Abstract
Carbohydrate response element-binding protein (ChREBP) is a basic helix-loop-helix/leucine zipper transcription factor that binds to the carbohydrate response element in the promoter of certain lipogenic and glycolytic genes. High glucose can activate ChREBP by releasing an intramolecular inhibition within the glucose-sensing module (GSM) that occurs in low glucose. We report here that the glucose response of GSM is mediated by cooperation between five conserved submodules known as Mondo conserved regions (MCRs) I through V within GSM. Deletion of individual MCRs leads to complete (for MCR II, III, and IV) or partial (MCR I) loss of glucose response of ChREBP. MCR IV is necessary and sufficient for inhibiting the transcriptional activity of ChREBP under low glucose. The roles of MCR II and III in glucose response of ChREBP are independent of and distinct from their function in controlling subcellular localization. We further demonstrate that, instead of inhibiting ChREBP activity as would be predicted from its cytoplasmic retentive function, 14-3-3 binding with MCR III is essential for the glucose responsiveness of ChREBP. The interaction between 14-3-3 and ChREBP is constitutive, indicating a permissive role of 14-3-3 in the glucose response of ChREBP. We further uncovered an unconventional 14-3-3 binding motif (residues 116–135) lacking phosphor-serine/threonine within MCR III, a predicted α-helix highly conserved in all Mondo proteins. We conclude that individual subdomains in the GSM (MCR I through V) play diverse but crucial roles in cooperation with essential trans-acting cofactors such as 14-3-3 proteins to mediate the glucose response of ChREBP.
HIGHER MULTICELLULAR ORGANISMS have developed some sophisticated mechanisms to cope with fluctuations in food supply. Excess dietary carbohydrates can be converted into fat, the major form of energy storage, from which energy can be released in times of need. An important player involved in this process is carbohydrate response element-binding protein (ChREBP), a basic helix-loop-helix/leucine zipper (bHLH/ZIP) transcription factor that is activated by glucose and turns on the transcription of several genes responsible for lipogenesis and glycolysis, such as acetyl-coenzyme A carboxylase (ACC), fatty acid synthase (FAS), and liver pyruvate kinase (L-PK) (1,2,3,4). ChREBP knockout mice display reduced glycolysis and lipogenesis and severe simple sugar intolerance, indicating that ChREBP plays an important role in glucose and lipid homeostasis (5).
ChREBP, also known as WBSCR14 and MondoB in different contexts (6,7), belongs to the Mondo family of transcription factors, which encompasses homologs existing in species from nematodes to mammals (7,8). A paralog of ChREBP known as MondoA is also found in vertebrates (7). Although ChREBP is highly expressed in the liver (9), MondoA is predominantly expressed in skeletal muscle (7). The transcriptional activity of most Mondo proteins can be activated by an increase in glucose concentration in the environment (10). Activation of ChREBP by glucose involves a two-step process: nuclear translocation and DNA binding through dephosphorylation of ChREBP by protein phosphatase 2A (3) and transactivation of ChREBP by glucose through a mechanism to be clarified (11). By systematic serial deletions, we found that the latter mechanism is mediated by an evolutionarily conserved domain located at the N terminus of Mondo proteins, which has been named the glucose-sensing module (GSM). The GSM is composed of a low-glucose inhibitory domain (LID) and a transactivation domain called glucose-response activation conserved element (GRACE). The glucose responsiveness of ChREBP is mediated by a dynamic intramolecular inhibition between LID and GRACE. Under low glucose concentration, the transcriptional activity of GRACE is inhibited by the LID, whereas high glucose releases the inhibition and turns on the transcriptional activity of GRACE (10). However, the exact mechanism of this intramolecular inhibition and its release by glucose stimulation is not known.
The GSM in Mondo proteins coincides with a region conserved in sequence across species from Caenorhabditis elegans to human. This region was called protein amino-terminal domain of repression (PADRE) (8) or Mondo conserved regions (MCR I through V) (7,12) in different contexts. The region has been suggested to be responsible for the cytoplasmic localization of ChREBP and MondoA through the action of a nuclear export signal (NES) and a 14-3-3 binding site, located in MCR II and III, respectively (12,13). The function of the MCRs other than conferring cytoplasmic retention to the Mondo proteins was unknown until we associated them with the glucose-stimulated gene transcription (10). Although it has been suggested that changes in subcellular localization may underlie the activation of Mondo proteins by extracellular signals (7), we found that change of nuclear abundance of ChREBP after glucose stimulation does not fully account for the activation of ChREBP by glucose, and the dynamic actions of the GSM mentioned earlier play a major role in the glucose responsiveness of ChREBP (10).
In this report, we have examined the structural determinants of glucose response in the GSM and other trans-acting factors required for activation of ChREBP by glucose. We performed serial deletions in the GSM region and evaluated the glucose response of individual deletion mutants. Here we report that MCR I through V cooperate with one another to mediate the glucose-stimulated transactivation of ChREBP, and all except MCR I are essential for glucose responsiveness. Moreover, MCR IV is necessary and sufficient for the inhibition of the transcriptional activity of ChREBP under low glucose. We further showed that MCR II is a bona fide NES; notwithstanding, its role in glucose response seems to be separate from that of regulation of subcellular localization, and its function cannot be replaced with a classic NES from the Rev protein of HIV-1. On the other hand, we found that MCR III interaction with 14-3-3 proteins is essential for the glucose response of ChREBP; either mutating the 14-3-3 binding site or overexpressing a 14-3-3 antagonist abrogates the glucose response of ChREBP. We further mapped the 14-3-3 binding motif in MCR III to a predicted 20-residue α-helical region. However, the function of MCR III in glucose response extends beyond 14-3-3 binding, because the region outside the 14-3-3 binding motif is found to be also essential for the glucose response. Through these studies, we have identified novel functions that can be assigned to specific regions within the GSM that mediate the glucose responsiveness of ChREBP.
RESULTS
Systematic Structure-Function Analysis of the GSM
We have previously established a sensitive reporter system for glucose activation of ChREBP based on GAL4-ChREBP fusion protein and UASGAL-driven luciferase reporter in 832/13 insulinoma cells (10). Using this system, we uncovered at the N terminus of ChREBP a structure called GSM, which is both necessary and sufficient for executing the glucose response of ChREBP. GSM encompasses a regulatory domain called LID and a transactivation domain called GRACE. The glucose response of ChREBP is mediated by a reversible intramolecular inhibition of the transactivation activity of GRACE by LID. The mechanism that underlies the reversible inhibition, however, remains unclear. To further dissect the processes involved in the glucose response, we performed a detailed structure-function analysis of GSM.
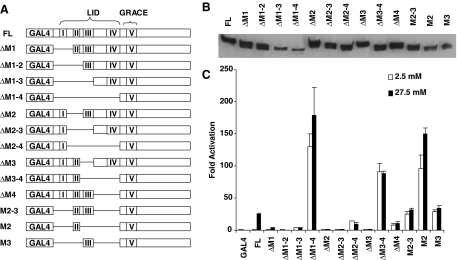
GSM coincides in location with the evolutionarily conserved MCRs I through V, which are separated by nonconserved sequences (see Ref. 10 for alignment), making it possible to perform fine mapping of individual MCRs without disrupting these functional units. We showed that the transactivation domain GRACE overlaps MCR V, whereas the regulatory domain LID encompasses MCRs I–IV. Because we have previously demonstrated that GRACE is a transactivation domain essential for the glucose response of ChREBP (10), we focused our analysis on LID, i.e. MCRs I–IV. We made a series of GAL4-ChREBP deletion mutants in which we truncated the LID at different positions involving either its N- or C-terminal region or both (Fig. 1A). After we transfected these mutant constructs into 832/13 cells, the respective mutant proteins were expressed with the expected molecular weights at comparable levels (see immunoblot, Fig. 1B). We then tested the ability of these mutants to activate the UASGAL-driven luciferase reporter. A normal glucose response of wild-type ChREBP consists of an inhibition of transactivation activity under low glucose and the release of the inhibition under high glucose. When we examined these parameters in individual mutants, we found that all deletion mutants with intact MCR IV, including one missing MCR I–III in the LID with only MCR IV remaining (ΔM1–3), suppress the transactivation activity; and all mutants missing MCR IV (ΔM1–4, ΔM2–4, ΔM3–4, ΔM4, M2–3, M2, and M3) exhibit significantly enhanced basal activity (Fig. 1C). These data suggested that MCR IV is sufficient and necessary for the inhibition of the transcriptional activity of ChREBP under low glucose and may be the key module involved in this process. Other MCRs seem to also contribute to the inhibition, as can be seen in the case of ΔM4, where the transcriptional activity was still markedly lower than that in ΔM1–4, which has the entire LID region removed (Fig. 1C). We note that the physical location of MCR IV immediately next to the transactivation domain (GRACE or MCR V) makes MCR IV well suited to perform this switch-like function in response to glucose stimulation (Fig. 1A).
Figure 1.
Structure-Function Analysis of the GSM
A, Schematic illustration of the deletion constructs (not to scale). Each MCR is labeled as I through V, and regions corresponding to LID and GRACE are bracketed. B, Expression of above constructs after being transfected into 832/13 cells for 24 h as examined by Western blot using anti-c-myc antibody. C, Luciferase assay. pG5-luc and pRL-TK were cotransfected with indicated plasmids into 832/13 cells. Transfected cells were treated with 2.5 mm (white bar) or 27.5 mm (black bar) glucose for 24 h.
On the other hand, with the exception of MCR I deletion (ΔM1), which retained a meaningful glucose response (∼3-fold compared with ∼25-fold response in the full-length construct), all deletion mutants lost their response to glucose (Fig. 1C). Although some minor variation was occasionally seen in mutants with extremely high background, e.g. M2, the minimal response (∼0.5-fold) was likely artifactual. Even if we assumed that such a minor response was real, given the high basal activity, the mechanism would be very different from that mediated by GSM, which works by release of transactivation repression from a minimal level under low glucose. These data suggest that all MCRs play important roles in eliciting the glucose response, probably by removing the inhibition mediated by MCR IV in the GSM.
MCR II and III Mediate Glucose Responsiveness of ChREBP through Mechanisms Independent of Subcellular Translocation
Before they were found to be associated with the glucose response of ChREBP (10), the function of MCRs had been attributed to be the regulation of the subcellular localization of Mondo proteins through a NES in MCR II and a 14-3-3 binding site in MCR III (12,13). Both Chromosome Region Maintenance 1 (CRM1)-mediated nuclear export through the NES and cytoplasmic 14-3-3 binding have been thought to contribute to the cytoplasmic localization of MondoA and ChREBP (12,13). It was suggested that extracellular signals may activate MondoA by inactivating the cytoplasmic retentive function of MCRs, allowing the molecule to translocate into the nucleus (7,12). Therefore, deletion of MCR II or III would be expected to lead to nuclear accumulation and transcriptional activation of ChREBP if the sole function of these MCRs is cytoplasmic retention. On the contrary, however, we found that deletion of MCR II and/or III leads to complete silencing of the transactivation activity of ChREBP under both low and high glucose concentrations (Fig. 1C). Although this observation is contrary to previous predictions (7,12), it is consistent with our previous data that activation of ChREBP by glucose is largely independent of changes in subcellular localization (10).
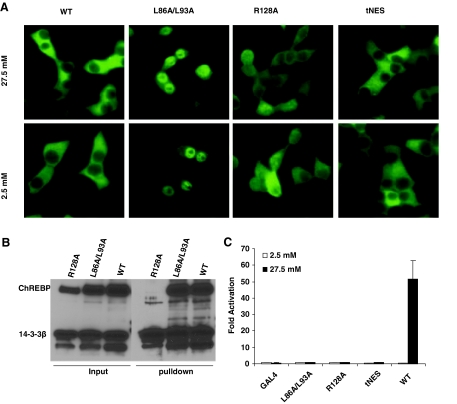
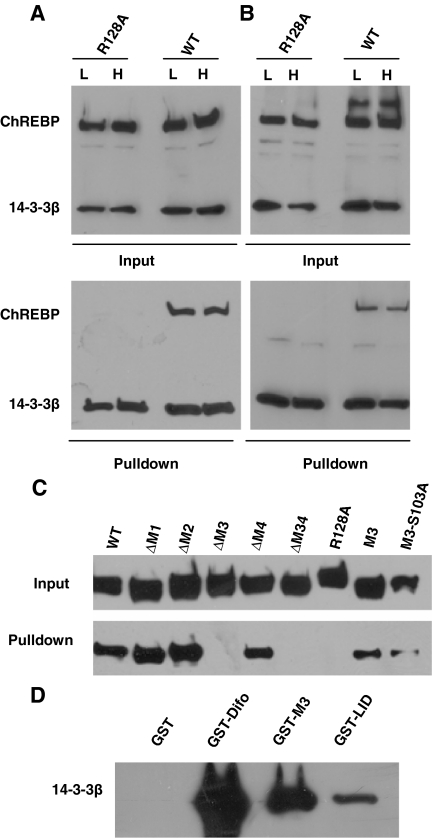
To further understand the role of MCR II and III in response to high glucose, we generated point mutations in the NES (L86A/L93A) in MCR II or the 14-3-3 binding site (R128A) in MCR III, analogous to similar mutations reported earlier in MondoA (12) and ChREBP (13), to disable their putative individual functions. To assess whether these mutations affect the subcellular localization of ChREBP in 832/13 cells, we generated 832/13 cells stably expressing c-myc-tagged ChREBP or its L86A/L93A or R128A mutants by retroviral infection. Immunofluorescence indicated that, consistent with previous results, wild-type ChREBP was located mainly in the cytoplasm. As predicted, the L86A/L93A double mutant was located mainly in the nucleus in both low- and high-glucose conditions (Fig. 2A). The R128A mutant, which completely lost its interaction with 14-3-3β in a poly-His pull-down assay (Fig. 2B), also displayed significantly increased nuclear abundance under both low- and high-glucose concentration, as expected (Fig. 2A).
Figure 2.
Subcellular Localization and Glucose Response of ChREBP and Its Mutants
A, Immunofluorescent staining. The 832/13 cells stably expressing c-myc-tagged wild-type or indicated mutant ChREBP were treated with low (2.5 mm) or high (27.5 mm) glucose for 12 h before staining with anti-c-myc antibody. B, Poly-His pull-down assay. pCHM-14-3-3β was cotransfected with plasmids expressing indicated wild-type or mutant ChREBP into 832/13 cells. Twenty-four hours after transfection, we used Western blot with anti-c-myc antibody to examine the levels of expressed proteins in the cell lysate (input) or in the precipitate after pull-down assay. C, Luciferase assay. pG5-luc and pRL-TK were cotransfected with indicated plasmids into 832/13 cells that were treated with 2.5 or 27.5 mm glucose for 24 h. WT, Wild type.
If nuclear translocation is the major mechanism for glucose-activated gene transcription, one would predict an enhanced basal (constitutive) transcriptional activity for the L86A/L93A and R128A mutants, because both exhibited enhanced nuclear abundance under low glucose. However, we found that GAL4 fusion proteins with both L86A/L93A and R128A mutants were transcriptionally silent, regardless of the glucose concentration (Fig. 2C). These data suggest that subcellular localization is not the only or major mechanism controlling the transcription activity of ChREBP, and the function of MCR II and III in the glucose response of ChREBP may be independent of their role in regulating the subcellular localization of ChREBP.
To further test this hypothesis, we replaced residues 88–99 in ChREBP, which overlap MCR II, with a classic NES from the HIV-1 Rev protein to generate mutant tNES. This mutant was labeled with a c-myc tag and stably expressed in 832/13 cells via retroviral infection, and its subcellular localization was determined by immunofluorescence staining (Fig. 2A). As compared with the substantial nuclear level of L86A/L93A and R128A mutants, tNES replacement largely restored the predominantly cytoplasmic localization of ChREBP. The GAL4 fusion protein with this tNES mutant was also transcriptionally inactive under either low or high glucose concentration in the luciferase assay (Fig. 2C). Therefore, even though the cytoplasmic localization of ChREBP can be restored by a classic NES, it failed to restore the glucose response of ChREBP. These data indicate that the function of MCR II in glucose-induced transcription activation is distinct from its role as a NES controlling subcellular localization, and change in subcellular localization is not the major mechanism whereby glucose regulates transcriptional activity of ChREBP.
As we were preparing this manuscript for publication, Davies et al. (14) presented their data on mutations in NES in ChREBP in the 2007 Experimental Biology meeting. Interestingly, they produced a number of NES mutants with either abrogated or intact response to high glucose, none of which displayed the predicted constitutive activation of ChREBP, suggesting that nucleocytoplasmic shuttling is not essential for the glucose-responsive activation of ChREBP (14).
Interaction with 14-3-3 Proteins Is Essential for the Glucose Responsiveness of ChREBP
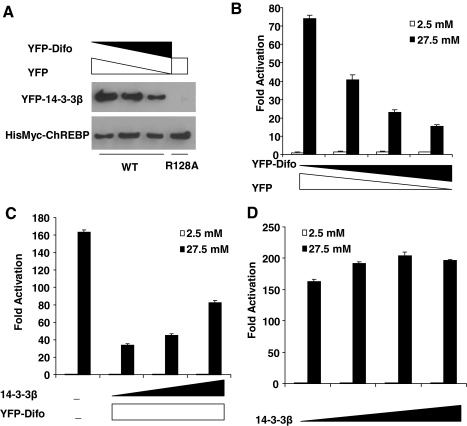
Previously, we showed that the R128A mutation that abolished interaction between 14-3-3 and ChREBP also causes loss of transcriptional activity of ChREBP despite the enrichment of this mutant in the nucleus under both low and high glucose (Fig. 2). However, the lack of glucose responsiveness is not necessarily caused by loss of 14-3-3 interaction. To examine this issue, we determined the effects of 14-3-3 silencing on the glucose responsiveness of ChREBP. Given the existence of multiple isoforms of 14-3-3 and high abundance of 14-3-3 proteins in mammalian cells (15), the common silencing tool such as RNA interference is not an option. We therefore took advantage of the availability of a 14-3-3 antagonist, difopein, which has been shown to competitively inhibit the function of 14-3-3 proteins (16). Indeed, overexpression of difopein, as a yellow fluorescent protein (YFP) fusion protein, in 832/13 cells dose-dependently decreased interaction between 14-3-3 and ChREBP (Fig. 3A). Interestingly, we found that overexpression of difopein also dose-dependently inhibited the glucose responsiveness of GAL4-ChREBP (Fig. 3B). Importantly, the suppression of glucose responsiveness of ChREBP by difopein was largely rescued by overexpression of 14-3-3β, confirming that the inhibitory effect of difopein indeed results from competitive binding to 14-3-3 proteins (Fig. 3C). In contrast, in the absence of difopein, overexpression of 14-3-3β had only a minimal effect on the glucose responsiveness of ChREBP, suggesting that under normal conditions, there are abundant 14-3-3 proteins present to support the glucose response of ChREBP (Fig. 3D).
Figure 3.
14-3-3 Proteins Are Essential for Glucose Response of ChREBP
A, Poly-His pull-down assay. pCHM-ChREBP or its R128A mutant, pYFP-14-3-3β and different doses of pYFP and pYFP-difo as indicated, were transfected into 832/13 cells. We collected the cell lysate after 24 h and performed poly-His pull-down assay. The amount of YFP-14-3-3β and ChREBP pulled down were measured with Western blot using anti-GFP and anti-c-myc antibodies, respectively. B–D, Luciferase assays. B, pG5-luc, pRL-TK, and pGAMPAC-ChREBP were cotransfected with indicated amounts of pYFP-Difo or pYFP into 832/13 cells that were treated with 2.5 or 27.5 mm glucose for 24 h. C, pG5-luc and pRL-TK were cotransfected with plasmids expressing indicated proteins into 832/13 cells that were treated with 2.5 or 27.5 mm glucose for 24 h. The total amounts of transfected DNA for each group were held the same with stuffer plasmid pcDNA3.1. D, pG5-luc and pRL-TK were cotransfected with different amounts of pCHM-14-3-3β into 832/13 cells that were treated with 2.5 or 27.5 mm glucose for 24 h. The total amounts of transfected DNA for each group were held the same with stuffer plasmid pcDNA3.1. WT, Wild type.
14-3-3 Proteins Are Important for Physiological Regulation of ChREBP Target Genes by Glucose
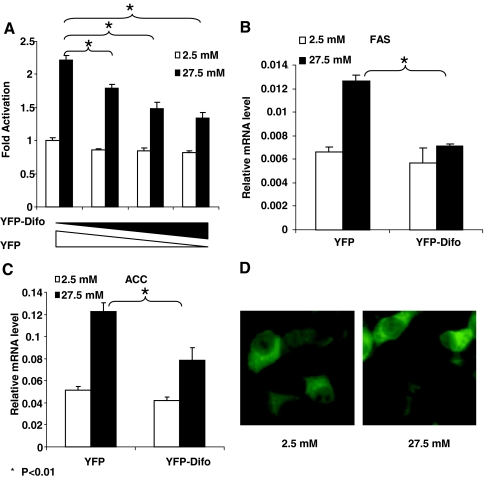
Because the above experiments were carried out with the artificial system of GAL4-ChREBP and a UAS-driven reporter, we wished to confirm our findings on the role of 14-3-3 in glucose-responsive transcription in a relatively more physiological setting. Therefore, we examined the effects of difopein overexpression in 832/13 insulinoma cells on the promoter activity of L-PK in a luciferase assay; again we found that glucose-activated transcription of L-PK promoter was dose-dependently inhibited by difopein overexpression (Fig. 4A). We further measured by quantitative RT-PCR the endogenous mRNA expression levels of two other glucose/ChREBP target genes, ACC and FAS, in response to glucose stimulation in the presence or absence of difopein. The results indicate that difopein significantly blunted the high-glucose-stimulated increase in the mRNA levels of these two genes but had no effect on the basal mRNA levels under low glucose conditions (Fig. 4, B and C). These data suggest that 14-3-3 proteins play an important role in the control of high-glucose-regulated target gene expression.
Figure 4.
The 14-3-3 Proteins Are Required for Glucose-Stimulated Expression of ChREBP Target Genes
A, Luciferase assay. pLPK-luc and pRL-TK were cotransfected with the indicated amounts of pYFP-Difo or pYFP into 832/13 cells that were treated with 2.5 or 27.5 mm glucose for 24 h. B and C, Quantitative RT-PCR. The 832/13 cells overexpressing YFP or YFP-Difo were treated with 2.5 or 27.5 mm glucose for 48 h. The levels of FAS (B) and ACC (C) were measured with real-time RT-PCR. *, P < 0.01. D, Subcellular localization of YFP-14-3-3β fusion protein. The 832/13 cells were infected with retrovirus expressing YFP-14-3-3β fusion protein and treated with 2.5 or 27.5 mm glucose for 12 h.
To further examine the role of 14-3-3 proteins in the regulation of glucose-responsive gene transcription by ChREBP, we studied the subcellular localization of YFP-14-3-3β fusion protein in 832/13 cells. Consistent with a previous report (13), the distribution pattern of 14-3-3β is very similar to that of ChREBP, i.e. mainly located in cytoplasm, and glucose has little effect on its distribution (Fig. 4D), suggesting that 14-3-3 and ChREBP colocalize and function in similar subcellular compartments. The relative abundance of 14-3-3, however, appears to be higher in nuclei compared with that of ChREBP (Fig. 4D). It is not surprising because 14-3-3 is involved in many cellular processes in mammalian cells other than ChREBP regulation.
Interaction between 14-3-3 Proteins and ChREBP Is Not Regulated by Glucose and Phosphorylation
The detailed mechanism for involvement of 14-3-3 in the glucose responsiveness of ChREBP remains to be determined. The 14-3-3 proteins have been involved in a vast number of cellular processes (15). Most ligands for 14-3-3 proteins encompass protein motifs with phosphoserine or -threonine, although some nonphosphorylated motifs have also been found to bind to 14-3-3 (15). It is a plausible hypothesis that glucose stimulation may be related to phosphorylation of ChREBP by an unknown kinase.
To test this hypothesis, we first determined whether in vivo interaction between ChREBP and 14-3-3 proteins is affected by glucose treatment. We stably expressed 6×His/c-myc-tagged 14-3-3β and c-myc-tagged wild-type ChREBP or R128A mutant via retroviral infection in 832/13 cells, treated them with low or high glucose, and used poly-His pull-down assay to analyze the interaction between ChREBP and 14-3-3β. Contrary to what is predicted by the hypothesis, robust interaction occurred between 14-3-3β and ChREBP regardless of the glucose concentration in the medium (Fig. 5A). In case the binding between ChREBP and 14-3-3β in vivo was changed by the lysis buffer despite the presence of a sufficient amount of phosphatase inhibitors, we also used a cell-permeable protein cross-linker dithiobis succinimidylpropionate (DSP) to capture the intracellular interaction between ChREBP and 14-3-3β before cell lysis. The results again demonstrated that glucose did not affect the in vivo interaction between 14-3-3β and ChREBP (Fig. 5B). Therefore, it is highly likely that the glucose response of ChREBP fails to happen at the level of its interaction with 14-3-3 proteins, and 14-3-3 may play a permissive role by allowing glucose-regulated events to occur.
Figure 5.
Characterization of Interaction between ChREBP and 14-3-3
A and B, Poly-His pull-down assay. The 832/13 cells stably expressing 6×His/c-myc-tagged 14-3-3β and c-myc-tagged ChREBP or its R128 mutant were treated with low (L, 2.5 mm) or high (H, 27.5 mm) glucose for 4 h and then either directly lysed for the pull-down assay (A) or treated with DSP before lysis for pull-down assay (B). C, Poly-His pull-down assay. pCHM-14-3-3β was cotransfected with plasmids expressing indicated wild-type or mutant ChREBP into 832/13 cells. Cells were lysed after 24 h for poly-His pull-down assay. D, GST pull-down assay. Glutathione-Sepharose beads coated with the indicated recombinant protein were incubated with recombinant c-myc-tagged 14-3-3β. The amount of 14-3-3β retained on these beads was examined by Western blot with anti-c-myc antibody. WT, Wild type.
The above results do not support the hypothesis that glucose-induced phosphorylation of ChREBP is required for interaction with 14-3-3. To further delineate the interaction between 14-3-3 and ChREBP, we took advantage of the truncation constructs generated earlier (Fig. 1A) to confirm whether MCR III is responsible for interaction with 14-3-3 in ChREBP, as it is in its paralog MondoA (12). In a poly-His pull-down assay, all truncation mutants with intact MCR III (ΔM1, ΔM2, and ΔM4) could be pulled down by 14-3-3β, whereas in the case of ΔM3 and ΔM34 mutants, where the entire MCR III is missing, interaction was completely abrogated. Furthermore, a construct in which all MCRs in the LID except MCR III were deleted (M3) exhibited good interaction with 14-3-3 (Fig. 5C). These data indicate that MCR III is sufficient and necessary for interaction with 14-3-3 proteins.
Armed with this knowledge, we tested whether the interaction between MCR III and 14-3-3 proteins is dependent on serine/threonine phosphorylation, because there are only two such residues located in this region, S103 and S140, of which S140 is not conserved in MondoA and mutation of S140 to alanine has been shown previously not to alter its interaction with 14-3-3 proteins (13). On the other hand, S103 is conserved in other Mondo proteins and is likely involved in interaction with 14-3-3, although it is not predicted to be a potential phosphorylation site by the NetPhos program (17). To determine experimentally whether interaction between 14-3-3 and MCR III depends on serine/threonine phosphorylation of MCR III, we generated the S103A mutant based on the M3 truncation construct (M3-S103A) so that it could no longer be phosphorylated at this residue. We studied the interaction of this mutant with 14-3-3β by poly-His pull-down assay and found that the S103A mutation does not affect the interaction between MCR III and 14-3-3β; in contrast, the R128A mutation completely abolished its interaction with 14-3-3β (Fig. 5C). Therefore, we can conclude that interaction between ChREBP and 14-3-3 is not mediated by serine/threonine phosphorylation.
An alternative hypothesis was proposed that Mondo protein interaction with 14-3-3 could be indirectly mediated by a bridging protein yet to be identified (12,13). To test this hypothesis, we used 14-3-3β and glutathione S-transferase (GST) fusion protein with MCR III (GST-M3), LID (GST-LID), or difopein (GST-Difo, as a positive control) produced in and purified from bacteria, which obviates the presence of the hypothetical bridging protein, and performed GST pull-down assay to study the interaction between the two sets of recombinant proteins in vitro. The results showed that GST alone did not interact with 14-3-3β, whereas GST fusion proteins with MCR III, LID, and difopein all bound strongly to 14-3-3β (Fig. 5D). These data confirm that MCR III contains a 14-3-3-binding motif, which directly interacts with 14-3-3 proteins.
The Discovery of a Novel 14-3-3-Interacting Motif
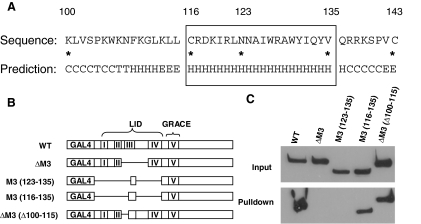
To understand the putative structural basis of MCR III interaction with 14-3-3, we analyzed the secondary structure of the MCR III using the SSpro8 program (18), which predicts in MCR III the presence of a stretch of α-helix spanning residues 116–136 (Fig. 6A). Because in this region Q136 is not conserved in other Mondo proteins (see multiple alignment in Refs. 10 and 12), we focused our attention on the conserved part of the helix (residues 116–135, Fig. 6A). Interestingly, mutations and deletion within this region have previously been shown to lead to loss of 14-3-3 binding by MondoA (12) and ChREBP (13) (Fig. 2B), supporting an essential role of the helix in 14-3-3 binding.
Figure 6.
Identification of the 14-3-3-Interacting Motif in ChREBP
A, Secondary structure prediction with SSpro8 program. Key residues were marked by asterisks and their number in the ChREBP amino acid sequence. The conserved α-helix (116–135) is boxed. Codes for secondary structure: C, coil; H, α-helix; T, turn; E, extended strand. B, Schematic illustration of the ChREBP deletion constructs (not to scale). C, Poly-His pull-down assay. pCHM-14-3-3β was cotransfected with indicated plasmids shown in B into 832/13 cells. Cells were lysed 24 h after transfection for poly-His pull-down assay.
To further analyze the role of the α-helix (116–135) in MCR III-14-3-3 interactions, we created additional mutants with fine deletions in the MCR III, including M3 (123–135) in which the LID region of ChREBP was replaced with the C terminus of the α-helix (123–135), M3 (116–135) where only the α-helix portion of the LID was included, and ΔM3(Δ100–115) in which the N-terminal portion of the MCR III up to the α-helix (100–115) was deleted (Fig. 6B).
We tested interaction of individual mutants with 14-3-3β using a poly-His pull-down assay and found that all constructs with an intact α-helical region [WT, M3 (116–135) and ΔM3 (Δ100–115)] retained their capacity to bind to 14-3-3β; furthermore, the α-helical region itself [M3 (116–135)] was sufficient for binding to 14-3-3β (Fig. 6C). Importantly, although construct M3 (123–135) contains the I126/W127/R128 residues previously found to be essential for 14-3-3 binding (12), lack of the N-terminal half of the α-helix in this construct led to complete loss of interaction with 14-3-3β (Fig. 6C). Taken together, the data indicate that the intact α-helical region (116–135) of MCR III is necessary and sufficient for interaction with 14-3-3 proteins and can be defined as the 14-3-3-interacting motif in ChREBP.
The Glucose Responsiveness of ChREBP Involves Other Functions of MCR III in Addition to 14-3-3 Binding
We showed that 14-3-3 binding is an essential function of MCR III (Fig. 3). MCR III encompasses a highly conserved motif of 44 residues in Mondo proteins. Mutations or deletions in residues outside the 14-3-3 interacting motif in MCR III had been shown earlier not to affect binding of 14-3-3 with ChREBP or MondoA (12,13), implying that these residues may serve functions other than 14-3-3 binding, and the function of MCR III in glucose responsiveness of ChREBP is likely not limited to interaction with 14-3-3.
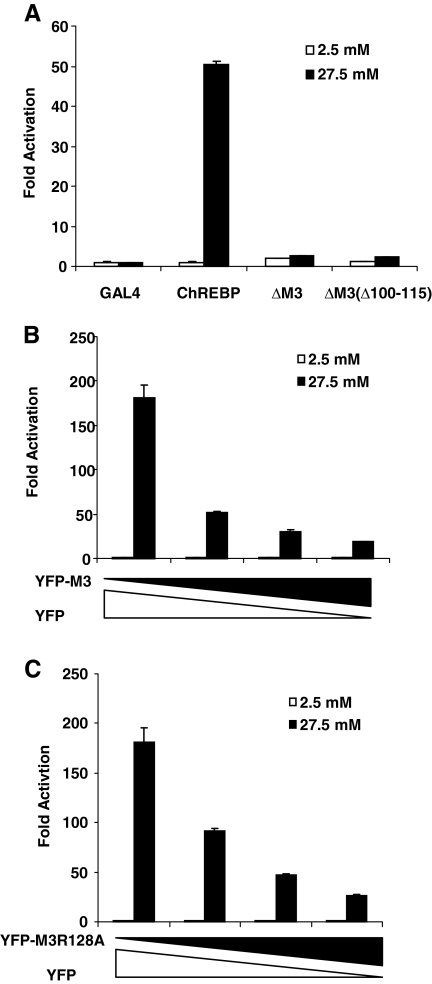
After defining the 14-3-3-binding motif in MCR III, we were in a position to test whether 14-3-3 binding is the sole function of MCR III in glucose response of ChREBP. The mutant ΔM3(Δ100–115) has the N terminus of MCR III deleted but retains the 14-3-3-interacting α-helix and therefore remains competent to bind to 14-3-3 (Fig. 6C). We used a luciferase reporter assay to determine the glucose responsiveness of this mutant in 832/13 cells and observed only slight and negligible response to glucose in this mutant ChREBP despite unperturbed 14-3-3 binding (Fig. 7A). These data indicate that sequences outside the α-helix, although not essential for binding to 14-3-3 proteins, are nonetheless required for the glucose responsiveness of ChREBP, suggesting that the function of MCR III extends beyond 14-3-3 binding.
Figure 7.
The 14-3-3-Independent Function of MCR III
A–C, Luciferase assay: A, pG5-luc and pRL-TK were cotransfected with plasmids expressing indicated proteins into 832/13 cells that were treated with 2.5 or 27.5 mm glucose for 24 h; B, pG5-luc, pRL-TK and pGAMPAC-ChREBP were cotransfected with different amounts of pYFP-M3 or pYFP into 832/13 cells that were treated with 2.5 or 27.5 mm glucose for 24 h. C, pG5-luc, pRL-TK, and pGAMPAC-ChREBP were cotransfected with different amounts of pYFP-M3R128A or pYFP into 832/13 cells that were treated with 2.5 or 27.5 mm glucose for 24 h.
To further examine this thesis, we tested the effects of overexpression of YFP fusion proteins with MCR III (YFP-M3) or its R128A mutant (YFP-M3R128A) on the glucose response of GAL4-ChREBP, under the premise that overexpression of these proteins can sequester essential proteins to which ChREBP may bind additionally according to our hypothesis. In this experiment, the overall structure of ChREBP remains intact; therefore, we can avoid disruption of ChREBP structure that may have resulted from deletions of key residues. Like difopein, overexpression of MCR III dose-dependently suppressed the glucose response of GAL4-ChREBP, which may at least partially be caused by sequestration of 14-3-3 proteins in a similar fashion as addition of difopein (Fig. 7B). Significantly, the R128A mutant of MCR III (M3R128A), which does not interact with 14-3-3 at all (Figs. 2B and 5, A–C), also suppressed the glucose response of GAL4-ChREBP with comparable efficiency as wild-type MCR III (Fig. 7C). These data strongly suggest that the role of MCR III is not limited to binding to 14-3-3 proteins but involves also trans-acting factors other than 14-3-3 in the glucose response of ChREBP.
DISCUSSION
ChREBP is a key transcription factor for the regulation of glucose and lipid metabolism and their cross talk. ChREBP is a typical thrift gene in that it facilitates the conversion of extra carbohydrate into a storage form of energy such as triglyceride by up-regulation of genes involved in lipogenesis. These actions of ChREBP are physiologically important, particularly in times when the organism lives in basic subsistence and food resources are scarce. However, these same actions of ChREBP may also lead to deleterious effects in times of abundance and may be responsible for disorders related to overnutrition. The important role of ChREBP is clearly illustrated in the phenotype of ChREBP knockout mice, which display impaired glycolytic and lipogenic activity (5), whereas in the ob/ob obesity mouse model, ChREBP ablation results in improvement in plasma glucose control and prevention of obesity (19). ChREBP has also been implicated in the development of glucotoxicity in pancreatic islet β-cells (20) and hepatosteatosis (21). Therefore, modulation of ChREBP activity is a promising strategy for treating these disorders, whose importance is underscored by the global epidemic of obesity, diabetes, and other nutrition-related health problems.
To realize ChREBP as a drug target, we must thoroughly understand its mechanism of action and its activation by glucose. Unfortunately, although we have appreciated the importance of ChREBP in the regulation of lipogenesis and glycolysis through experiments on gene knockout mice, we have only a limited knowledge of how glucose activates the transcription activity of ChREBP. The activation of ChREBP occurs via two separate steps (11): first, dephosphorylation at S196 and T666 in rat ChREBP, mediated by glucose-activated protein phosphatase 2A, is responsible for translocation of ChREBP into the nucleus and the binding of ChREBP to DNA (3,22), whereas another mechanism independent of phosphorylation/dephosphorylation is responsible for transactivation of ChREBP after glucose stimulation (11). We previously presented evidence that this latter mechanism is mediated by a conserved GSM domain at the N-terminal region of ChREBP through the release of a LID (the regulatory domain)-mediated intramolecular inhibition on the transactivation activity of GRACE (the transactivation domain) (10). However, the molecular details of this intramolecular inhibition remain to be clarified.
Molecular Dissection of the GSM Domain
The glucose response of ChREBP is composed of the inhibition of the transactivation activity under low glucose concentration as well as the release of the inhibition under high glucose. To define the functional entities responsible for each of these activities, we created serial GSM deletion mutants to conduct a detailed structure-function analysis of the GSM. Our results suggest that different MCRs may be responsible for different aspects of the glucose response: the major function of MCR IV seems to be inhibition of the transactivation activity, whereas the other MCRs are responsible for removal of the inhibition when stimulated by glucose (Fig. 1). It is worth pointing out that, being located immediately adjacent to the transactivation domain GRACE, MCR IV is well suited to perform this switch-like function in controlling the transactivation activity of ChREBP. We also documented that all individual MCRs with the exception of MCR I, which greatly diminishes the glucose response but falls short of complete abolition, are needed for the glucose response of ChREBP. We caution, however, that a better knowledge of the structure of GSM and its associated proteins is required for a more complete picture of the glucose-regulated ChREBP activation. Nonetheless, the information we provided will be useful in guiding the direction of further research, because it has led to our discovery of 14-3-3 proteins as an essential player in glucose response of ChREBP.
The Relationship between Subcellular Localization and Transcriptional Activity of ChREBP
One interesting finding in the mapping experiment is that deletions of MCR II or MCR III lead to complete silencing of the transactivation activity of ChREBP. These two regions were previously suggested to be responsible for cytoplasmic retention of Mondo proteins through the NES in MCR II and the 14-3-3 binding site in MCR III. If the transcriptional activity of ChREBP were solely dependent on its nuclear abundance, we would expect these two mutants to be constitutively active, instead of being totally inactive as we observed in our experiment. These findings were bolstered by our analysis of the NES and 14-3-3 binding site mutants (L86A/L93A and R128A, respectively). Although the latter two mutants (especially the L86A/L93A mutant) were enriched in the nucleus compared with wild-type ChREBP, they both were transcriptionally inactive in low- or high-glucose medium. These data contradict the original hypothesis of the all-important function of nucleus/cytoplasmic distribution in the action of Mondo proteins (7,12,13); they are, however, consistent with our previous results (10) and those of the Towle laboratory (11,14) that transactivation activity of ChREBP is not solely determined by its nuclear abundance but also subject to other glucose concentration-dependent regulatory effects on ChREBP. Our data also provide important clues as to how the glucose response of ChREBP is effected. The functions of MCR II and III are not only involved in regulation of subcellular localization, as was believed previously (12), but are also essential for transcriptional activation of ChREBP, and as such need to be further studied for their respective roles in this process.
MCR II: a Multipurpose NES
MCR II is a short fragment of 13 amino acids (residues 85–97) in the LID that fits the consensus NES sequence L-x (2,3)-[LIVFM]-x(2,3)-L-x-[LI] (13,23). Like other NES with similar sequences, MCR II reportedly mediates cytoplasmic localization of MondoA through the action of CRM1 and is subject to regulation by leptomycin, a fungicide that inhibits CRM1 (12). It was proposed that MCR II functions as an NES, i.e. it excludes the protein from the nucleus. However, our observation that both deletion of MCR II and the L86A/L93A mutation lead to complete loss of transactivation activity instead of constitutive activation suggests that MCR II is also involved in the transcriptional activation of ChREBP by glucose. This came as a surprise, because MCR II conforms completely to the NES consensus sequence. In a domain-swap experiment, we further showed that MCR II is indeed more than an NES because it cannot be replaced by a classic NES from an unrelated protein (Fig. 2). This interpretation is further supported by the Davies et al. (14) report that one of their NES mutants actually retained glucose response, again proving that nuclear export and glucose response are two separate events in regulation of ChREBP activity.
The 14-3-3 Proteins Are Essential for Glucose Response of ChREBP
Interaction between MCR III and 14-3-3 proteins has been suggested to contribute to the cytoplasmic retention of ChREBP (12). The fact that R128A mutant enhances its nuclear abundance (Fig. 2A) supports this notion. However, the increase in nuclear abundance does not translate into higher transactivation activity of ChREBP, as would be predicted whether the sole function of 14-3-3 binding is to regulate subcellular localization. This finding suggests to us that, similar to MCR II, MCR III also carries other functions related to the glucose response of ChREBP. With a length of 44 amino acids (residues 100–143), MCR III is a more complex domain than MCR II (13 amino acid residues), which perhaps would allow it to perform more complex functions in ChREBP. What was unexpected was that 14-3-3 binding seems to play a permissive and positive role in the transactivation of ChREBP rather than inhibitory, despite its confirmed function in helping exclude ChREBP from the nucleus (12,13), because both the mutation to the 14-3-3 binding site (R128A) and the overexpression of a 14-3-3 antagonist in trans can shut down glucose- induced transactivation of ChREBP (Figs. 2C and 3). Counterintuitive as it may seem, there are precedents that 14-3-3 proteins up-regulate the activity of transcription factors through enhanced transactivation, as in the case of the glucocorticoid receptor (24), or increased DNA binding activity, as for p53 (25). However, in the case of ChREBP, the role of 14-3-3 binding appears not to fit into either category, because the 14-3-3 binding motif (in MCR III) is located outside both the DNA-binding domain and the transactivation domain (GRACE or MCR V). Rather, it is most likely involved in removing the inhibitory effects of the LID, possibly mediated by MCR IV. The exact mechanism for the inhibition and its release, however, remains to be elucidated. To date, 14-3-3 proteins are the only trans-acting regulatory factor that has been found to be required for the glucose responsiveness of ChREBP. Our data showing that an antagonist for 14-3-3 proteins can efficiently shut down the activity of ChREBP (Fig. 3), down-regulate glucose-stimulated L-PK promoter activity, and inhibit glucose-stimulated synthesis of ACC and FAS (Fig. 4) provide a proof-of-principle that 14-3-3 is a possible therapeutic target for modulation of ChREBP activity.
The 14-3-3 Proteins Play a Supportive Role in the Glucose Response of ChREBP by Interacting with a Novel Binding Motif in ChREBP
In the two examples of the positive regulation of transcription factors by 14-3-3 proteins cited earlier, i.e. glucocorticoid receptor and p53 (24,25), the environmental stimuli that activate these transcription factors (glucocorticoid binding and irradiation, respectively) enhance the binding between the transcription factors and 14-3-3 proteins. It is interesting that the binding between ChREBP and 14-3-3 is not affected by glucose (Fig. 5, A and B), which supports the interpretation that 14-3-3 proteins are not a direct activator of ChREBP; instead, they most likely play a permissive role by providing a platform on which glucose-regulated events can take place to activate transcription. This premise is consistent with the fact that interaction between 14-3-3 and ChREBP is independent of serine/threonine phosphorylation (Fig. 5, C and D). This mode of action by 14-3-3 is unique compared with other known examples of 14-3-3 ligands whose interaction with 14-3-3 is modulated by environmental stimuli. We also showed that MCR III directly binds to 14-3-3 without any bridging molecules (Fig. 5D), even though the classic 14-3-3 binding motif could not be found in this region. Instead, a previously uncharacterized 14-3-3 binding motif in ChREBP is located within a 20-residue α-helix (residues 116–135) (Fig. 6). This motif differs fundamentally from other previously identified consensus sequences in that its interaction with 14-3-3 does not involve phosphorylation of serine or threonine. In fact, this motif does not contain any serine or threonine residues at all and, instead, is highly enriched with hydrophobic and basic amino acids (Fig. 6A). The fact that difopein can competitively inhibit the interaction between ChREBP and 14-3-3 suggests, however, that the binding site of ChREBP in 14-3-3 is similar to that of classic 14-3-3 ligands. It remains to be determined whether binding of this motif to 14-3-3 induces similar biological changes to those induced by classic 14-3-3 ligands and whether their interaction is subject to any modifications other than phosphorylation. Acquisition of such knowledge in the future will add to our understanding of 14-3-3, a seemingly all-purpose adaptor protein, and pave the way to the discovery of compounds to modify their interaction.
Function of MCR III
Essential as 14-3-3 is to the glucose response of ChREBP, the 14-3-3-interacting motif in ChREBP covers only half of the MCR III. It is therefore not surprising that the highly conserved MCR III is playing additional role(s) in the glucose response of ChREBP, as reflected by the finding that deletion of the non-14-3-3-binding region of MCR III leads to almost complete loss of ChREBP action (Fig. 7A), and overexpression of the R128A mutant of MCR III dose-dependently shuts down the glucose responsiveness of ChREBP (Fig. 7C). This observation further underscores the highly complex nature of the activation of ChREBP by glucose. The process requires cooperation of the cis-acting structural motifs, such as MCR I through V, and the trans-acting cofactors such as 14-3-3 proteins among other unknown ones. Missing any of these components of the glucose-sensing machinery would lead to loss of its ability to respond to glucose. The elucidation of how these motifs and cofactors interact would be essential for further advancing our understanding of the mechanism of glucose-responsive gene transcription mediated by ChREBP.
In conclusion, we have performed a detailed structure-function analysis on the GSM of ChREBP and have localized the inhibitory function of LID to the MCR IV while demonstrating that the rest of LID helps with the removal of this inhibitory effect under high glucose stimulation. This study has also shed new light on the function of the MCR II and III in the transactivation of ChREBP; these are subdomains whose sole function had previously been thought to enable Mondo proteins to stay in the cytoplasm. Not only did we characterize and confirm their function in nuclear export, but we also found that both MCR II and III are essential for the glucose-induced transactivation of ChREBP. The role of MCR II in the glucose response of ChREBP is independent of its function as an NES, whereas the function of MCR III in glucose response is at least partially mediated by interaction with 14-3-3, which is essential for glucose-induced transactivation of ChREBP. Moreover, we defined in MCR III a novel type of 14-3-3-binding motif, which directly interacts with 14-3-3 in a manner that is independent of glucose regulation or serine/threonine phosphorylation. Thus, 14-3-3 may play a permissive role in the process of glucose response by allowing other glucose-regulated events to occur. This knowledge would help us further understand the mechanism of the glucose response of ChREBP and may enable the development of novel therapeutic strategies, e.g. by antagonizing 14-3-3 binding, for modulating ChREBP activity and its downstream effects on glucose and lipid homeostasis.
MATERIALS AND METHODS
Plasmid Construction
The GSM serial deletion mutants of ChREBP (Fig. 1A) were made on the basis of the pGAMPAC-ChREBP plasmid described earlier (10), which encodes a GAL4-ChREBP fusion protein with a c-myc/protein-A double tag fused to its C terminus. We synthesized the cDNAs for serial deletions in the LID region by PCR and cloned them into the appropriate regions in pGAMPAC-ChREBP. The amino acid residues deleted for each construct in Fig. 1A are as follows: ΔM1, residues 1–71; ΔM1–2, 1–99; ΔM1–3, 1–143; ΔM1–4, 1–196; ΔM2, 72–99; ΔM2–3, 72–143; ΔM2–4, 72–196; ΔM3, 100–143; ΔM3–4, 100–196; ΔM4, 144–196; M2–3, 1–71 and 144–196; M2, 1–71 and 100–196; and M3, 1–99 and 144–196. Adaptor containing NES sequence from HIV-1 Rev protein was synthesized by annealing two oligonucleotides NES5 and NES3 (Table 1). The annealed NES adaptor was used to replace the residues 88–99 in ChREBP to generate the tNES mutant. Point mutations of ChREBP including L86A, L93A, R128A, and S103A were generated by PCR-based DNA mutagenesis. We cloned the cDNA for enhanced YFP into mammalian expression vector pCMX (26) to generate plasmid pYFP. The cDNA for difopein was synthesized by annealing four pairs of oligonucleotides (Difo51/31, 52/32, 53/33, and 54/34) (Table 1) that were treated with T4 polynucleotide kinase (Invitrogen, Carlsbad, CA) according to the manufacturer’s instruction and cloned in-frame to 3′ of enhanced YFP cDNA in pYFP to generate plasmid pYFP-Difo. We also cloned the cDNAs for MCR III and its R128A mutant in the same fashion to pYFP to generate plasmids pYFP-M3 and pYFP-M3R128A. We subcloned the cDNA of mouse 14-3-3β (GenBank accession no. AF058797) by RT-PCR from mouse liver in a modified pCMX plasmid (pCHM) whose multiple cloning site is flanked by a 6×His/c-myc double tag at the N terminus, to generate pCHM-14-3-3β for transient expression of 14-3-3β. Similarly, we also generated pCHM-ChREBP plasmid to transiently express 6×His/c-myc double-tagged ChREBP for poly-His pull-down experiments. The 14-3-3β cDNA was also cloned into pYFP plasmid to generate pYFP-14-3-3β to express fusion protein YFP-14-3-3β. For retroviral infection and stable expression, cDNAs of c-myc/protein-A double-labeled ChREBP and its mutants (tNES, L86A/L93A, and R128A) were cloned into pMSCVpuro (Clontech, Mountain View, CA), whereas the 6×His/c-myc double-tagged 14-3-3β was cloned into pMSCVhygro (Clontech). We also subcloned the cDNA of tetracycline transactivator (tTA) into pMSCVhygro to generate plasmid pMSCVhygro-tTA. For stable overexpression of YFP-difopein fusion protein or YFP via retroviral infection, corresponding cDNAs were subcloned into retroviral vector pSRQT (Li, M.V., and L. Chan, unpublished data), which supports tTA-driven overexpression in the presence of tTA to generate plasmids pSRQT-YFP and pSRQT-YFP-Difo. Detailed cloning procedures are available upon request.
Table 1.
Oligonucleotide Sequences for Adaptors
| Oligonucleotide | Sequence (5′–3′) |
|---|---|
| NES5 | tcgacgtgcctctgcagctaccaccgcttgagagacttactcttgattgtaacgaga |
| NES3 | cgcgtctcgttacaatcaagagtaagtctctcaagcggtggtagctgcagaggcacg |
| Difo51 | aattcagcgccgacggcgctccccactgcgtgcctagagacctgtcctg |
| Difo31 | cagccaggacaggtctctaggcacgcagtggggagcgccgtcggcgctg |
| Difo52 | gctggacctggaggccaacatgtgtctgcctggcgccgccggcctggaca |
| Difo32 | gcgctgtccaggccggcggcgccaggcagacacatgttggcctccaggtc |
| Difo53 | gcgccgacggcgccccccactgcgtgcctagggatctgtcctggctggac |
| Difo33 | ccaggtccagccaggacagatccctaggcacgcagtggggggcgccgtcg |
| Difo54 | ctggaggccaacatgtgtctgcctggcgccgccggcctggagg |
| Difo34 | tcgacctccaggccggcggcgccaggcagacacatgttggcct |
Sequences for cut restriction sites are underlined(SalI and MluI for NES and EcoRI and SalI for difopein adaptors).
Cell Culture and Transfection
The 832/13 cells (gift of Dr. C. Newgard, Duke University, Durham, NC) were cultured as described (27). Lipofectamine 2000 (Invitrogen) was used for transfection according to the manufacturer’s instructions.
Retrovirus Production and Infection
We cotransfected indicated retroviral plasmids with packaging vector pCL-eco (Imgenex, Sorrento Valley, CA) into packaging cells BOSC23 (American Type Culture Collection, Manassas, VA). Twenty-four hours after transfection, we collected the culture medium containing viral particles and mixed it 1:1 with INS-1 cell culture medium. Polybrene was added at a final concentration of 6 μg/ml to the mixture, which was then applied to target 832/13 cells for infection. Twenty-four hours after infection, infected cells were selected with antibiotics (1 μg/ml puromycin or 200 μg/ml hygromycin), depending on vector selection.
Luciferase Assay
We cotransfected UASGAL-driven firefly luciferase reporter pG5-luc (Promega, Madsion, WI) or L-PK promoter-driven firefly luciferase reporter (pLPK-Luc) (10) with internal control pRL-TK (Promega) and specified plasmids into 832/13 cells and treated them with the indicated concentration of glucose for 24 h. Dual-luciferase kit (Promega) was used for luciferase assay. All results were shown as fold activation over reporter activity at the basal condition (the first column in each figure).
Immunofluorescence Staining and Fluorescence Imaging
We stably expressed c-myc-tagged ChREBP and its derivatives by retroviral infection in 832/13 cells, which were seeded on glass coverslips. After treating infected cells with low (2.5 mm) or high (27.5 mm) glucose, we fixed the cells on glass coverslips and permeabilized and sequentially incubated them with primary (9B11 anti-c-myc monoclonal antibody; Cell signaling, Danvers, MA) and secondary (fluorescein isothiocyanate-conjugated goat anti-mouse Ig; Invitrogen) antibodies, before mounting them with 4′,6-diamidino-2-phenylindole-containing mounting medium (Vector Labs, Burlingame, CA) for nuclear counter stain. Alternatively, for imaging of YFP fusion proteins, cells infected with virus expressing YFP-14-3-3 fusion protein were treated with low or high glucose. After fixation, the coverslips were directly mounted for imaging. Fluorescent images were captured with Zeiss Axioplan 2 fluorescent microscope (Zeiss, Göttingen, Germany).
Poly-His Pull-Down Assay
We expressed both 6×His/c-myc tagged 14-3-3β and c-myc-labeled ChREBP or its mutants in 832/13 cells through transient transfection or retroviral infection as indicated. We then isolated 6×His/c-myc-tagged 14-3-3β and its associated proteins with Ni-NTA agarose beads (QIAGEN, Valencia, CA) according to manufacturer’s instructions and examined their levels in the lysates (input) or the precipitates (pull down) by Western blot with anti- c-myc antibody 9B11 (Cell Signaling). For in vivo cross- linking, cells were treated with DSP (Pierce, Rockford, IL) according to the manufacturer’s description before cell lysis and pull-down assay. Alternatively, 6×His/c-myc-tagged ChREBP (or R128A mutant) was also used to pull down YFP-14-3-3β fusion protein, and Living Colors A.v. monoclonal antibody (JL-8; CLONTECH, Mountain View, CA) was used for Western blot of YFP fusion proteins.
Generation of Recombinant c-myc-Tagged 14-3-3β
We subcloned the cDNA for 14-3-3β, which is tagged at the C terminus by a c-myc epitope tag, into pGEX-4T1 vector (Amersham, Piscataway, NJ). The resulting plasmid, pGEX-14-3-3β, was transformed into DH5α Escherichia coli cells (Invitrogen) to generate a recombinant GST fusion protein with c-myc-labeled 14-3-3β, which was further purified with glutathione-Sepharose beads (Amersham) according to the manufacturer’s instructions. We then released the c-myc-tagged 14-3-3β from the beads and GST by digestion with thrombin (Amersham) and removed the thrombin from the elution with p-aminobenzamidine agarose (Sigma Chemical Co., St. Louis, MO). The purified protein was stored in 50% glycerol at −20 C.
GST Pull-Down Assay
We subcloned the cDNAs for difopein, MCR III, and the LID into pGEX-4T1 vector. We then transformed the resulting plasmids or the vector pGEX-4T1 into DH5α cells for generation of recombinant GST protein or GST fusion proteins with difopein, MCR III, or LID, respectively, which were in turn immobilized on glutathione-Sepharose beads (Amersham) according to the manufacturer’s instruction. After washing with PBS, these beads were incubated with recombinant c-myc-tagged 14-3-3β protein at 4 C for 1 h. The amount of 14-3-3β retained on these beads after washing with PBS was then examined by Western blot with anti-c-myc antibody 9B11 (Cell Signaling).
Quantitative RT-PCR
We overexpressed YFP or YFP-difopein in 832/13 cells by coinfecting retroviruses produced from pSRQT-YFP or pSRQT-YFP-Difo with those produced from pMSCVhygro-tTA. After selection with both puromycin and hygromycin, positive cells were treated with 2.5 or 27.5 mm glucose for 48 h. We then extracted mRNA from these cells with RNeasy RNA miniprep kit (QIAGEN) and synthesized cDNA with Omniscript RT kit (QIAGEN). The levels of FAS (GenBank accession no. X62888) and ACC (NM_022193) in these samples were determined by real-time PCR with cDNA from these samples as templates. The primer sequences for ACC and internal control eEF-1G (XM_215165) has been reported earlier (10), and the primer sequences for FAS is as follows: sense, 5′-acattcccctctggagaag-3′, and antisense, 5-ggatttggtggagccaattaa-3′.
Statistical Analysis
Student’s t test was used to evaluate statistical significance in two groups of data with Microsoft Excel software.
Acknowledgments
We thank Dr. Christopher Newgard for providing the 832/13 cells.
Footnotes
This work was supported by National Institutes of Health Grants DK68037 and HL51586, the Rutherford Chair for Diabetes Research from St. Luke’s Episcopal Hospital, the T.T. & W.F. Chao Foundation (to L.C.), and a Mentor-Based Fellowship Grant (to W.C. and L.C.).
Disclosure Statement: All authors have nothing to declare.
First Published Online April 24, 2008
Abbreviations: ACC, Acetyl-coenxyme A carboxylase; ChREBP, carbohydrate response element-binding protein; CRM1, chromosome region maintenance 1; DSP, dithiobis succinimidylpropionate; FAS, fatty acid synthase; GRACE, glucose-response activation conserved element; GSM, glucose-sensing module; GST, glutathione S-transferase; LID, low-glucose inhibitory domain; L-PK, liver pyruvate kinase; MCR, Mondo conserved region; NES, nuclear export signal; tTA, tetracycline transactivator; YFP, yellow fluorescent protein.
References
- Thompson KS, Towle HC 1991 Localization of the carbohydrate response element of the rat L-type pyruvate kinase gene. J Biol Chem 266:8679–8682 [PubMed] [Google Scholar]
- Bergot MO, Diaz-Guerra MJ, Puzenat N, Raymondjean M, Kahn A 1992 Cis-regulation of the L-type pyruvate kinase gene promoter by glucose, insulin and cyclic AMP. Nucleic Acids Res 20:1871–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi T, Takenoshita M, Kabashima T, Uyeda K 2001 Glucose and cAMP regulate the L-type pyruvate kinase gene by phosphorylation/dephosphorylation of the carbohydrate response element binding protein. Proc Natl Acad Sci USA 98:13710–13715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii S, Iizuka K, Miller BC, Uyeda K 2004 Carbohydrate response element binding protein directly promotes lipogenic enzyme gene transcription. Proc Natl Acad Sci USA 101:15597–15602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iizuka K, Bruick RK, Liang G, Horton JD, Uyeda K 2004 Deficiency of carbohydrate response element-binding protein (ChREBP) reduces lipogenesis as well as glycolysis. Proc Natl Acad Sci USA 101:7281–7286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Luis O, Valero MC, Jurado LA 2000 WBSCR14, a putative transcription factor gene deleted in Williams-Beuren syndrome: complete characterisation of the human gene and the mouse ortholog. Eur J Hum Genet 8:215–222 [DOI] [PubMed] [Google Scholar]
- Billin AN, Eilers AL, Coulter KL, Logan JS, Ayer DE 2000 MondoA, a novel basic helix-loop-helix-leucine zipper transcriptional activator that constitutes a positive branch of a max-like network. Mol Cell Biol 20:8845–8854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairo S, Merla G, Urbinati F, Ballabio A, Reymond A 2001 WBSCR14, a gene mapping to the Williams-Beuren syndrome deleted region, is a new member of the Mlx transcription factor network. Hum Mol Genet 10:617–627 [DOI] [PubMed] [Google Scholar]
- Yamashita H, Takenoshita M, Sakurai M, Bruick RK, Henzel WJ, Shillinglaw W, Arnot D, Uyeda K 2001 A glucose-responsive transcription factor that regulates carbohydrate metabolism in the liver. Proc Natl Acad Sci USA 98:9116–9121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MV, Chang B, Imamura M, Poungvarin N, Chan L 2006 Glucose-dependent transcriptional regulation by an evolutionarily conserved glucose-sensing module. Diabetes 55:1179–1189 [DOI] [PubMed] [Google Scholar]
- Tsatsos NG, Towle HC 2006 Glucose activation of ChREBP in hepatocytes occurs via a two-step mechanism. Biochem Biophys Res Commun 340:449–456 [DOI] [PubMed] [Google Scholar]
- Eilers AL, Sundwall E, Lin M, Sullivan AA, Ayer DE 2002 A novel heterodimerization domain, CRM1, and 14-3-3 control subcellular localization of the MondoA-Mlx heterocomplex. Mol Cell Biol 22:8514–8526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merla G, Howald C, Antonarakis SE, Reymond A 2004 The subcellular localization of the ChoRE-binding protein, encoded by the Williams-Beuren syndrome critical region gene 14, is regulated by 14-3-3. Hum Mol Genet 13:1505–1514 [DOI] [PubMed] [Google Scholar]
- Davies MN, O'Callaghan BL, Towle HC 2007 Glucose activation of carbohydrate response element binding protein (ChREBP) requires a nuclear event independent of nucleocytoplasmic shuttling. FASEB J 21:A1041-b [Google Scholar]
- Fu H, Subramanian RR, Masters SC 2000 14-3-3 proteins: structure, function, and regulation. Annu Rev Pharmacol Toxicol 40:617–647 [DOI] [PubMed] [Google Scholar]
- Masters SC, Fu H 2001 14-3-3 proteins mediate an essential anti-apoptotic signal. J Biol Chem 276:45193–45200 [DOI] [PubMed] [Google Scholar]
- Blom N, Gammeltoft S, Brunak S 1999 Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J Mol Biol 294:1351–1362 [DOI] [PubMed] [Google Scholar]
- Pollastri G, Przybylski D, Rost B, Baldi P 2002 Improving the prediction of protein secondary structure in three and eight classes using recurrent neural networks and profiles. Proteins 47:228–235 [DOI] [PubMed] [Google Scholar]
- Iizuka K, Miller B, Uyeda K 2006 Deficiency of carbohydrate-activated transcription factor ChREBP prevents obesity and improves plasma glucose control in leptin-deficient (ob/ob) mice. Am J Physiol Endocrinol Metab 291:E358–E364 [DOI] [PubMed] [Google Scholar]
- da Silva Xavier G, Rutter GA, Diraison F, Andreolas C, Leclerc I 2006 ChREBP binding to fatty acid synthase and L-type pyruvate kinase genes is stimulated by glucose in pancreatic β-cells. J Lipid Res 47:2482–2491 [DOI] [PubMed] [Google Scholar]
- Dentin R, Benhamed F, Hainault I, Fauveau V, Foufelle F, Dyck JR, Girard J, Postic C 2006 Liver-specific inhibition of ChREBP improves hepatic steatosis and insulin resistance in ob/ob mice. Diabetes 55:2159–2170 [DOI] [PubMed] [Google Scholar]
- Kabashima T, Kawaguchi T, Wadzinski BE, Uyeda K 2003 Xylulose 5-phosphate mediates glucose-induced lipogenesis by xylulose 5-phosphate-activated protein phosphatase in rat liver. Proc Natl Acad Sci USA 100:5107–5112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- la Cour T, Gupta R, Rapacki K, Skriver K, Poulsen FM, Brunak S 2003 NESbase version 1.0: a database of nuclear export signals. Nucleic Acids Res 31:393–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakui H, Wright AP, Gustafsson J, Zilliacus J 1997 Interaction of the ligand-activated glucocorticoid receptor with the 14-3-3 η-protein. J Biol Chem 272:8153–8156 [DOI] [PubMed] [Google Scholar]
- Waterman MJ, Stavridi ES, Waterman JL, Halazonetis TD 1998 ATM-dependent activation of p53 involves dephosphorylation and association with 14-3-3 proteins. Nat Genet 19:175–178 [DOI] [PubMed] [Google Scholar]
- Forman BM, Umesono K, Chen J, Evans RM 1995 Unique response pathways are established by allosteric interactions among nuclear hormone receptors. Cell 81:541–550 [DOI] [PubMed] [Google Scholar]
- Hohmeier HE, Mulder H, Chen G, Henkel-Rieger R, Prentki M, Newgard CB 2000 Isolation of INS-1-derived cell lines with robust ATP-sensitive K+ channel-dependent and -independent glucose-stimulated insulin secretion. Diabetes 49:424–430 [DOI] [PubMed] [Google Scholar]