Abstract
The function of local protein synthesis in synaptic plasticity and its dysregulation in fragile X syndrome (FSX) is well studied, however the contribution of regulated mRNA transport to this function remains unclear. We report a function for the fragile X mental retardation protein (FMRP) in the rapid, activity-regulated transport of mRNAs important for synaptogenesis and plasticity. mRNAs were deficient in glutamatergic signaling-induced dendritic localization in neurons from FMRP KO mice, and single mRNA particle dynamics in live neurons revealed diminished kinesis. Motor-dependent translocation of FMRP and cognate mRNAs involved the C-terminus of FMRP and kinesin light chain, and KO brain showed reduced kinesin-associated mRNAs. Acute suppression of FMRP and target mRNA transport in WT neurons resulted in altered filopodia-spine morphology that mimicked the FXS phenotype. These findings highlight a mechanism for stimulus-induced dendritic mRNA transport and link its impairment in a mouse model of FXS to altered developmental morphologic plasticity.
Keywords: FMRP, mGluR, mRNA transport, fragile X syndrome, kinesin, filopodia, spine
An emerging theme in the study of molecular mechanisms of learning and memory suggests an important function for the spatio-temporal control of gene expression at the synapse. Interestingly, altered expression of a single RNA-binding protein, FMRP, results in Fragile X syndrome (FXS), a highly prevalent form of inherited mental retardation, and contributes significantly to autism spectrum disorders(Penagarikano et al., 2007). FMRP demonstrates selective affinity for mRNA, including its own transcript and other targets important for neuronal development and plasticity(Penagarikano et al., 2007), and is transported in a stimulus-induced manner within dendrites and at spine-synapses (Antar et al., 2004) where it is associated with polyribosomes and represses translation(Penagarikano et al., 2007). FMRP is also itself translated within isolated synaptic fractions(Weiler et al., 1997) and in turn, affects translation of mRNA targets, suggesting potential for fine-tuning of local translational control at synapses. FMRP knockout (KO) neurons display excess basal translation, yet lack of stimulus-induced translation(Lu et al., 2004; Muddashetty et al., 2007; Zalfa et al., 2003). KO mice exhibit excessive immature dendritic spines, altered learning and behavior, and increased group I metabotropic glutamate receptor (mGluR)-dependent LTD, a form of plasticity that requires protein synthesis(Bagni and Greenough, 2005). Since altered translation of mRNAs has not been observed in situ, our knowledge of how FMRP affects translation in neurons remains limited to biochemically heterogeneous synapse fractions. One mechanism by which FMRP could affect translation is through selective delivery of mRNAs to dendritic translation sites in response to synaptic activity, however the contribution of mRNA-binding proteins to localization remains poorly understood.
Diverse neuronal signaling pathways can induce the localization of mRNAs(Bramham and Wells, 2007; Dictenberg and Singer, 2008), and mRNA binding proteins may serve to link subcellular trafficking of mRNA targets to local translation sites (Kiebler and Bassell, 2006). While details of FMRP function in translational regulation are known(Vanderklish and Edelman, 2005), a potential role for FMRP in mRNA localization has not been directly addressed. Previous low resolution, non-quantitative in situ histological methods failed to detect gross changes in mRNA localization to dendrites in neurons lacking FMRP(Steward et al., 1998). However a more sensitive FISH approach revealed alterations in the steady-state abundance of two FMRP target mRNAs in the molecular layer of hippocampus derived from KO brain, suggesting a role for FMRP in either mRNA transport or stability (Miyashiro et al., 2003) (Zalfa et al., 2007).
To explore a role for FMRP in dendritic mRNA localization we quantitatively examined several FMRP target mRNAs in dendrites of hippocampal neurons in response to neuronal activity. We report a deficiency in mGluR-induced mRNA localization in neurons derived from KO mice, and show that the motility of select FMRP mRNA target mRNAs important for synapse function is altered. This was measured directly using both fixed cells and living neurons by visualizing GFP-labeled mRNAs. We describe a molecular mechanism whereby FMRP acts as an adapter for kinesin light chain to promote stimulus-induced mRNA transport, and observe a widespread uncoupling of FMRP target mRNAs from kinesin in KO brains. Acute suppression of FMRP transport in WT neurons resulted in both diminished mRNA transport and a significant increase in the length and number of dendritic filopodia-spine protrusions that is similar to the phenotype observed in the mouse model and in humans with FXS. We propose that an alteration in stimulus-induced synaptic localization and transport kinetics of FMRP target mRNAs involved in synaptogenesis and plasticity may contribute to translational and synaptic defects observed in FXS.
Results
FMRP functions in stimulus-induced dendritic mRNA localization
We previously observed increased dendritic transport of FMRP and associated mRNAs upon stimulation of mGluR with DHPG (Antar et al., 2004; Antar et al., 2005). Since FMRP colocalized with known target mRNAs, we have investigated its role in mRNA transport. Here, the localization of several mRNA targets of FMRP, as well as non-targets, were compared in both wild-type (WT) and KO hippocampal cultures.
In response to mGluR stimulation, total polyadenylated (polyA) mRNA was increased in dendrites by ∼1.5 to 2-fold in both WT and KO cultures, indicating no gross defects in mRNA transport in KO (Suppl. Fig. 1a, p<0.01, n=12). This was in contrast to the localization of MAP1b and CaMKIIa mRNAs, which are both translationally regulated by FMRP and encode proteins critical for neurite outgrowth and synapse function(Lu et al., 2004; Muddashetty et al., 2007). MAP1b mRNA (Fig. 1a) was increased ∼4-fold (p<0.001, n=16) in response to DHPG in WT, while CaMKIIa mRNA (Fig. 1b) increased ∼2-fold (p<0.01, n=15). However, in KO neurons the mRNAs did not increase above unstimulated levels (MAP1b, p>0.25, n=15; CaMKIIa, p>0.25, n=17). We did not observe significantly decreased dendritic levels of CaMKIIa or MAP1b mRNAs in unstimulated KO neurons compared to WT. However, in KO neurons the signal appeared more diffuse compared to granules observed in WT (Fig. 1a, b), suggesting altered mRNP structure in the absence of FMRP (Aschrafi et al., 2005). The specificity of the localization response to mGluR was underscored by a lack of differences observed in dendritic beta-actin mRNA abundance using this paradigm (Fig. 1c, p>0.15, n=11-13), both in WT and KO cells, as beta-actin mRNA localization was previously shown to respond to neurotrophin (Zhang et al., 2001) and NMDA (Tiruchinapalli et al., 2003) receptor stimulation, and does not associate with FMRP (Muddashetty et al., 2007).
Fig. 1. Fluorescence in situ hybridization (FISH) of hippocampal neurons.
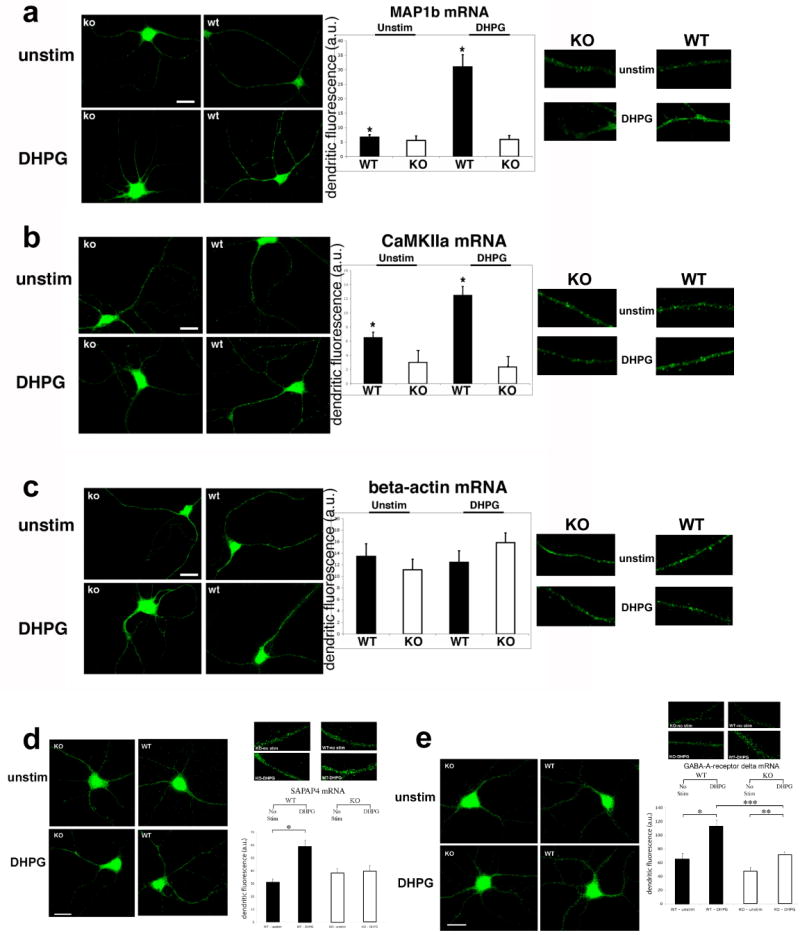
(a-d) Neurons from wild-type (WT, right panels) or FMRP-knockout (KO, left panels) were cultured for 10 days and either not stimulated (‘unstim’, upper panels) or stimulated with 50uM DHPG for 15 minutes (‘DHPG’, lower panels). Corresponding histograms showing dendritic quantification of FISH experiments are shown to the right. Results for WT (black bars) and KO (white bars) are labeled on the x-axis. Close-up images of dendritic signals are shown to the right (in same order). (a) MAP1b mRNA (n=15-16; *p<0.001 for unstimulated vs. DHPG in WT, P>0.25 for KO). (b) CaMKIIa mRNA (n=15-17; *p<0.01 for WT, p>0.2 for KO). There was no significant difference in CaMKIIa abundance of unstimulated neurons between WT and KO (n=15, p>0.1). (c) Beta-actin mRNA (n=11-13; p>0.2). Scale bars=12um. (d-e) FISH for other mRNA targets of FMRP in both WT (right image panels) and KO (left image panels). Representative images shown for unstimulated (upper panels; ‘unstim’) and stimulated (lower panels; DHPG, 50uM), and close-up images of dendrites are displayed above the histograms (in same order). Histograms (right) showing dendritic fluorescence quantification are shown for WT (green) and KO (red) using probes to (d) SAPAP4 (n=16-17; p<0.01 for WT, p>25 for KO) and (e) GABA-A receptor delta (n=13-15; *, ***p<0.01 for unstimulated vs. DHPG in WT and for DHPG in WT and KO, **p<0.05 for unstimulated vs. DHPG in KO). Scale bars=12um. Values are mean±SEM.
Further inspection of mRNA targets of FMRP showed an interesting panel of mRNA localization defects similar to that observed for MAP1b and CaMKIIa. The mRNAs for RGS5 and SAPAP4, both implicated in the regulation of post-synaptic receptor signaling (Gold et al., 1997; Takeuchi et al., 1997) and whose mRNAs associate with FMRP (Brown et al., 2001; Miyashiro et al., 2003), were also significantly increased (∼2-fold; RGS5, p<0.05, n=15; SAPAP4, p<0.01, n=16) by mGluR activation in WT but not KO neurons (Suppl. Fig. 1b, Fig. 1d; RGS5, p>0.20, n=15; SAPAP4, p>0.25, n=17). The mRNA for GABA-A-receptor delta subunit (GABAR-d)(Miyashiro et al., 2003) was significantly increased by ∼2-fold in WT dendrites in response to DHPG (p<0.01, n=13), but not in KO (Fig. 1e; p<0.05, n=15). Although we also observed an increase in GABAR-d mRNA in KO upon DHPG, the magnitude was small (26% increase) compared to the WT response (81% increase, p<0.01, n=13-15). However, another proposed target of FMRP, Arc mRNA, was not significantly increased in either WT or KO neurons under these conditions (Suppl. Fig. 1c, p>0.20, n=14-15). To further ensure that the mRNA signals were not due to individual cell-to-cell variation or culture conditions, we normalized the SAPAP4 mRNA in situ signals to both dendritic MAP2 fluorescence and synapsin puncta number, which similarly showed significant increases in mRNA levels after stimulation in WT but not KO neurons (Suppl. Fig. 1d, e; n=16-17, p<0.05 for SAPAP/MAP2, p<0.0001 for SAPAP/synapsin).
The in situ results suggested that either mRNA targets of FMRP were diminished in dendritic transport or the stability of mRNAs was decreased upon DHPG stimulation. To directly test the loss of FMRP on mRNA transport kinetics, we used the MS2-GFP system to visualize movements of a reporter for the CaMKIIa mRNA in live neurons cultured from both WT and KO brain. This reporter construct contains the whole 3′UTR, which is necessary for dendritic localization of CaMKIIa mRNA in vivo (Miller et al., 2002) and sufficient for localization in cultured neurons (Rook et al., 2000). Neurons were transfected with CaMKIIa-MS2-GFP and imaged in the presence of DHPG. mRNA-GFP particles were tracked over two-minute intervals and the mean square displacement (MSD), a measure of the average distance of travel, of each particle along with the percent motile particles was calculated. Strikingly, there was a significant difference in the kinetics of mRNA movements between the two genotypes (Fig. 2a; p<0.02, n=307 particles; Suppl. Movie 1, 2). On average 23% of CaMKIIa mRNA particles were motile in WT neurons, while only 5% of mRNA particles in KO neurons exhibited vectorial trajectories (Fig. 2b). Consistent with a known biochemical interaction of CaMKIIa and FMRP, we observed CaMKIIa-MS2-GFP mRNA particles colocalized with endogenous FMRP in WT neurons using FISH to the MS2 mRNA sequence (Fig. 2c; Suppl. Fig. 1f).
Figure 2. Time-lapse analysis of CaMKIIa reporter mRNA transport in live neurons from WT or Fmr1-KO.
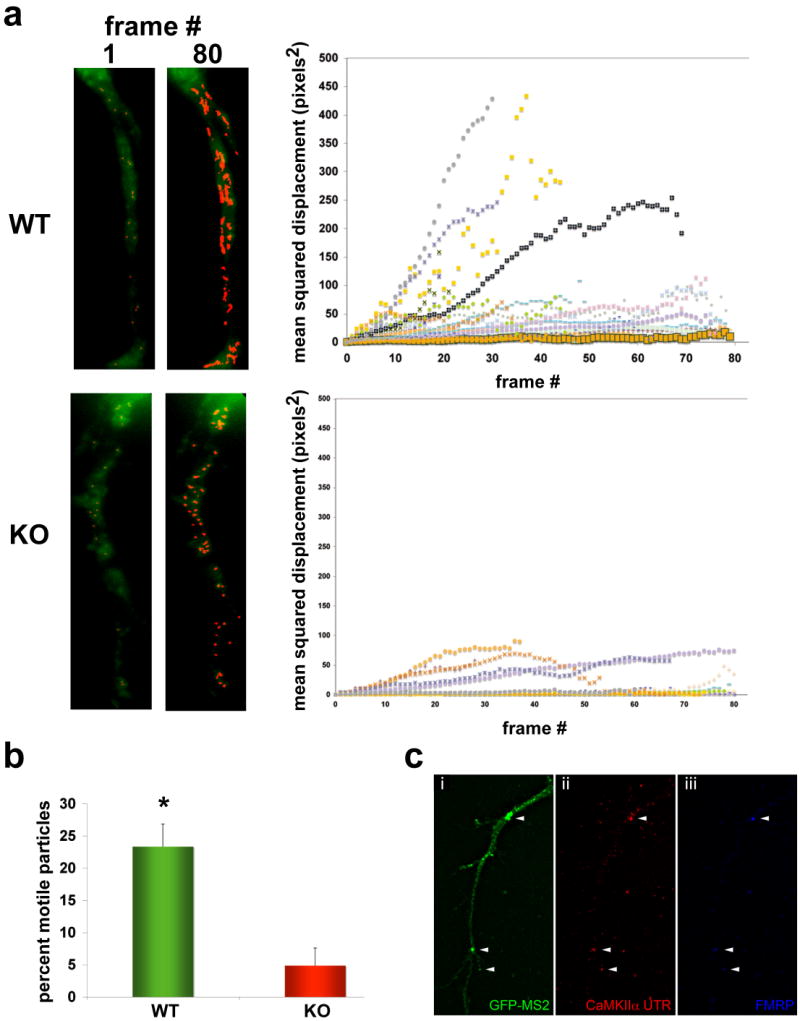
Neurons (10DIV) were transfected with a GFP-MS2-CaMKIIa mRNA reporter, and exposed to DHPG (50uM, 15 min.). mRNA movements were imaged over a two-minute interval, with each frame captured every 1.5s. (a) Images (left panels) show the first frame (1) and last frame (80) of the time-lapse series of GFP-MS2-CaMKIIa (green) and highlight the tracked mRNA particles (red). The mean-squared displacement (MSD) of individual particle trajectories were analyzed (graph, right) in WT (upper panels) and KO (lower panels) neurons, and an example of the MSD analysis graphs showing 24 particle trajectories for each genotype are shown at right (24 trajectories on graph; many are overlapping at bottom of graph and obscured by icons). The slope of the individual trajectories approximates the particle velocity, and the MSD measures the trajectory length over time.
(b) Histogram showing the average number of mRNA particles among several movies (n=6 neurons, 307 particles total, *p<0.02, mean±SEM) from both WT and KO neurons that were motile.
(c) Colocalization of CaMKIIa 3′UTR-MS2-GFP with endogenous FMRP. Fluorescence images of a hippocampal dendrite showing MS2-GFP (i, green), CaMKIIa-MS2 reporter mRNA (ii, red) and FMRP (iii, blue). Arrowheads show three granules in dendrites of this representative neuron triple-labeled where all colocalize, which was determined using 3D deconvolution and image reconstruction (see Suppl. Fig. 1f). Scale bar = 5um.
mGluR activates dendritic FMRP transport through kinesin light chain
To better understand the mechanism of FMRP mRNP transport in neurons, we analyzed motor interactions. Classic pelleting experiments revealed a nucleotide-dependent association of FMRP with taxol-stabilized microtubules (Suppl. Fig. 2a). Rapid imaging of living neurons expressing FMRP-GFP revealed bi-directional transport of granules at rates up to 1.8um/s in dendrites, suggesting involvement of both kinesin and dynein-based movements (Fig. 3a; Suppl. Movie 3). This was consistent with co-precipitation data showing an association of FMRP with these motors (Suppl. Fig. 2b) and specifically KLC (see below), which is a major cargo-binding subunit of the Kif5 holoenzyme (KHC and KLC). However another form of kinesin, Kif3 was not significantly associated with FMRP in brain. As a direct test for the role of kinesin in FMRP association with the cytoskeleton in situ we utilized a highly specific small molecule inhibitor of kinesin (adociasulfate, AS-2), which binds to the motor domain and releases it from microtubules (Sakowicz et al., 1998). Neurons exposed to AS-2 for 15 seconds showed a significant loss of FMRP in dendrites (Suppl. Fig. 2c; extract with AS-2) compared with either mock extracted (extract only; †p<0.01, n=15) or unextracted (no extract; *p<0.005, n=15) neurons. This same treatment caused specific removal of Kif5 (p<0.01, n=8) with no significant effect on cytoplasmic dynein (Suppl. Fig. 2d, p>0.3, n=7) or on overall MT organization (not shown).
Fig. 3. The C-terminus of FMRP is involved in kinesin-dependent stimulus-induced transport.
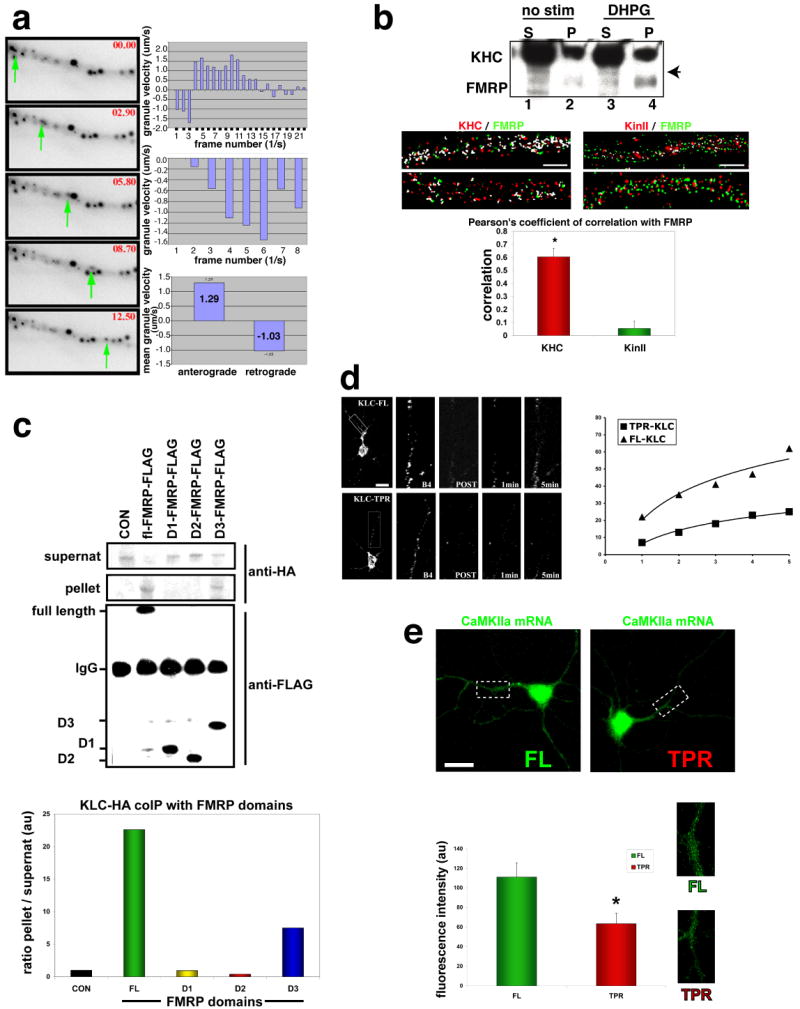
(a) Rapid FMRP-GFP movements in live hippocampal neurons (10DIV). (Images) 5 consecutive image panels (top to bottom, left side) show time points (seconds, red) indicated in the upper-right corner and a green arrow tracing the particle movement. (Histograms) Individual particle velocities (top 2 histograms) were traced on a frame-per-frame basis (granule velocities, um/sec, + is anterograde, - is retrograde). Average velocities (bottom histogram) were measured for multiple granules (lower graph, right; n=8, mean values shown).
(b) (Upper panel) Western showing KHC and FMRP coIP from cortical cultures. Either nothing (no stim) or DHPG was added to cultures for 15 min and then KHC IP performed. Supts (S) and pellets (P) were analyzed by SDS-PAGE. Arrowhead denotes FMRP band only seen in IP of KHC (other bands in SUPT are likely cross-reacting fragile X related proteins or post-translationally modified FMRP) (Lower Panel) Super-resolution 3-D colocalization analysis of kinesin heavy chain and FMRP in dendrites. Hippocampal neurons were cultured (12DIV), fixed and stained for FMRP (green) and either Kif5 (KHC, left two panels, red) or Kif3 (right two panels, red), and IF images were captured and processed for deconvolution. Images are maximum projections of 3-D stacks, and white pixels represent overlap of the two signals. The histogram shows the average Pearson's coefficient of correlation for each antigen pair as calculated (green bar is for KHC/FMRP and red bar is for Kif3/FMRP; n=14, p<0.0005, mean±SEM). Scale bar, 2um.
(c) Domain analysis of FMRP for kinesin interaction. (Top (first) panel - supernat) Quantitative western blot of KLC (anti-HA) shows the supts after the immunoprecipitation (IP) of FMRP domains, with both full-length (FL)-FMRP and C-terminal domain of FMRP (D3, aa386-585) able to significantly deplete the supt compared to the control (CON, no FMRP protein), N-terminal domain (D1, aa1-208) or the central domain (D2, aa290-387) of FMRP. (Second panel- pellet) Western blot against HA shows pellets contain KLC-HA that associate with FMRP by coIP. (Third panel-FMRP pellets) Western blot against FLAG shows the control (CON, no FMRP protein expressed) or IP FL-, D1-, D2-, or D3-FMRP proteins that were used to pull-down KLC-HA. (Histogram) HA-KLC bands pulled-down for each pellet and the corresponding supt were digitally quantified using the LiCOR quantitative blot system and expressed as a ratio of pellet to supt for each FMRP domain.
(d) Hippocampal neurons (7DIV) showing FRAP of FMRP-GFP in response to DHPG. (Left Panel) Images of dendrites subjected to FRAP: Upper series show a low magnification image of a control cell (co-transfected KLC-FL) and the dendritic area bleached (inset box), a higher mag image of the subregion just before bleach (B4, second panel) and after (third panel), 1 minute (fourth panel) and 5 minutes later (fifth panel). Lower series show the same sequence for KLC-TPR co-transfected with FMRP-GFP. Scale bar=15um.
(Right panel) Graph showing the percent recovery (y-axis) measured over 300 seconds (x-axis) is shown. Squares represent the KLC-TPR transfected cells while the triangles represent FL-KLC (n=6; p<0.005 at 300 second time point). Calculated time constants of recovery (τ, 50%) are shown on the graph.
(e) (Upper panels) Representative images showing CaMKIIa mRNA FISH for neurons transfected with either FL-KLC (left) or TPR-KLC (right) constructs and stimulated with DHPG for 15 minutes. Scale bar=15um. (Lower panel) Histogram of CaMKIIa mRNA dendritic abundance in KLC transfected neurons (n=17-22; *p<0.02; mean±SEM). Images to right show close-ups of dendrites from upper-panel images.
(f) (Upper panels) Representative images showing MAP2 mRNA FISH for neurons transfected with either FL-KLC (upper) or TPR-KLC (lower) constructs and stimulated with DHPG for 15 minutes. Scale bar=15um. (Lower panel) Histogram of MAP2 mRNA dendritic abundance in KLC transfected neurons (n=12-14; p>0.22; mean±SEM. Images to right show close-ups of MAP2 mRNA in dendrites from upper panel images.
An activity-induced increase in dendritic FMRP localization (Antar et al., 2004) suggested two potential mechanisms involving kinesin: one being an increase in the processivity of kinesin associated with FMRP; an alternative being an increase the fraction of FMRP associated with kinesin. To test the latter possibility the fraction of FMRP mRNP granules associated with Kif5 was measured. Cortical neurons cultured from WT mice were extracted either in basal states or after 15 minutes of mGluR activation, and then probed for FMRP association with Kif5 by immunoprecipitation (IP). Activation of mGluR caused a noticeable increase in the amount of FMRP associated with Kif5 (Fig. 3b, western blot). Cultured neurons were also stimulated with DHPG and processed for super-resolution microscopy using image deconvolution (Carrington et al., 1995), and the reconstructed images were used to compare the degree of overlap within the same volume pixels for two distinct isoforms of kinesin. The Kif5 signal correlated significantly with FMRP in dendritic shafts in comparison to Kif3, which did not show a robust correlation (Fig. 3b, image panels and histogram). This was not due to changes in total FMRP levels in response to DHPG (Antar et al., 2004). The stimulus-induced increase in Kif5 association was also observed for a homolog of FMRP, FXR1p, which is known to interact biochemically with the FMRP mRNP(Bagni and Greenough, 2005)(Supp. Fig. 3).
The molecular mechanism of motor attachment to RNA-binding protein and specifically FMRP remains unknown. Therefore several domains of FMRP were tested for KLC binding in neuronal cell lines (Fig. 3c) since we have observed this interaction in brain (see Supp. Fig. 2b). Importantly these cells express type I mGluR and were stimulated with DHPG for 15 minutes, which increases FMRP association with KLC. Quantitation showed that only the C-terminal domain of FMRP (D3), but not the N-terminal or central domain, could significantly interact with KLC (Fig. 3c, histogram). To test the role of Kif5 in FMRP transport in neurons, we used a dominant negative strategy (Verhey et al., 2001). We quantified FMRP-GFP transport rates in neurons co-transfected with either full-length (FL) KLC (control) or the TPR domain, which is able to bind to cargo but lacks the KHC-binding domain. Fluorescence recovery after photobleaching (FRAP) analysis (Fig. 3d, image panels) showed that the TPR domain significantly reduced transport kinetics by ∼5-fold, with an average time constant (50% recovery) of 1165 seconds, compared with the control time constant of 237 seconds (Fig. 3d, graph, n=6, p<0.005 at 300 second time point). TPR-KLC had no significant effect on another proposed Kif5 dendritic cargo, the glutamate-receptor interacting protein (GRIP1), which has been shown to interact with KHC. We confirmed that transfection of dominant negative KLC-TPR into neurons resulted in a ∼4-6-fold increase in cargo-binding domain in both the cell body (n=15, p<0.0001) and in dendrites (n=15, p<0.005) compared with endogenous KLC in untransfected controls (Suppl. Fig. 4a).
To investigate the role of Kif5 in dendritic transport of FMRP mRNA targets, TPR-KLC transfected neurons were stimulated and processed for FISH. Neurons transfected with KLC-TPR showed significantly diminished CaMKIIa mRNA localization compared to controls (Fig. 3e, 44% lower in TPR-KLC (n=22) vs. FL-KLC (n=17), p<0.02). We found a similar effect on the Fmr1 mRNA itself, which is a known target of FMRP (Ceman et al., 1999) (Supp. Fig. 4b, p<0.005, n=13-15 cells per treatment). This is in contrast to MAP2 mRNA, which was not altered by TPR-KLC (n=12) overexpression compared to controls (n=14, FL-KLC) (Suppl. Fig. 4c, p>0.22). Transfection of neurons with KLC constructs was confirmed for all cells and quantification showed that expression levels were similar (Suppl. Fig. 4d, e).
FMRP mediates association of target mRNAs with kinesin
The loss of stimulus-induced mRNA localization in KO neurons suggested that FMRP might play a structural role between mRNAs and molecular motors. One prediction from this model is that loss of FMRP could result in altered target mRNA coupling to kinesin. To test this, WT and KO brains were used to isolate Kif5 and analyze target mRNAs that co-purified (Fig. 4). IP of KHC with semi-quantitative PCR showed that SAPAP4 mRNA was significantly reduced in KO brains compared to WT (Fig. 4a, WT vs. KO lane for KHC-IP), in the range of 50-70% (also see Suppl. Fig. 5a). Real-time Q-PCR was performed on a large panel of target mRNAs that revealed different levels (∼20-50%) to which mRNAs were reduced in association with Kif5 in KO brains (Fig. 4b, *p<0.05, **p<0.01, ***p<0.005, ****p<0.0001, all n=8). The mRNA targets reduced in Kif5 association included genes involved in actin remodeling at synapses (cofilin phosphatase (PP2Ac); p116-RIP), synapse-associated signaling (DAG1; RGS5) and synapse structure (SAPAP4; CaMKIIa; MAP1b). Not all mRNAs were significantly reduced in Kif5 IPs, as OCRL1 mRNA was similar in KO brain (P>0.05, n=8). In addition two negative control mRNAs known not to associate with FMRP, MAP2 and beta-actin mRNA, were not altered in their association with Kif5 (P>0.4, n=8). A positive control for the assay was Fmr1 mRNA itself, which showed almost zero abundance in KO brain using this method (the first few exons (1-5) of Fmr1 are still expressed at the 5′ end of the gene in the KO where the PCR primers are located). Other controls consisted of an mRNA restricted to the soma, SM51, and GAPDH which were both not different in Kif5 IPs from KO compared to WT (Suppl. Fig. 5b), and were detected at low levels in association with Kif5. Quantification of input mRNA (as well as supernatants after IP, not shown) by Q-PCR demonstrated that the Kif5-associated mRNAs represented only a fraction of the total mRNA, which was not significantly altered in KO brain for any of the targets examined (Fig. 4c).
Fig. 4. FMRP target mRNAs associated with kinesin.
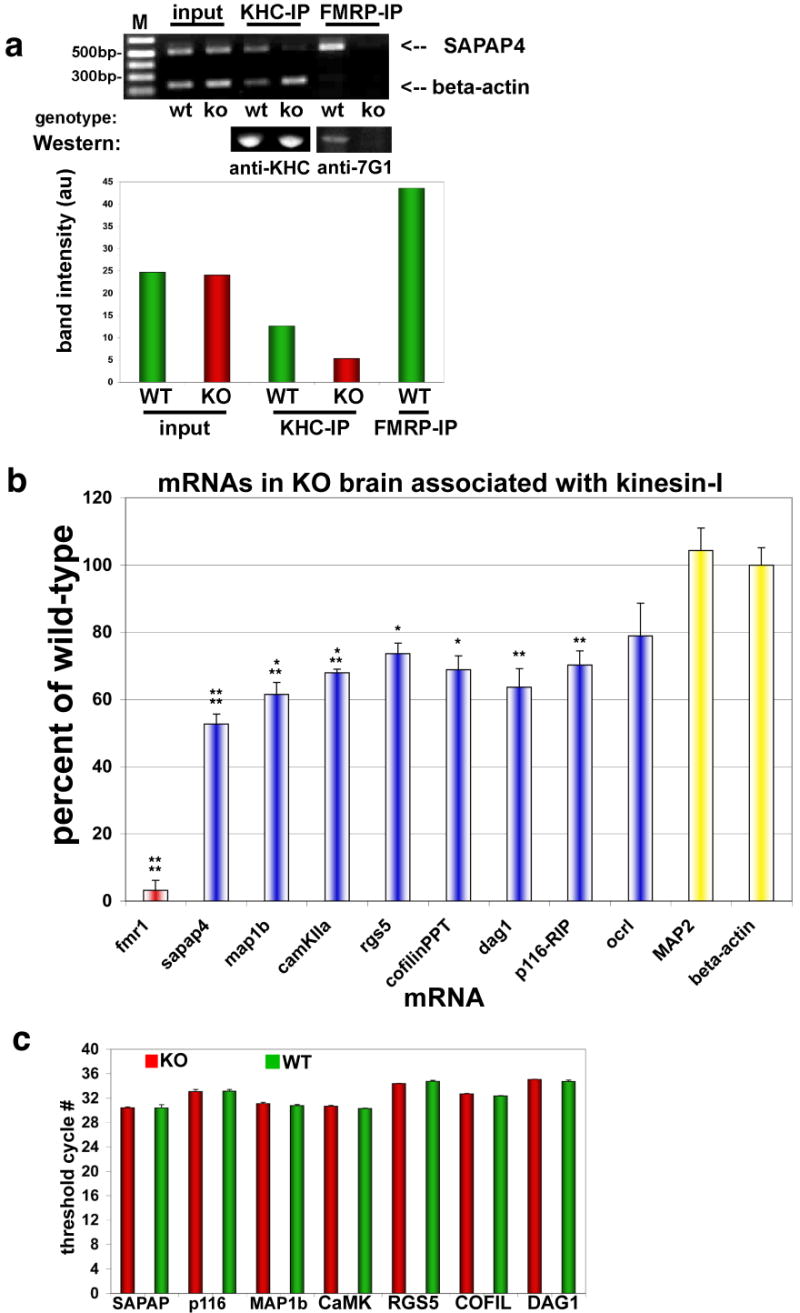
Immunoprecipitation (IP) of KHC was used to analyze associated mRNAs by PCR and determine differences between WT and FMRP KO brain.
(a) (Upper panel) Ethidium gel for semi-quantitative RT-PCR products from KHC and FMRP IP pellets using primers for beta-actin mRNA (lower bands) and SAPAP4 mRNA (upper bands). ‘Input’ is the input lysate showing starting material for both WT and KO brain. IPs of both kinesin heavy chain (KHC) and FMRP are indicated above the gel, and genotype is indicated below. FMRP IP in KO brain was used to subtract background from other bands, and was ∼4% of WT IP. (Middle panel) Western blot of protein pellets showing that the KHC IPs are identical from WT and KO brain. FMRP antibodies (anti-7G1) also reveal that FMRP is able to precipitate in WT but not KO brain (Lower panel - histogram) Quantification of mRNA bands from PCR (upper panel) by digital fluorescence analysis.
(b) Real-time Q-PCR of indicated mRNAs associated with KHC by IP, expressed as a percent of mRNA isolated from KHC in KO compared to WT brain (n=8; *p<0.05; **p<0.01; ***p<0.005; ****p<0.0001; mean±SEM).
(c) Real-time Q-PCR analysis of mRNA abundance. Histogram showing that several mRNA targets of FMRP were analyzed for total mRNA levels in both WT (green bars) and KO (red bars) brain derived from P7 mice (n=8, p≥0.4 (minimum)).
Transport of FMRP and target mRNAs affects the length and number of dendritic protrusions
Since the loss of FMRP caused pronounced defects in the association of Kif5 with mRNAs important for synapse function and their localization to dendrites, we investigated whether the dendritic protrusion phenotype of FXS was directly affected by target mRNA transport. To test the physiological effect of acute loss of FMRP transport on dendritic morphology in situ, we established a dominant negative approach by overexpression of the KLC-binding C-terminal domain of FMRP (D3) (see Fig. 3c), which can compete with endogenous FMRP for KLC binding. Neurons were transfected with either the full-length (FL) FMRP (control) or the D3 construct, together with trace amounts of FL-FMRP-GFP driven by the endogenous promoter as a marker for dendritic transport. Since FL-FMRP can compete with FMRP-GFP for binding to kinesin, we determined a ratio of construct stoichiometry that demonstrated significant FMRP-GFP dendritic localization (it is also likely that multimerization of FMRP diminishes this potentially competitive effect). Compared to the FL protein, overexpression of D3 caused a significant reduction (∼7-fold) of FMRP-GFP transport into dendrites (Fig. 5a, p<0.002, n=23), while controls (FL-FMRP; n=23) or overexpression of the N-terminal (aa1-208) or central domain (aa209-386) of FMRP (see Fig. 3c) caused no significant differences (Suppl. Fig. 6a, p>0.2, n=15-19). Under these conditions we have verified that the expression of both constructs were not significantly different (Suppl. Fig. 6b). To analyze endogenous FMRP similarly, we utilized a monoclonal antibody that recognizes a region outside of the C-terminus (D3-FMRP). Immunofluorescence staining of D3-FMRP neurons (with CFP as marker) showed diminished dendritic FMRP staining compared to control neurons (FL-FMRP) (Fig. 5b, p<0.001, n=15-22). We believe that blocking FMRP transport did not significantly affect other mRNA binding proteins since the overall staining pattern for RNA was not grossly altered (Fig. 5b). An antibody to the C-terminus of FMRP showed that transfection of D3-FMRP constructs resulted in a ∼5-fold increase in expression levels in both the cell body (n=9, p<0.001) and dendrites (n=18, p<0.0005) compared with endogenous FMRP in untransfected neurons (Suppl. Fig. 6c), confirming its ability to act in a dominant manner.
Fig. 5. FMRP and target mRNA transport regulates the length and number of dendritic filopodia-spine protrusions.
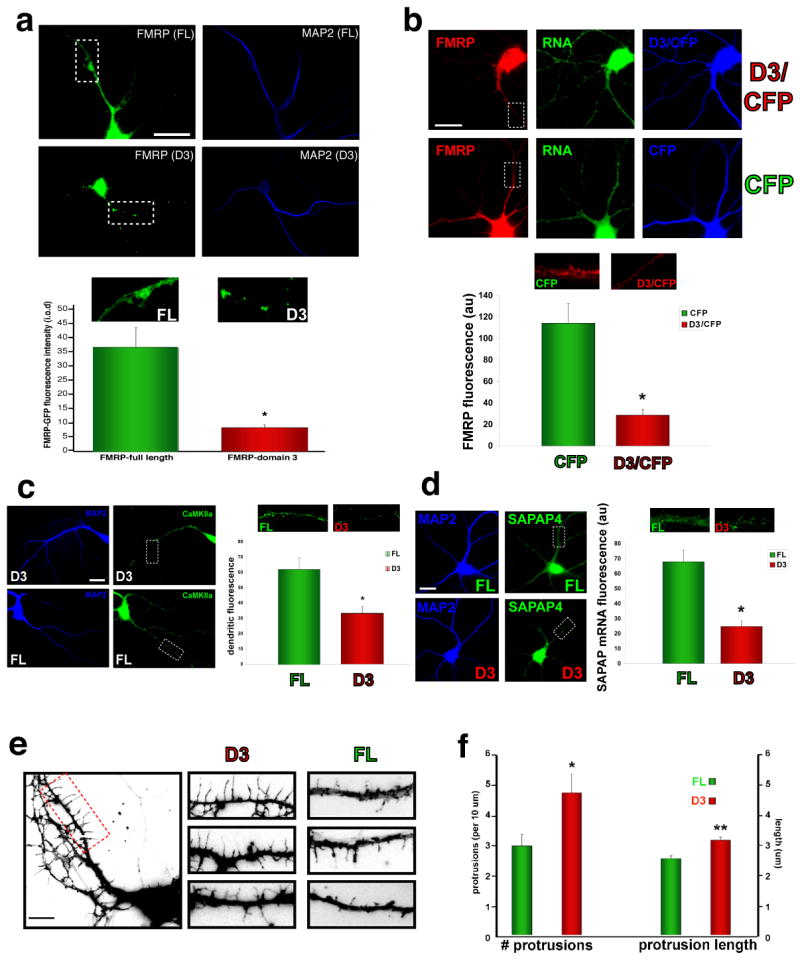
(a, b) Overexpression of the C-terminal domain (D3) of FMRP in hippocampal neurons.
(a) Cultured neurons (11 DIV) were transfected with constructs bearing either FL-FMRP (upper panels) or the C-terminal domain (D3, lower panels) as FLAG-tagged proteins along with FMRP-GFP as a reporter for dendritic transport. GFP fluorescence (left panels) and MAP2 staining (right panels) are shown for representative images. The histogram (below) indicates the average fluorescence intensities of FMRP-GFP in dendrites of treated cells (n=20-23; *p<0.002), with images showing blow-up regions of corresponding dendritic FMRP-GPF outlined in the whole cell images (above). Scale bar = 15uM.
(b) Staining of endogenous FMRP after overexpression of D3-FMRP (with CFP as marker). Neurons were stained for endogenous FMRP (red images; using an antibody that does not recognize domain 3 (D3) of FMRP), total RNA (green images; SYTO-Select), and co-transfected CFP is shown (blue). FMRP IF was quantified in dendrites (histogram, below) for both D3 neurons (upper panels, D3/CFP) and control transfected neurons (lower panels, CFP). (n=15-22; *p<0.001). Blow-up images above the histogram bars show the corresponding outlined regions of the whole cell images (above). Scale bar=15um.
(c) Dendritic CaMKIIa mRNA localization in response to DHPG (11DIV). Representative images are shown (left) for MAP2 staining (blue) and CaMKIIa mRNA FISH (green) for both D3 (upper panels) and FL FMRP (lower panels) overexpression. Histogram (right) shows quantification of dendritic fluorescence of CaMKIIa (n=13; *p<0.01), with blow-up images above bars showing dendritic regions outlined in the whole cell images (above). Scale bar=10um.
(d) Dendritic SAPAP4 mRNA localization in response to DHPG (11DIV). Representative images are shown (left) for MAP2 staining (blue) and SAPAP4 mRNA FISH (green) for both D3 (lower panels) and FL FMRP (upper panels) overexpression. Histogram (right) shows quantification of dendritic fluorescence of SAPAP4 (n=14-15; *p<0.001), with blow-up images above bars showing outlined dendritic regions outlined in whole cell images (above). Scale bar=10um.
(e, f) Dendritic filopodia-spine protrusion analysis in response to DHPG. (e) Actin-CFP staining (10 DIV) co-transfected with either D3 or FL FMRP highlights dendritic filopodia-spine protrusion morphology. A representative neuron with D3 overexpression is shown at left, with blow-up panels from representative dendrites at right for both treatments. Scale bar=5um. (f) Histogram showing quantification of dendritic filopodia-spine protrusion number and lengths in D3 or FL FMRP neurons (n=749-852 protrusions measured; *p<0.005 for # protrusions; **p<0.0001 for protrusion length).
Having established a dominant negative transport assay, we used this approach to assess the effect on dendritic mRNA localization and in turn, spine morphology. The abundance of FMRP target mRNAs was assessed by FISH after activation of mGluR. Examination of transfected neurons showed that CaMKIIa mRNA was reduced in D3-FMRP dendrites by approximately half compared to controls (FL-FMRP) after only 36 hours post-transfection (Fig. 5c, p<0.01, n=13). This pattern was similar for another FMRP target mRNA that was reduced almost 3-fold in DHPG-stimulated localization in KO neurons, SAPAP4 (Fig. 5d, p<0.001, n=14-15). However neither MAP2 (not an FMRP mRNA target), nor Arc (not a well-established mRNA target) were significantly affected by overexpression of D3-FMRP (Suppl. Fig. 6d). In all cases we verified the transfection of FL- and D3-FMRP and determined that expression levels were not significantly different between treatments on average (Suppl. Fig. 6e, f).
To examine the effect of decreased target mRNA transport on dendritic spine-like protrusion morphology, D3-FMRP was transfected into WT neurons for 36 hours and stimulated with DHPG. Morphology was monitored by co-transfection of trace amounts of CFP-beta-actin, and neurons were processed for IF staining of MAP2 (not shown) to unequivocally identify filopodia and spine protrusions. The morphological similarity between filopodia and spines at this stage made it difficult to distinguish the two simply based on morphology. Therefore protrusions having a long and thin morphology were designated as filopodia-spines as has been described previously (Prange and Murphy, 2001), and represent the major protrusion type during this developmental stage of cultured neurons (Bonhoeffer and Yuste, 2002). Surprisingly, D3-overexpression caused a significant increase in both the length (p<0.0001, n=749-852) and number (p<0.005) of filopodia-spine protrusions compared to control (FL-FMRP) transfected neurons (Fig. 5e, f). The average number of protrusions increased from 3.0 to 4.8/10um dendrite length in D3-FMRP neurons (60%, left side of histogram in Fig. 5f), while their average length increased from 2.5um in controls to 3.2um (28%, right side of histogram). Under these conditions there was no significant alteration in the total number of synapses in either group as assessed by synapsin-positive dendritic puncta (Suppl. Fig. 6g, p>0.35, n=12-14). The effect of reduced FMRP transport on filopodia-spine morphology in WT neurons was strikingly similar to that observed in neurons lacking FMRP, and suggest a mechanistic link between decreases in dendritic target mRNAs important for synaptic function and the dynamics of dendritic protrusions.
Discussion
We have uncovered an aspect of mRNA trafficking into dendrites upon mGluR stimulation that is deficient in neurons lacking FMRP. The mechanism by which this occurs indicates that the C-terminal domain of FMRP can associate with KLC in a dynamic fashion regulated by mGluR activation. These data support a model by which FMRP acts as a molecular adapter to bind mRNA targets and suppress their translation during kinesin-mediated transport to synaptic sites. In support of this model, a number of mRNA targets are diminished in their association with Kif5 isolated from KO brains. Further work is needed to determine whether FMRP or its partners directly bind KLC and whether these interactions are regulated by kinase/phosphatase signaling pathways downstream of mGluR activation. The stimulus-induced delivery of FMRP mRNPs may play a critical role in protein synthesis-dependent mGluR-mediated LTD, and deficits in mRNA transport may contribute to the LTD and/or spine phenotypes in KO neurons (Penagarikano et al., 2007). Indeed, we found that a relatively brief suppression of FMRP and target mRNA transport into dendrites caused an alteration of dendritic filopodia-spine protrusions during a period of active synaptogenesis.
Impaired motor-dependent delivery of dendritic mRNAs in the absence of FMRP
In the absence of FMRP we discovered marked alterations in activity-induced localization of a subset of mRNAs in neurons. To our knowledge, this study represents the first characterization of an mRNA localization defect in neurons from a mouse knockout for a specific RBP. A similar effect was reported for beta-actin mRNA in dendrites after knockdown of either ZBP1 or Staufen2 in neuron (Eom et al., 2003; Goetze et al., 2006). Our use of MS2-GFP tethered to the CaMKIIa 3′UTR, coupled with an automated particle tracking program, represents a significant advance in the ability to monitor mRNA dynamics in living neurons. In support of the FISH studies, the biophysical data solidify the finding of altered mRNA trafficking in dendrites of KO neurons in response to activation of mGluR. CaMKIIa is a particularly good example of a dendritic mRNA that associates with FMRP(Muddashetty et al., 2007; Zalfa et al., 2003), and its 3′UTR is sufficient for regulated dendritic trafficking(Rook et al., 2000). Previous reports implicate CaMKIIa protein function in the attenuation of DHPG-induced LTD in the CA1 region of hippocampus(Schnabel et al., 1999), and perhaps failed delivery of CaMKIIa mRNA into dendrites contributes to exaggerated LTD observed in the KO mouse. Interestingly, a mouse KO of the CaMKIIa mRBP translin exhibits defects in learning and memory and increased epilepsy similar to the KO mouse (Stein et al., 2006). Translin may function in the constitutive pathway for CaMKIIa mRNA localization that contributes to basal levels of CaMKIIa in dendrites since its disruption affects unstimulated neurons (Severt et al., 1999), whereas we observe no reduction in constitutively localized CaMKIIa mRNA in KO neurons, as previously reported (Steward et al., 1998). In addition, diminished synaptic CaMKIIa protein contributes to learning deficits and epilepsy in a mouse model of Angelman syndrome (Weeber et al., 2003), providing yet a further example that CaMKIIa may be a common target among diverse autism spectrum disorders.
Mislocalized FMRP target mRNAs have direct implications for altered synapse structure and signaling, such as MAP1b(Davidkova and Carroll, 2007) which can modify AMPA-R endocytosis and RGS5, which belongs to a family of G-protein regulators implicated in hyperactive mGluR signaling(Saugstad et al., 1998). SAPAP4 mRNA has not previously been shown to localize to dendrites, and may function in establishment of early post-synaptic structures that target AMPA receptors(Bresler et al., 2001). Diminished stimulus-induced localization of GABAR-d mRNA is consistent with the decreased protein measured in KO mouse brain (D'Hulst et al., 2006) and has relevant implications for the pathology of FXS (Spigelman et al., 2002). Another proposed mRNA target of FMRP is Arc (Zalfa et al., 2003), but its localization in KO neurons was apparently not different from WT, consistent with previous reports(Steward et al., 1998). Other RBPs may mediate mRNA transport via alternate signaling pathways in the absence of FMRP (Steward et al., 1998), such as translin (Severt et al., 1999) or CPEB (Huang et al., 2002) which are both implicated in CaMKIIa mRNA localization. Thus while excess mRNA translation at basal states contributes to altered synaptic function in FXS(Muddashetty et al., 2007), an additional deficit is the impaired delivery of specific mRNAs in response to mGluR.
Although microtubule motors have been implicated in mRNP trafficking to dendrites(Kanai et al., 2004), the mechanism of regulation remains unknown. The data here support a model placing the interactions between an RBP, such as FMRP, and Kif5 downstream of a glutamate receptor. The C-terminal domain of FMRP (D3; Figs. 1, 6, and Suppl. Fig. 2) associates with Kif5 through KLC, which may be a unique interaction given the divergence of the C-terminus compared to its close homologs FXR1p and FXR2p. This region also interacts with Ran-binding protein that directly binds to KLC (Menon et al., 2004). Although KLC may be dispensable for transport of the Drosophila homolog of FMRP (dFMR) in cultured S2 cells, KLC function is essential for Kif5-mediated transport in mammalian neurons(Rahman et al., 1999), and our data extend KLC function to neuronal mRNA granule transport.
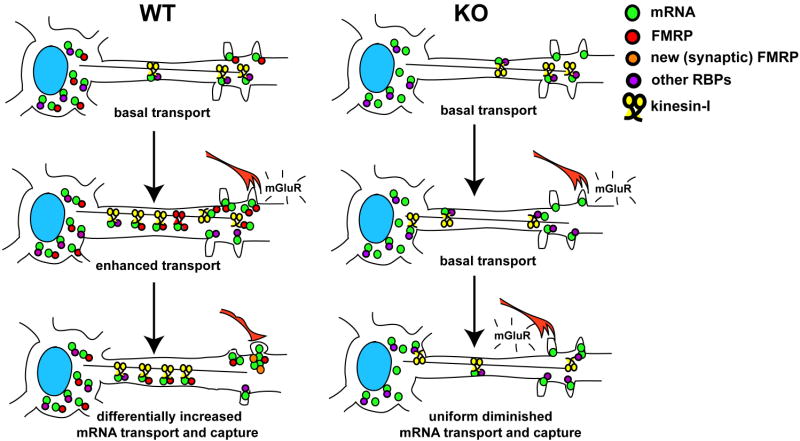
Figure 6. Model for FMRP function in linking stimulus-induced dendritic mRNA transport to local translation at synapses.
mGluR activation causes a differential accumulation and subsequent translation of mRNAs important for synapse formation and maturation during development in WT neurons; but in neurons lacking FMRP this selective transport-dependent increase in mRNAs is dysregulated.
Transport of dendritic mRNAs important for filopodial-spine dynamics
In support of a model whereby FMRP functions as a structural adapter, we found reduced association of FMRP-specific mRNAs with Kif5 in FMRP KO brain (Fig. 4). However none of the mRNAs were completely dissociated, demonstrating a role for other RBPs in this function. We speculate that defects in the regulated dynamics of mRNA transport may contribute, in part, to the spine phenotype in FXS. For example, since regulators of actin dynamics mediate spine shrinkage during LTD, RIP (p116) function in F-actin regulation through Rho (Mulder et al., 2004) may induce newly formed long filopodia or spines, while diminished targeting of RIP mRNA could lead to destabilization of actin and block spine maturation. Localized cofilin phosphatase (PP2Ac) mRNA (Castets et al., 2005) may also be critical to regulate local actin dynamics and spine morphology in response to synaptic activity.
In an attempt to link mRNA localization defects to alterations in spine morphology in FXS, we disrupted transport in WT neurons during a period of development enriched in filopodial protrusions that may be precursors to spines (Bonhoeffer and Yuste, 2002). Acute disruption of FMRP (36 hours) caused a significant increase in the length and density of dendritic filopodia-spine protrusions (Fig. 5) that was strikingly similar to FXS. Since filopodia-spines can elongate rapidly in response to the same glutamatergic stimulus that induces FMRP and target mRNA localization (Vanderklish and Edelman, 2002), our results suggest that the exaggerated morphologic response in KO neurons is directly related to mRNA delivery. We speculate that an acute loss of stimulus-induced CaMKIIa and SAPAP4 mRNA localization to synapses (Fig. 5), for example, could account for the persistence of filopodial protrusions that fail to mature into spine synapses(Jourdain et al., 2003).
FMRP links mGluR-induced dendritic mRNA transport to local translation
Taken together, our results on mGluR-regulated mRNA localization defects in KO neurons, in combination with documented defects in translation, suggest a new model that couples stimulus-induced transport to localized translation (Fig. 6). In this model, neurons under basal conditions constitutively target mRNAs that transport mostly via alternative RBPs and arrive at the synapse where they are normally repressed by FMRP, but in its absence (as in FXS) are translated precociously. Upon enhanced mGluR stimulation in WT neurons synaptic derepression of FMRP allows mRNAs to be translated while somatic FMRP becomes highly engaged in transport of additional mRNAs into dendrites. Over longer periods of time recurrent glutamatergic stimulation driving synapse formation during development may lead to preferential capture of mRNAs at certain synapses. In neurons lacking FMRP mistargeting of mRNAs may result in insufficient protein synthesis at a subset of synapses, while others may retain relatively normal or elevated levels of proteins. In turn, the inability to differentially augment target mRNA distribution may result in dysregulated synapse-specific and neuron-wide plasticity in response to stimulation. This scenario may be similar to other defects in homeostatic synaptic control that are hallmarks of related neurological syndromes, such as Rett (Dani et al., 2005). The present study has identified several key FMRP targets that are dysregulated in dendrites and may provide insight into new therapeutics for the treatment of FXS that target the mRNA localization pathway and its signaling components.
Experimental Procedures
Animals, Cell Culture, and Drug Treatments
Wild-type and KO FMR1 mice (C57Bl6) were from Jackson Labs. Hippocampal neurons from P0 mice were cultured as described(Antar et al., 2004). Cortical neurons from P0 mice were plated at a density of ∼2×106 (10cm dish). DHPG (Tocris) was added to the medium for the indicated times before fixation. For cytoskeletal disruption, either nocodazole (Sigma; 2ug/mL) or latrunculin (Sigma; 150uM) were added to media for 30 minutes before fixation. For kinesin removal from microtubules, AS-2 (gift, Dr. L. Goldstein, U.C.S.D.) was diluted (50uM final) into prewarmed (37 degrees) microtubule-stabilizing buffer (MSB; 80mM PIPES, pH 6.8, 10% glycerol, 2mM MgCl2, 1mM EGTA, 1mM GTP, 0.1% saponin) and immediately used to extract cells for 15 seconds before fixation in −20 degrees MeOH.
IF and Antibodies
IF was performed as described(Antar et al., 2004). Kinesin heavy chain (KHC; mouse) and dynein (mouse; clone 74.1) antibodies were from Chemicon. FMRP monoclonal (IC3; Chemicon) and polyclonal (H-120; Santa Cruz Biotech) antibodies were used. Antibodies to MAP2 (mouse), FLAG (rabbit, mouse) and HA (rabbit, mouse) were from Sigma. Antibodies against FXR1p (rabbit) were obtained (gift, Dr. E. Khandjian (Laval U.). Dendritic RNA was stained with SytoSelect and F-actin was detected with Alexa-488-conjugated phalloidin (Invitrogen). Secondary antibodies against mouse (Cy3) and rabbit (Cy5) were used (Jackson ImmunoResearch). Images captured in three-dimensions were taken using 0.1um Z-steps, and deconvolved using AutoQuant (Media Cybernetics). Restored images were analyzed in 3D using Imaris (Bitplane, Inc).
Fluorescence in situ hybridization (FISH)
Probes were designed using DNAsis and Oligo 4 against the 3′UTR region of mouse MAP1b and b-actin mRNA as described (Bassell et al., 1998). Coding region probes were designed to the Fmr1 mRNA as described(Antar et al., 2004). Probes were checked for specificity using BLAST on NCBI. Amino-modified oligos were made on a DNA synthesizer and labeled directly with Cy3 or with digoxigenin succinimide ester(Bassell et al., 1998). Some probes (MAP2, RGS5, SAPAP4, GABA-A-R-delta, Arc, CaMKIIa) were designed to the 3′UTR and directly labeled with Cy3 as described(Shav-Tal et al., 2004). All in situs were carried as out as previously described (Antar et al., 2004). For indirect labeling, hybridized probes were detected by IF using a Cy3-conjugated mouse anti-dig antibody and a Cy3-conjugated anti-mouse IgG antibody (Jackson). For negative controls, sense or scrambled probes were used. For analysis of KLC-TPR on Fmr1 mRNA, 6 DIV neurons were transfected with the KLC-TPR-HA construct (or FL-KLC-HA as control) for 16 hours before fixation and FISH. For MS2 FISH experiments neurons were co-transfected with MS2-GFP and CaMKIIα 3′ UTR-MS2 at 7DIV. After 24 hours, the neurons were subjected to FISH with MS2-Cy3 probes and immunostained for endogenous FMRP.
Microtubule Co-sedimentation experiments
Embryonic brain extracts were prepared in MSB with GTP (1mM) and taxol was added (10uM) before incubation at RT for 10 minutes. Extracts were centrifuged and supernatants (SUPS) were used for co-sedimentation assays. Taxol microtubules were made as suggested (Cytoskeleton) and added (0.2mg/mL) to extracts, supplemented with either ATP (1mM), AMP-PNP (0.5mM), or hexokinase (1uM) plus glucose (1mM, ATP-depleted) and incubated at 37 degrees for 15 minutes before centrifugation through a sucrose cushion (20%) for 20 minutes at 25,000g,.
Live cell imaging of FMRP-GFP and FRAP
For live cell particle tracking, CMV-pEGFP-FMRP was transfected into cultured rat hippocampal neurons and imaged 12-16 hours later. Images were captured with a Nikon Eclipse inverted microscope with 1 sec shutter intervals using IP Lab Software (Scanalytics) and analyzed as described(Tiruchinapalli et al., 2003). For FRAP analysis, neurons were transfected with EGFP-FMRP driven by the FMR1 promoter (gift, Dr. J. Darnell, Rockefeller U.) and either FL-KLC or TPR-KLC as HA-fusion proteins (gift, Dr. B. Schnapp, O.H.S.U.) and imaged 12-16 hours later. Dendritic FMRP was subjected to FRAP analysis as described previously(Antar et al., 2004) and calculated recovery rates determined.
Neuronal Transfections and in vivo domain analysis
Neurons were transfected between 3-11 DIV using Lipofectamine 2000 as suggested (Invitrogen). For analysis of kinesin interacting domains, FLAG-FMRP constructs were co-transfected with full length-EGFP-FMRP (FMR1 promoter) at a ratio of 5:1 (FLAG:GFP constructs) to determine the effect of domain overexpression on transport into dendrites. For analysis of dendritic filopodia-spine protrusions, neurons were transfected both with CFP-beta-actin and either FMRP-D3-FLAG or FMRP-FL-FLAG at a ratio of 1:5. Neurons were subjected to IF to identify dendrites using anti-MAP2 antibody staining.
FMRP domain constructs
Domains of FMRP as FLAG-tagged fusion proteins were subcloned by PCR using primers that contained EcoRI (5′) and SalI (3′) sites in the pcDNA3.1(+) vector (Invitrogen) driven by the CMV promoter containing a FLAG-fusion at the N-terminus. FMRP domains contained amino acids 1-208 (domain 1-FMRP), 209-386 (domain 2-FMRP) and 387-615 (domain 3-FMRP).
Dendritic filopodia-spine protrusion analysis
Quantification of dendritic filopodia-spines was performed on CFP-beta-actin transfected images that were chosen randomly from a series of collected neurons (at least 13 per treatment) from coverslips obtained from at least 3 separate hippocampal cultures. After transfection of CFP-beta-actin and either D3-FMRP or FL-FMRP and fixation (36 hours), neurons were co-stained for MAP2 and synapsin to identify dendrites and synapses. Quantification of protrusion length was done using IPlab software, and measurements for total protrusion density were performed by diving the total number of protrusions by the length of total dendrite lengths measured, expressed ultimately per 10um of dendrite. All protrusions, either in contact with presynaptic terminals or not, were counted as filopodia-spines as previously described (Prange and Murphy, 2001).
Immunoprecipitations (IP) and western blotting
For FMRP domain analysis, KLC-HA and either full-length FMRP-FLAG or domains of FMRP-FLAG constructs were co-transfected into human embryonic kidney (HEK-293) cell lines for 48-72 hours, lysed in COIP buffer (50mM Tris-Cl, pH 7.3, 100mM NaCl, 2mM MgCl2, 1mM EDTA, 5% glycerol) and 1mM ATP for 20 minutes on ice, and then supernatants (SUPS) isolated by centrifugation (16,000g) for 10 minutes at 4 degrees. SUPS were IP by mouse anti-FLAG beads (Sigma) for 2-12 hours at 4 degrees. To resolve FLAG-FMRP-domains from co-migrating IgG light chains on SDS-PAGE, blots were probed with mouse anti-HA and rabbit anti-FLAG (Sigma).
Cortical neurons were either not stimulated or stimulated with DHPG for 15 minutes before solubilization in COIP buffer and subjected to centrifugation. SUPS were IP with anti-KHC antibody coupled to protein G-sepharose at 4 degrees, isolated and processed similar to FLAG proteins (above) for SDS-PAGE analysis. IPs of kinesin and FMRP from mouse brain used an established IP protocol for FMRP(Brown et al., 2001) with some modification. Two ml of lysate (w/2mM ATP) was IP with 10ug 7G1-1 (mouse anti-FMRP; gift, Dr. S. Warren, Emory U.) or 10ug anti-KHC conjugated to 60ul of protein A sepharose (Pharmacia). Total brain IP of KHC, Kif3, and dynein (anti-74.1) were performed similarly with control (non-specific IgG) antibodies. Lysates were IP for 3-12 hours and washed with excess lysis buffer. RNAse-free DNAse I (50U) was added during the last wash. Western blots used alpha-tubulin (Sigma), FLAG (Sigma; mouse, rabbit), Kif5 (Covance) and dynein IC (anti-74.1; Sigma) at 1:3000, 1:1000, 1:500 and 1:1000, respectively.
RT-PCR and Quantitative Real-Time PCR (Q-PCR)
For semi-Q RT-PCR analysis of motor-associated RNAs, the IP pellets (see kinesin IPs above) were resuspended in DEPC-H20 supplemented with RNAse inhibitor, a fraction was set aside for protein analysis (10%), and the remaining pellet was extracted using Trizol (Gibco BRL). RNA pellets were resuspended in RNAse-free H2O, supplemented with RNAse inhibitor, and then used for RT (SuperScript First Strand Synthesis, Invitrogen) coupled with PCR using specific primers for the genes indicated. For the RT step, 1.1ug RNA was used for each sample, and oligo-dT was used as primer. All amounts of RNA or cDNA were determined by UV-spec analysis (NanoDrop). For RT-PCR, 10% of the RT product (cDNA; 2uL) was used with gene-specific primers (see Suppl. Meth.) and performed as described previously(Zhang et al., 2001). For Q-PCR, 400ng of each cDNA was used for each reaction. Reactions used Taqman PCR Master Mix (Appl. Biosys.), each cDNA tested in quadruplicate in two experiments. Taqman gene probes (Appl. Biosys.) were used as suggested. Real-time analysis was performed at the AECOM DNA Facility (Appl. Biosys. 7500), and each mRNA (cycle-time) was analyzed at a threshold value (0.2) where all samples were in the linear range of amplification. Quantification of relative amounts of mRNA was performed using the ΔΔCt method, with background subtracted using values from anti-FMRP pellets in KO brain.
Quantification and image analysis
Dendritic quantification of fluorescence was performed on images taken with the same exposure times and within the same experiment using IP lab (Scanalytics) software. Total fluorescence was measured by tracing a defined region of interest (ROI) and the fluorescence normalized for area. Dendrites were chosen by DIC and traces were transferred to the corresponding fluorescence images to remove bias; dendrites segments (1-3 per neuron) were taken from regions at least 10um away from the cell body and up to 80um away from the cell body. Background (normalized for area) from regions on the coverslips outside the cell were subtracted from each dendritic measurement to attain an average dendritic intensity. 3D reconstructions and colocalizations were performed using Imaris software (Tiruchinapalli et al., 2003). Pearson's correlation coefficient was calculated using Costes' algorithm (to minimize noise contribution; ImageJ, NIH) for each dendritic segment obtained from the 3D-reconstructed images for each antigen (FMRP, Kif5, Kif2). All histograms show mean values with error bars reflecting the calculated standard error of the mean. The indicated total number of cells (n) analyzed for each experiment are shown in the figure legends, and all statistical measurements were performed using Student's t-test (unpaired). Data were derived from at least 3 separate experiments unless stated otherwise.
MS2-GFP mRNA tracking experiments
MS2-GFP was transfected into neurons along with the lacZ-plasmid containing 8 repeats of MS2-hairpin binding sites fused to the 3′UTR of CaMKIIa mRNA, as previously described (Rook et al., 2000). Briefly, 6 ug of MS2-CaMKIIa-lacZ (gift, K. Kosik, U.C.S.B.) and 2ug of MS2-GFP were transfected and neurons were imaged 12-16 hours later (see live cell imaging of FMRP-GFP). Transfected cells were identified and dendritic granules imaged for 2-minute intervals at 1.5 seconds per frame (80 frames total). Wild-type and KO neurons were imaged similarly, and movies were submitted to the Localize program (R.H.Singer Lab), which tracked particles above background using identical thresholding parameters for all cells. Mean-squared displacement was calculated and each trajectory graphed accordingly. The computer-based program Localize was used to calculate dynamics based on the algorithm as described (Thompson et al., 2002). Particles were defined as motile only if they met strict criteria of active transport based on previously calculated rates of diffusion of an MS2-GFP mRNA of similar mass (Shav-Tal et al., 2004). For our purposes, we assumed that these rates would be at least in excess of 2-fold of the diffusion rates for at least 6 frames.
Supplementary Material
Suppl. Movie 1: Time-lapse microscopic analysis of CaMKIIa reporter mRNA transport in living dendrites of neurons cultured from wild-type mouse hippocampus. WT mouse hippocampal neurons were cultured (10DIV), transfected with a GFP-MS2-CaMKIIa mRNA reporter, and exposed to DHPG (50uM) to observe mRNA movements over a two-minute interval, 80 frames with each frame captured every 1.5s. A computer-based tracking program was used to highlight and trace each mRNA particle (red) through the series. Each position of a particle is traced and maintained over the subsequent frames so that the final frame contains the trajectory of the particle displayed as a continuous trace over the history of each particle.
Suppl. Movie 2: Time-lapse microscopic analysis of CaMKIIa reporter mRNA transport in living dendrites of neurons cultured from Fmr1-KO mouse hippocampus. FMRP-KO mouse hippocampal neurons were cultured (10DIV), transfected with a GFP-MS2-CaMKIIa mRNA reporter, and exposed to DHPG (50uM) to observe mRNA movements over a two-minute interval, 80 frames with each frame captured every 1.5s. A computer-based tracking program was used to highlight and trace each mRNA particle (red) through the series. Each position of a particle is traced and maintained over the subsequent frames so that the final frame contains the trajectory of the particle displayed as a continuous trace over the history of each particle.
Suppl. Movie 3: FMRP-GFP movements in dendrites of hippocampal neurons grown (10 DIV) in culture. FMRP-GFP was transfected and neurons imaged to capture fast movements of particles. Several particles are shown to move (arrows) within the dendrite in both anterograde (right) and retrograde (left) directions. Some particles move first in one direction, then stop and oscillate, and then move again in either the same direction or switch directions. The movie is inverted to better visualize particles on the background.
Acknowledgments
We thank Dan Larson for help with Localize and single mRNA particle analysis, Shailesh Shenoy for help with microscopy and image analysis, Ines Petersen for FMRP domain analysis in KLC binding, Daniel Cuzzone for technical help and Chanxia Li for Fmr1 KO cultures. This work was supported by FRAXA (J.B.D., L.N.A. and G.J.B.), a NFXF Basic Science Grant to J. Dictenberg, NIH AR-41480 to R.H. Singer, and NIH NS051127 and Dana Foundation to G.J. Bassell. J.B. Dictenberg is a recipient of the Albert Einstein Scholar Award.
Abbreviations
- FMRP
fragile X mental retardation protein
- FXS
fragile X syndrome
- mRNP
messenger ribonucleoprotein
- RBP
RNA-binding protein
- mGluR
metabotropic glutamate receptor
- KHC
kinesin heavy chain
- KLC
kinesin light chain
- WT
wild-type
- KO
knockout
- FRAP
fluorescence recovery after photobleaching
- Kif
kinesin superfamily protein
- IP
immunoprecipitation
- IF
immunofluorescence
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Antar LN, Afroz R, Dictenberg JB, Carroll RC, Bassell GJ. Metabotropic glutamate receptor activation regulates fragile x mental retardation protein and FMR1 mRNA localization differentially in dendrites and at synapses. J Neurosci. 2004;24:2648–2655. doi: 10.1523/JNEUROSCI.0099-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antar LN, Dictenberg JB, Plociniak M, Afroz R, Bassell GJ. Localization of FMRP-associated mRNA granules and requirement of microtubules for activity-dependent trafficking in hippocampal neurons. Genes Brain Behav. 2005;4:350–359. doi: 10.1111/j.1601-183X.2005.00128.x. [DOI] [PubMed] [Google Scholar]
- Aschrafi A, Cunningham BA, Edelman GM, Vanderklish PW. The fragile X mental retardation protein and group I metabotropic glutamate receptors regulate levels of mRNA granules in brain. Proc Natl Acad Sci U S A. 2005;102:2180–2185. doi: 10.1073/pnas.0409803102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagni C, Greenough WT. From mRNP trafficking to spine dysmorphogenesis: the roots of fragile X syndrome. Nat Rev Neurosci. 2005;6:376–387. doi: 10.1038/nrn1667. [DOI] [PubMed] [Google Scholar]
- Bassell GJ, Zhang H, Byrd AL, Femino AM, Singer RH, Taneja KL, Lifshitz LM, Herman IM, Kosik KS. Sorting of beta-actin mRNA and protein to neurites and growth cones in culture. J Neurosci. 1998;18:251–265. doi: 10.1523/JNEUROSCI.18-01-00251.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonhoeffer T, Yuste R. Spine motility. Phenomenology, mechanisms, and function. Neuron. 2002;35:1019–1027. doi: 10.1016/s0896-6273(02)00906-6. [DOI] [PubMed] [Google Scholar]
- Bramham CR, Wells DG. Dendritic mRNA: transport, translation and function. Nat Rev Neurosci. 2007;8:776–789. doi: 10.1038/nrn2150. [DOI] [PubMed] [Google Scholar]
- Bresler T, Ramati Y, Zamorano PL, Zhai R, Garner CC, Ziv NE. The dynamics of SAP90/PSD-95 recruitment to new synaptic junctions. Mol Cell Neurosci. 2001;18:149–167. doi: 10.1006/mcne.2001.1012. [DOI] [PubMed] [Google Scholar]
- Brown V, Jin P, Ceman S, Darnell JC, O'Donnell WT, Tenenbaum SA, Jin X, Feng Y, Wilkinson KD, Keene JD, et al. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell. 2001;107:477–487. doi: 10.1016/s0092-8674(01)00568-2. [DOI] [PubMed] [Google Scholar]
- Carrington WA, Lynch RM, Moore ED, Isenberg G, Fogarty KE, Fay FS. Superresolution three-dimensional images of fluorescence in cells with minimal light exposure. Science. 1995;268:1483–1487. doi: 10.1126/science.7770772. [DOI] [PubMed] [Google Scholar]
- Castets M, Schaeffer C, Bechara E, Schenck A, Khandjian EW, Luche S, Moine H, Rabilloud T, Mandel JL, Bardoni B. FMRP interferes with the Rac1 pathway and controls actin cytoskeleton dynamics in murine fibroblasts. Hum Mol Genet. 2005;14:835–844. doi: 10.1093/hmg/ddi077. [DOI] [PubMed] [Google Scholar]
- Ceman S, Brown V, Warren ST. Isolation of an FMRP-associated messenger ribonucleoprotein particle and identification of nucleolin and the fragile X-related proteins as components of the complex. Mol Cell Biol. 1999;19:7925–7932. doi: 10.1128/mcb.19.12.7925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Hulst C, De Geest N, Reeve SP, Van Dam D, De Deyn PP, Hassan BA, Kooy RF. Decreased expression of the GABAA receptor in fragile X syndrome. Brain Res. 2006;1121:238–245. doi: 10.1016/j.brainres.2006.08.115. [DOI] [PubMed] [Google Scholar]
- Dani VS, Chang Q, Maffei A, Turrigiano GG, Jaenisch R, Nelson SB. Reduced cortical activity due to a shift in the balance between excitation and inhibition in a mouse model of Rett syndrome. Proc Natl Acad Sci U S A. 2005;102:12560–12565. doi: 10.1073/pnas.0506071102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidkova G, Carroll RC. Characterization of the role of microtubule-associated protein 1B in metabotropic glutamate receptor-mediated endocytosis of AMPA receptors in hippocampus. J Neurosci. 2007;27:13273–13278. doi: 10.1523/JNEUROSCI.3334-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dictenberg JB, Singer RH. Dendritic RNA transport: dynamic spatio-temporal control of neuronal gene expression. In: Squire LR, editor. Encyclopedia of Neuroscience. Oxford: Academic Press; 2008. [Google Scholar]
- Eom T, Antar LN, Singer RH, Bassell GJ. Localization of a beta-actin messenger ribonucleoprotein complex with zipcode-binding protein modulates the density of dendritic filopodia and filopodial synapses. J Neurosci. 2003;23:10433–10444. doi: 10.1523/JNEUROSCI.23-32-10433.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetze B, Tuebing F, Xie Y, Dorostkar MM, Thomas S, Pehl U, Boehm S, Macchi P, Kiebler MA. The brain-specific double-stranded RNA-binding protein Staufen2 is required for dendritic spine morphogenesis. J Cell Biol. 2006;172:221–231. doi: 10.1083/jcb.200509035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold SJ, Ni YG, Dohlman HG, Nestler EJ. Regulators of G-protein signaling (RGS) proteins: region-specific expression of nine subtypes in rat brain. J Neurosci. 1997;17:8024–8037. doi: 10.1523/JNEUROSCI.17-20-08024.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YS, Jung MY, Sarkissian M, Richter JD. N-methyl-D-aspartate receptor signaling results in Aurora kinase-catalyzed CPEB phosphorylation and alpha CaMKII mRNA polyadenylation at synapses. Embo J. 2002;21:2139–2148. doi: 10.1093/emboj/21.9.2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jourdain P, Fukunaga K, Muller D. Calcium/calmodulin-dependent protein kinase II contributes to activity-dependent filopodia growth and spine formation. J Neurosci. 2003;23:10645–10649. doi: 10.1523/JNEUROSCI.23-33-10645.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai Y, Dohmae N, Hirokawa N. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron. 2004;43:513–525. doi: 10.1016/j.neuron.2004.07.022. [DOI] [PubMed] [Google Scholar]
- Kiebler MA, Bassell GJ. Neuronal RNA granules: movers and makers. Neuron. 2006;51:685–690. doi: 10.1016/j.neuron.2006.08.021. [DOI] [PubMed] [Google Scholar]
- Lu R, Wang H, Liang Z, Ku L, O'Donnell WT, Li W, Warren ST, Feng Y. The fragile X protein controls microtubule-associated protein 1B translation and microtubule stability in brain neuron development. Proc Natl Acad Sci U S A. 2004;101:15201–15206. doi: 10.1073/pnas.0404995101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon RP, Gibson TJ, Pastore A. The C terminus of fragile X mental retardation protein interacts with the multi-domain Ran-binding protein in the microtubule-organising centre. J Mol Biol. 2004;343:43–53. doi: 10.1016/j.jmb.2004.08.024. [DOI] [PubMed] [Google Scholar]
- Miller S, Yasuda M, Coats JK, Jones Y, Martone ME, Mayford M. Disruption of dendritic translation of CaMKIIalpha impairs stabilization of synaptic plasticity and memory consolidation. Neuron. 2002;36:507–519. doi: 10.1016/s0896-6273(02)00978-9. [DOI] [PubMed] [Google Scholar]
- Miyashiro KY, Beckel-Mitchener A, Purk TP, Becker KG, Barret T, Liu L, Carbonetto S, Weiler IJ, Greenough WT, Eberwine J. RNA cargoes associating with FMRP reveal deficits in cellular functioning in Fmr1 null mice. Neuron. 2003;37:417–431. doi: 10.1016/s0896-6273(03)00034-5. [DOI] [PubMed] [Google Scholar]
- Muddashetty RS, Kelic S, Gross C, Xu M, Bassell GJ. Dysregulated metabotropic glutamate receptor-dependent translation of AMPA receptor and postsynaptic density-95 mRNAs at synapses in a mouse model of fragile X syndrome. J Neurosci. 2007;27:5338–5348. doi: 10.1523/JNEUROSCI.0937-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder J, Ariaens A, van den Boomen D, Moolenaar WH. p116Rip targets myosin phosphatase to the actin cytoskeleton and is essential for RhoA/ROCK-regulated neuritogenesis. Mol Biol Cell. 2004;15:5516–5527. doi: 10.1091/mbc.E04-04-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penagarikano O, Mulle JG, Warren ST. The pathophysiology of fragile x syndrome. Annual review of genomics and human genetics. 2007;8:109–129. doi: 10.1146/annurev.genom.8.080706.092249. [DOI] [PubMed] [Google Scholar]
- Prange O, Murphy TH. Modular transport of postsynaptic density-95 clusters and association with stable spine precursors during early development of cortical neurons. J Neurosci. 2001;21:9325–9333. doi: 10.1523/JNEUROSCI.21-23-09325.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman A, Kamal A, Roberts EA, Goldstein LS. Defective kinesin heavy chain behavior in mouse kinesin light chain mutants. J Cell Biol. 1999;146:1277–1288. doi: 10.1083/jcb.146.6.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rook MS, Lu M, Kosik KS. CaMKIIalpha 3′ untranslated region-directed mRNA translocation in living neurons: visualization by GFP linkage. J Neurosci. 2000;20:6385–6393. doi: 10.1523/JNEUROSCI.20-17-06385.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakowicz R, Berdelis MS, Ray K, Blackburn CL, Hopmann C, Faulkner DJ, Goldstein LS. A marine natural product inhibitor of kinesin motors. Science. 1998;280:292–295. doi: 10.1126/science.280.5361.292. [DOI] [PubMed] [Google Scholar]
- Saugstad JA, Marino MJ, Folk JA, Hepler JR, Conn PJ. RGS4 inhibits signaling by group I metabotropic glutamate receptors. J Neurosci. 1998;18:905–913. doi: 10.1523/JNEUROSCI.18-03-00905.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnabel R, Palmer MJ, Kilpatrick IC, Collingridge GL. A CaMKII inhibitor, KN-62, facilitates DHPG-induced LTD in the CA1 region of the hippocampus. Neuropharmacology. 1999;38:605–608. doi: 10.1016/s0028-3908(98)00229-9. [DOI] [PubMed] [Google Scholar]
- Severt WL, Biber TU, Wu X, Hecht NB, DeLorenzo RJ, Jakoi ER. The suppression of testis-brain RNA binding protein and kinesin heavy chain disrupts mRNA sorting in dendrites. J Cell Sci. 1999;112(Pt 21):3691–3702. doi: 10.1242/jcs.112.21.3691. [DOI] [PubMed] [Google Scholar]
- Shav-Tal Y, Darzacq X, Shenoy SM, Fusco D, Janicki SM, Spector DL, Singer RH. Dynamics of single mRNPs in nuclei of living cells. Science. 2004;304:1797–1800. doi: 10.1126/science.1099754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spigelman I, Li Z, Banerjee PK, Mihalek RM, Homanics GE, Olsen RW. Behavior and physiology of mice lacking the GABAA-receptor delta subunit. Epilepsia. 2002;43 5:3–8. doi: 10.1046/j.1528-1157.43.s.5.8.x. [DOI] [PubMed] [Google Scholar]
- Stein JM, Bergman W, Fang Y, Davison L, Brensinger C, Robinson MB, Hecht NB, Abel T. Behavioral and neurochemical alterations in mice lacking the RNA-binding protein translin. J Neurosci. 2006;26:2184–2196. doi: 10.1523/JNEUROSCI.4437-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steward O, Bakker CE, Willems PJ, Oostra BA. No evidence for disruption of normal patterns of mRNA localization in dendrites or dendritic transport of recently synthesized mRNA in FMR1 knockout mice, a model for human fragile-X mental retardation syndrome. Neuroreport. 1998;9:477–481. doi: 10.1097/00001756-199802160-00022. [DOI] [PubMed] [Google Scholar]
- Takeuchi M, Hata Y, Hirao K, Toyoda A, Irie M, Takai Y. SAPAPs. A family of PSD-95/SAP90-associated proteins localized at postsynaptic density. J Biol Chem. 1997;272:11943–11951. doi: 10.1074/jbc.272.18.11943. [DOI] [PubMed] [Google Scholar]
- Thompson RE, Larson DR, Webb WW. Precise nanometer localization analysis for individual fluorescent probes. Biophysical journal. 2002;82:2775–2783. doi: 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiruchinapalli DM, Oleynikov Y, Kelic S, Shenoy SM, Hartley A, Stanton PK, Singer RH, Bassell GJ. Activity-dependent trafficking and dynamic localization of zipcode binding protein 1 and beta-actin mRNA in dendrites and spines of hippocampal neurons. J Neurosci. 2003;23:3251–3261. doi: 10.1523/JNEUROSCI.23-08-03251.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderklish PW, Edelman GM. Dendritic spines elongate after stimulation of group 1 metabotropic glutamate receptors in cultured hippocampal neurons. Proc Natl Acad Sci U S A. 2002;99:1639–1644. doi: 10.1073/pnas.032681099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderklish PW, Edelman GM. Differential translation and fragile X syndrome. Genes Brain Behav. 2005;4:360–384. doi: 10.1111/j.1601-183X.2005.00134.x. [DOI] [PubMed] [Google Scholar]
- Verhey KJ, Meyer D, Deehan R, Blenis J, Schnapp BJ, Rapoport TA, Margolis B. Cargo of kinesin identified as JIP scaffolding proteins and associated signaling molecules. J Cell Biol. 2001;152:959–970. doi: 10.1083/jcb.152.5.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeber EJ, Jiang YH, Elgersma Y, Varga AW, Carrasquillo Y, Brown SE, Christian JM, Mirnikjoo B, Silva A, Beaudet AL, et al. Derangements of hippocampal calcium/calmodulin-dependent protein kinase II in a mouse model for Angelman mental retardation syndrome. J Neurosci. 2003;23:2634–2644. doi: 10.1523/JNEUROSCI.23-07-02634.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiler IJ, Irwin SA, Klintsova AY, Spencer CM, Brazelton AD, Miyashiro K, Comery TA, Patel B, Eberwine J, Greenough WT. Fragile X mental retardation protein is translated near synapses in response to neurotransmitter activation. Proc Natl Acad Sci U S A. 1997;94:5395–5400. doi: 10.1073/pnas.94.10.5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalfa F, Eleuteri B, Dickson KS, Mercaldo V, De Rubeis S, di Penta A, Tabolacci E, Chiurazzi P, Neri G, Grant SG, et al. A new function for the fragile X mental retardation protein in regulation of PSD-95 mRNA stability. Nat Neurosci. 2007;10:578–587. doi: 10.1038/nn1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalfa F, Giorgi M, Primerano B, Moro A, Di Penta A, Reis S, Oostra B, Bagni C. The fragile X syndrome protein FMRP associates with BC1 RNA and regulates the translation of specific mRNAs at synapses. Cell. 2003;112:317–327. doi: 10.1016/s0092-8674(03)00079-5. [DOI] [PubMed] [Google Scholar]
- Zhang HL, Eom T, Oleynikov Y, Shenoy SM, Liebelt DA, Dictenberg JB, Singer RH, Bassell GJ. Neurotrophin-induced transport of a beta-actin mRNP complex increases beta-actin levels and stimulates growth cone motility. Neuron. 2001;31:261–275. doi: 10.1016/s0896-6273(01)00357-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Suppl. Movie 1: Time-lapse microscopic analysis of CaMKIIa reporter mRNA transport in living dendrites of neurons cultured from wild-type mouse hippocampus. WT mouse hippocampal neurons were cultured (10DIV), transfected with a GFP-MS2-CaMKIIa mRNA reporter, and exposed to DHPG (50uM) to observe mRNA movements over a two-minute interval, 80 frames with each frame captured every 1.5s. A computer-based tracking program was used to highlight and trace each mRNA particle (red) through the series. Each position of a particle is traced and maintained over the subsequent frames so that the final frame contains the trajectory of the particle displayed as a continuous trace over the history of each particle.
Suppl. Movie 2: Time-lapse microscopic analysis of CaMKIIa reporter mRNA transport in living dendrites of neurons cultured from Fmr1-KO mouse hippocampus. FMRP-KO mouse hippocampal neurons were cultured (10DIV), transfected with a GFP-MS2-CaMKIIa mRNA reporter, and exposed to DHPG (50uM) to observe mRNA movements over a two-minute interval, 80 frames with each frame captured every 1.5s. A computer-based tracking program was used to highlight and trace each mRNA particle (red) through the series. Each position of a particle is traced and maintained over the subsequent frames so that the final frame contains the trajectory of the particle displayed as a continuous trace over the history of each particle.
Suppl. Movie 3: FMRP-GFP movements in dendrites of hippocampal neurons grown (10 DIV) in culture. FMRP-GFP was transfected and neurons imaged to capture fast movements of particles. Several particles are shown to move (arrows) within the dendrite in both anterograde (right) and retrograde (left) directions. Some particles move first in one direction, then stop and oscillate, and then move again in either the same direction or switch directions. The movie is inverted to better visualize particles on the background.