secreted NB1 in the supernatant. The two patterns of PR3 surface display detected by flow cytometry parallel the occurrence of proPR3 and PR3, respectively, in the cell culture supernatant. Thus, the data suggest that proPR3 accounts for the NB1-independent surface expression early during differentiation.
Fig. 4.
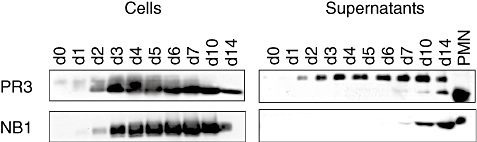
Proteinase 3 (PR3) and NB1 protein expression during neutrophilic differentiation. PR3 as detected by immunoblot was processed to a 29 kDa form by day 2 in the cells; in the cell culture supernatant, a 35 kDa proPR3 appeared on day 1; the 29 kDa PR3 was only detected from day 7. NB1 appeared in the cells on day 2 and in the supernatant on day 7. Lane loading was normalized for cell number and cell concentration respectively. PR3 was assessed by MCPR3-2 and polyclonal rabbit anti-PR3 giving similar results. A typical of three independent differentiations is shown.
Although PR3 was processed to a 29 kDa mature form in the neutrophil progenitors from day 2 PR3 display on the surface, in a form that was detected by all antibodies that recognize NB1-bound PR3 in mature neutrophils, appeared only several days later. To test if NB1–PR3 binding on the neutrophil progenitor surface was altered making PR3 inaccessible to two of the mAbs, we incubated the cells with purified neutrophil PR3. We found that the percentage of PR3 membrane positivity detectable with all antibodies increased to levels similar to NB1 positivity. This result is consistent with the idea that NB1 was functional as a receptor of mature PR3 already at early stages of differentiation (Fig. 5a). Fully processed PR3 is stored in granules and their translocation may be required for NB1-mediated PR3 display. We therefore tested translocation of markers for primary granules (CD66b), secondary/tertiary (CD11b) and secretory vesicles (CD35) in response to the chemotactic peptide N-formyl-Met-Leu-Phe during differentiation (Fig. 5b). NB1-dependent PR3 surface display developed in parallel to degranulation ability, suggesting that granule translocation machinery is involved in this process.
Fig. 5.
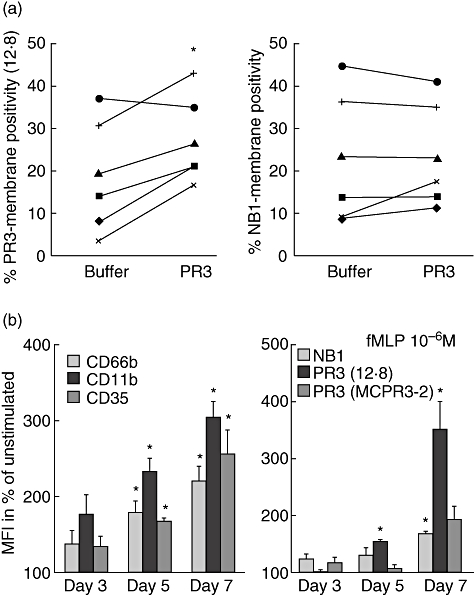
Proteinase 3 (PR3) binding to neutrophil progenitors and translocation to the plasma membrane. (a) Binding of purified neutrophil PR3 to neutrophilic progenitors (days 3–6 of differentiation) was assessed by flow cytometry. Percentages of membrane-positive cells for PR3 and NB1 from six independent experiments are shown (*P < 0·05). (b) Degranulation in response to 20 min stimulation with 10−6 M N-formyl-Met-Leu-Phe (fMLP) as percentage increase in membrane display of surface anchored marker molecules for primary granules (CD66b), secondary/tertiary (CD11b) and secretory vesicles (CD35) (MFI = mean fluorescence intensity). The increase in translocation after differentiation was especially marked for mature PR3 (*P < 0·05). Means of three independent differentiations are shown.
NB1-receptor expression is sufficient for PR3 surface presentation
Our previous data, showing that NB1 can act as a PR3 receptor, were obtained using either endogenous or purified neutrophil PR3. As PR3 processing is complex, we wanted to test if certain steps, especially N-terminal processing, influence PR3–NB1 interaction. When PR3 and proPR3 were transfected, similar amounts of both PR3 proteins were detected by immunoblot in cell lysates and supernatants (datanot shown). Single transfection of either form of PR3 didnot result in significant surface display. In contrast, co-transfection of NB1 with PR3, but not proPR3, led to PR3 membrane display (Fig. 6). Protease activity of PR3 was not required in this process. Results were similar for different anti-PR3mAbs (12·8, 4A5 or MCPR3-2). These results are the first to show that ectopic co-expression of PR3 and NB1 leads to PR3 surface presentation in a non-myeloid cell line. The data also underscore the role of NB1 as the specific receptor of mature PR3.
Fig. 6.
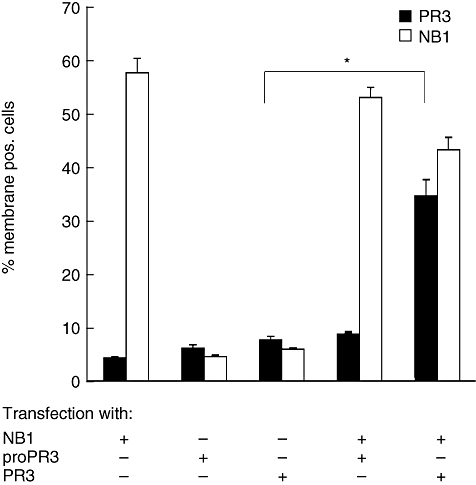
Surface presentation of NB1, pro proteinase 3 (proPR3) and PR3 expressed in HEK293 cells. Surface PR3 is exclusively up-regulated after co-transfection with NB1. There was no significant surface expression in single transfection of either proPR3 or PR3. Anti-PR3 12·8 was used in these experiments; similar data were obtained with MCPR3-2 and 4A5 (n = 5 independent transfections, *P < 0·05).
Discussion
Neutrophil surface display of the serine protease PR3 plays an important role for activation by autoantibodies. We found two distinct patterns of PR3 surface display during neutrophil differentiation. In addition to presentation via the NB1 receptor, as in mature neutrophils, there was NB1-independent membrane PR3 presentation early during differentiation that displayed only some of the PR3 epitopes detected on mature neutrophils. This up-regulation occurred concomitant with secretion of proPR3 into the supernatant and before PR3 secretion. The data suggest therefore that this early surface PR3 is proPR3. These data demonstrate a new mode of neutrophil autoantigen display. However, membrane display of proPR3 may also serve specific physiological purposes. Secreted proPR3 has an important role in myeloid differentiation. It inhibits proliferation and promotes differentiation [26]. Membrane-bound neutrophil serine proteases have altered activities [40–42]. Possibly, membrane-bound proPR3 exerts more stable and at the same time more localized signals to the direct progenitor environment. Similar membrane display during neutrophil differentiation was reported recently for another neutrophil serine protease, elastase [43].
How proPR3 surface presentation during neutrophil development is mediated on a molecular level remains to be elucidated. We did not observe similar amounts of proPR3 on the surface after transfection into HEK293 epithelial cells. This finding makes participation of another molecule in proPR3 trafficking or presentation likely. On peripheral blood neutrophils, our study confirms that no NB1-independent mode of PR3 presentation was present. As no proPR3 protein was detectable in cell lysates of mature neutrophils from peripheral blood, it cannot be differentiated if mature neutrophils still have the potential to membrane display proPR3. Subpopulations of membrane PR3-positive cells in a given individual are stable over years, even during chemotherapy [44,45]. However, recently an increase in membrane PR3-positive circulating neutrophils measured by a new mAb was reported in patients with a massive leucocyte left shift who had developed fatal sepsis [46]. Whether or not this presentation is NB1-dependent is unknown. If not, the presentation may have been a proPR3 display on neutrophil precursors mobilized during massive left shift.
Proteinase 3 surface display on a percentage of peripheral blood neutrophils mediated by the NB1 receptor is a risk factor in ANCA vasculitis [12,13,45]. Our second major finding is that a non-myeloid cell line develops PR3 surface expression when PR3 and NB1 are co-transfected. Neither protease activity nor other myeloid specific molecules were required for this interaction. This situation, together with our new data on parallel membrane presentation of NB1 and PR3 on neonatal neutrophils, increases further the evidence that NB1 acts as the PR3 receptor on human neutrophils [14]. PR3 surface presentation by the NB1-receptor was limited to mature PR3 without the N-terminal PR3-propeptide. This suggests that the PR3 N-terminus is an element of the PR3–NB1 binding site. Studies using recombinant PR3 demonstrated that nearly all ANCA sera from patients with vasculitis recognize PR3 irrespective of its N-terminus [30,31,47–49]. In fact, clinical data show that ANCA binding not only to mature PR3 but also to proPR3 have the same or even better predictive power for disease progression in patients with ANCA vasculitis [8,50]. This indicates that the PR3 N-terminus is not a pathophysiologically relevant ANCA binding site. Our findings suggest a mechanistic explanation for these clinical data. If PR3 presented on the neutrophil surface via the NB1 receptor is bound at its N-terminus, the N-terminus is less accessible to autoantibodies on the neutrophil surface. Therefore, anti-PR3 ANCA binding to other epitopes would have the greatest potential for neutrophil activation leading to tissue damage. Further analysis of the PR3–NB1 binding site is limited by the fact that the three-dimensional structure of NB1 or near family members is not known. This will be a prerequisite for better description of the PR3–NB1 interaction.
This study is the first to describe the time–course of neutrophil membrane PR3 presentation during neutrophil differentiation and found a novel mode of PR3 surface display. It also shows that the PR3 N-terminal region is essential for PR3–NB1 binding with implications for PR3 epitopes accessible to autoantibodies on the neutrophil surface.
Acknowledgments
The Deutsche Forschungsgemeinschaft and the Helmholtz Gemeinschaft supported the study with grants to R. K. and F. C. L. This study was presented in part at the American Society of Nephrology meeting, San Francisco, CA, 2007. We thank G. Tuennemann for help with microscopy.
References
- 1.Bosch X, Guilabert A, Font J. Antineutrophil cytoplasmic antibodies. Lancet. 2006;368:404–18. doi: 10.1016/S0140-6736(06)69114-9. [DOI] [PubMed] [Google Scholar]
- 2.Morgan MD, Harper L, Williams J, Savage C. Anti-neutrophil cytoplasm-associated glomerulonephritis. J Am Soc Nephrol. 2006;17:1224–34. doi: 10.1681/ASN.2005080882. [DOI] [PubMed] [Google Scholar]
- 3.Woywodt A, Streiber F, de Groot K, Regelsberger H, Haller H, Haubitz M. Circulating endothelial cells as markers for ANCA-associated small-vessel vasculitis. Lancet. 2003;361:206–10. doi: 10.1016/S0140-6736(03)12269-6. [DOI] [PubMed] [Google Scholar]
- 4.Xiao H, Heeringa P, Liu Z, et al. The role of neutrophils in the induction of glomerulonephritis by anti-myeloperoxidase antibodies. Am J Pathol. 2005;167:39–45. doi: 10.1016/S0002-9440(10)62951-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Falk RJ, Terrell RS, Charles LA, Jennette JC. Anti-neutrophil cytoplasmic autoantibodies induce neutrophils to degranulate and produce oxygen radicals in vitro. Proc Natl Acad Sci USA. 1990;87:4115–19. doi: 10.1073/pnas.87.11.4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Csernok E, Ludemann J, Gross WL, Bainton DF. Ultrastructural localization of proteinase 3, the target antigen of anti-cytoplasmic antibodies circulating in Wegener's granulomatosis. Am J Pathol. 1990;137:1113–20. [PMC free article] [PubMed] [Google Scholar]
- 7.Gilligan HM, Bredy B, Brady HR, et al. Antineutrophil cytoplasmic autoantibodies interact with primary granule constituents on the surface of apoptotic neutrophils in the absence of neutrophil priming. J Exp Med. 1996;184:2231–41. doi: 10.1084/jem.184.6.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finkielman JD, Merkel PA, Schroeder D, et al. Antiproteinase 3 antineutrophil cytoplasmic antibodies and disease activity in Wegener granulomatosis. Ann Intern Med. 2007;147:611–19. doi: 10.7326/0003-4819-147-9-200711060-00005. [DOI] [PubMed] [Google Scholar]
- 9.Wiik A. Autoantibodies in vasculitis. Arthritis Res Ther. 2003;5:147–52. doi: 10.1186/ar758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abdgawad M, Hellmark T, Gunnarsson L, Westman KW, Segelmark M. Increased neutrophil membrane expression and plasma level of proteinase 3 in systemic vasculitis are not a consequence of the −564 A/G promotor polymorphism. Clin Exp Immunol. 2006;145:63–70. doi: 10.1111/j.1365-2249.2006.03119.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Halbwachs-Mecarelli L, Bessou G, Lesavre P, Lopez S, Witko-Sarsat V. Bimodal distribution of proteinase 3 (PR3) surface expression reflects a constitutive heterogeneity in the polymorphonuclear neutrophil pool. FEBS Lett. 1995;374:29–33. doi: 10.1016/0014-5793(95)01073-n. [DOI] [PubMed] [Google Scholar]
- 12.Schreiber A, Otto B, Ju X, et al. Membrane proteinase 3 expression in patients with Wegener's granulomatosis and in human hematopoietic stem cell-derived neutrophils. J Am Soc Nephrol. 2005;16:2216–24. doi: 10.1681/ASN.2004070609. [DOI] [PubMed] [Google Scholar]
- 13.Rarok AA, Stegeman CA, Limburg PC, Kallenberg CG. Neutrophil membrane expression of proteinase 3 (PR3) is related to relapse in PR3–ANCA-associated vasculitis. J Am Soc Nephrol. 2002;13:2232–8. doi: 10.1097/01.asn.0000028642.26222.00. [DOI] [PubMed] [Google Scholar]
- 14.von Vietinghoff S, Tunnemann G, Eulenberg C, et al. NB1 mediates surface expression of the ANCA antigen proteinase 3 on human neutrophils. Blood. 2007;109:4487–93. doi: 10.1182/blood-2006-10-055327. [DOI] [PubMed] [Google Scholar]
- 15.Bauer S, Abdgawad M, Gunnarsson L, Segelmark M, Tapper H, Hellmark T. Proteinase 3 and CD177 are expressed on the plasma membrane of the same subset of neutrophils. J Leukoc Biol. 2007;81:458–64. doi: 10.1189/jlb.0806514. [DOI] [PubMed] [Google Scholar]
- 16.Friedman AD. Transcriptional regulation of granulocyte and monocyte development. Oncogene. 2002;21:3377–90. doi: 10.1038/sj.onc.1205324. [DOI] [PubMed] [Google Scholar]
- 17.Edvardsson L, Dykes J, Olsson ML, Olofsson T. Clonogenicity, gene expression and phenotype during neutrophil versus erythroid differentiation of cytokine-stimulated CD34+ human marrow cells in vitro. Br J Haematol. 2004;127:451–63. doi: 10.1111/j.1365-2141.2004.05227.x. [DOI] [PubMed] [Google Scholar]
- 18.Garwicz D, Lennartsson A, Jacobsen SE, Gullberg U, Lindmark A. Biosynthetic profiles of neutrophil serine proteases in a human bone marrow-derived cellular myeloid differentiation model. Haematologica. 2005;90:38–44. [PubMed] [Google Scholar]
- 19.Missen MA, Haylock D, Whitty G, Medcalf RL, Coughlin PB. Stage specific gene expression of serpins and their cognate proteases during myeloid differentiation. Br J Haematol. 2006;135:715–24. doi: 10.1111/j.1365-2141.2006.06360.x. [DOI] [PubMed] [Google Scholar]
- 20.Theilgaard-Monch K, Jacobsen LC, Borup R, et al. The transcriptional program of terminal granulocytic differentiation. Blood. 2005;105:1785–96. doi: 10.1182/blood-2004-08-3346. [DOI] [PubMed] [Google Scholar]
- 21.Stroncek DF. Neutrophil-specific antigen HNA-2a, NB1 glycoprotein, and CD177. Curr Opin Hematol. 2007;14:688–93. doi: 10.1097/MOH.0b013e3282efed9e. [DOI] [PubMed] [Google Scholar]
- 22.Brachemi S, Mambole A, Fakhouri F, et al. Increased membrane expression of proteinase 3 during neutrophil adhesion in the presence of anti proteinase 3 antibodies. J Am Soc Nephrol. 2007;18:2330–9. doi: 10.1681/ASN.2006121309. [DOI] [PubMed] [Google Scholar]
- 23.Jenne DE, Kuhl A. Production and applications of recombinant proteinase 3, Wegener's autoantigen: problems and perspectives. Clin Nephrol. 2006;66:153–9. doi: 10.5414/cnp66153. [DOI] [PubMed] [Google Scholar]
- 24.Specks U. What you should know about PR3–ANCA. Conformational requirements of proteinase 3 (PR3) for enzymatic activity and recognition by PR3-ANCA. Arthritis Res. 2000;2:263–7. doi: 10.1186/ar99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rao NV, Rao GV, Marshall BC, Hoidal JR. Biosynthesis and processing of proteinase 3 in U937 cells. Processing pathways are distinct from those of cathepsin G. J Biol Chem. 1996;271:2972–8. doi: 10.1074/jbc.271.6.2972. [DOI] [PubMed] [Google Scholar]
- 26.Skold S, Rosberg B, Gullberg U, Olofsson T. A secreted proform of neutrophil proteinase 3 regulates the proliferation of granulopoietic progenitor cells. Blood. 1999;93:849–56. [PubMed] [Google Scholar]
- 27.Fujinaga M, Chernaia MM, Halenbeck R, Koths K, James MN. The crystal structure of PR3, a neutrophil serine proteinase antigen of Wegener's granulomatosis antibodies. J Mol Biol. 1996;261:267–78. doi: 10.1006/jmbi.1996.0458. [DOI] [PubMed] [Google Scholar]
- 28.Skold S, Zeberg L, Gullberg U, Olofsson T. Functional dissociation between proforms and mature forms of proteinase 3, azurocidin, and granzyme B in regulation of granulopoiesis. Exp Hematol. 2002;30:689–96. doi: 10.1016/s0301-472x(02)00816-0. [DOI] [PubMed] [Google Scholar]
- 29.Specks U, Fass DN, Fautsch MP, Hummel AM, Viss MA. Recombinant human proteinase 3, the Wegener's autoantigen, expressed in HMC-1 cells is enzymatically active and recognized by c-ANCA. FEBS Lett. 1996;390:265–70. doi: 10.1016/0014-5793(96)00669-2. [DOI] [PubMed] [Google Scholar]
- 30.Witko-Sarsat V, Halbwachs-Mecarelli L, Almeida RP, et al. Characterization of a recombinant proteinase 3, the autoantigen in Wegener's granulomatosis and its reactivity with anti-neutrophil cytoplasmic autoantibodies. FEBS Lett. 1996;382:130–6. doi: 10.1016/0014-5793(96)00152-4. [DOI] [PubMed] [Google Scholar]
- 31.van der Geld YM, Oost-Kort W, Limburg PC, Specks U, Kallenberg CG. Recombinant proteinase 3 produced in different expression systems: recognition by anti-PR3 antibodies. J Immunol Methodsods. 2000;244:117–31. doi: 10.1016/s0022-1759(00)00261-1. [DOI] [PubMed] [Google Scholar]
- 32.Bories D, Raynal MC, Solomon DH, Darzynkiewicz Z, Cayre YE. Down-regulation of a serine protease, myeloblastin, causes growth arrest and differentiation of promyelocytic leukemia cells. Cell. 1989;59:959–68. doi: 10.1016/0092-8674(89)90752-6. [DOI] [PubMed] [Google Scholar]
- 33.Schreiber A, Luft FC, Kettritz R. Membrane proteinase 3 expression and ANCA-induced neutrophil activation. Kidney Int. 2004;65:2172–83. doi: 10.1111/j.1523-1755.2004.00640.x. [DOI] [PubMed] [Google Scholar]
- 34.Kjeldsen L, Sengelov H, Borregaard N. Subcellular fractionation of human neutrophils on Percoll density gradients. J Immunol Methods. 1999;232:131–43. doi: 10.1016/s0022-1759(99)00171-4. [DOI] [PubMed] [Google Scholar]
- 35.Van Der Geld YM, Limburg PC, Kallenberg CG. Characterization of monoclonal antibodies to proteinase 3 (PR3) as candidate tools for epitope mapping of human anti-PR3 autoantibodies. Clin Exp Immunol. 1999;118:487–96. doi: 10.1046/j.1365-2249.1999.01079.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carr R. Neutrophil production and function in newborn infants. Br J Haematol. 2000;110:18–28. doi: 10.1046/j.1365-2141.2000.01992.x. [DOI] [PubMed] [Google Scholar]
- 37.Le Cabec V, Cowland JB, Calafat J, Borregaard N. Targeting of proteins to granule subsets is determined by timing and not by sorting: the specific granule protein NGAL is localized to azurophil granules when expressed in HL-60 cells. Proc Natl Acad Sci USA. 1996;93:6454–7. doi: 10.1073/pnas.93.13.6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goldschmeding R, van Dalen CM, Faber N, et al. Further characterization of the NB 1 antigen as a variably expressed 56–62 kD GPI-linked glycoprotein of plasma membranes and specific granules of neutrophils. Br J Haematol. 1992;81:336–45. doi: 10.1111/j.1365-2141.1992.tb08237.x. [DOI] [PubMed] [Google Scholar]
- 39.Goldschmeding R, van der Schoot CE, ten Bokkel Huinink D, et al. Wegener's granulomatosis autoantibodies identify a novel diisopropylfluorophosphate-binding protein in the lysosomes of normal human neutrophils. J Clin Invest. 1989;84:1577–87. doi: 10.1172/JCI114335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Campbell EJ, Campbell MA, Owen CA. Bioactive proteinase 3 on the cell surface of human neutrophils: quantification, catalytic activity, and susceptibility to inhibition. J Immunol. 2000;165:3366–74. doi: 10.4049/jimmunol.165.6.3366. [DOI] [PubMed] [Google Scholar]
- 41.Korkmaz B, Attucci S, Jourdan ML, Juliano L, Gauthier F. Inhibition of neutrophil elastase by alpha1-protease inhibitor at the surface of human polymorphonuclear neutrophils. J Immunol. 2005;175:3329–38. doi: 10.4049/jimmunol.175.5.3329. [DOI] [PubMed] [Google Scholar]
- 42.Rice WG, Weiss SJ. Regulation of proteolysis at the neutrophil–substrate interface by secretory leukoprotease inhibitor. Science. 1990;249:178–81. doi: 10.1126/science.2371565. [DOI] [PubMed] [Google Scholar]
- 43.Tapper H, Kallquist L, Johnsson E, Persson AM, Hansson M, Olsson I. Neutrophil elastase sorting involves plasma membrane trafficking requiring the C-terminal propeptide. Exp Cell Res. 2006;312:3471–84. doi: 10.1016/j.yexcr.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 44.Schreiber A, Busjahn A, Luft FC, Kettritz R. Membrane expression of proteinase 3 is genetically determined. J Am Soc Nephrol. 2003;14:68–75. doi: 10.1097/01.asn.0000040751.83734.d1. [DOI] [PubMed] [Google Scholar]
- 45.Witko-Sarsat V, Lesavre P, Lopez S, et al. A large subset of neutrophils expressing membrane proteinase 3 is a risk factor for vasculitis and rheumatoid arthritis. J Am Soc Nephrol. 1999;10:1224–33. doi: 10.1681/ASN.V1061224. [DOI] [PubMed] [Google Scholar]
- 46.Matsumoto T, Kaneko T, Wada H, et al. Proteinase 3 expression on neutrophil membranes from patients with infectious disease. Shock. 2006;26:128–33. doi: 10.1097/01.shk.0000223122.11147.5a. [DOI] [PubMed] [Google Scholar]
- 47.Rarok AA, Huitema MG, van der Leij MJ, et al. Recombinant protein to analyze autoantibodies to proteinase 3 in systemic vasculitis. Am J Clin Pathol. 2003;120:586–95. doi: 10.1309/YTU2-FUHB-EXJL-WMLD. [DOI] [PubMed] [Google Scholar]
- 48.Sun J, Fass DN, Viss MA, et al. A proportion of proteinase 3 (PR3) -specific anti-neutrophil cytoplasmic antibodies (ANCA) only react with PR3 after cleavage of its N-terminal activation dipeptide. Clin Exp Immunol. 1998;114:320–6. doi: 10.1046/j.1365-2249.1998.00730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Farrag L, Pendergraft WF, Yang JJ, Jennette JC, Falk RJ, Preston GA. A study of conformational restraints on reactivity of human PR3-specific autoantibodies (ANCA) facilitated through protein folding manipulations of a new recombinant proteinase 3 protein. Autoimmunity. 2007;40:503–11. doi: 10.1080/08916930701680104. [DOI] [PubMed] [Google Scholar]
- 50.Russell KA, Fass DN, Specks U. Antineutrophil cytoplasmic antibodies reacting with the pro form of proteinase 3 and disease activity in patients with Wegener's granulomatosis and microscopic polyangiitis. Arthritis Rheum. 2001;44:463–8. doi: 10.1002/1529-0131(200102)44:2<463::AID-ANR65>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]


