Abstract
The glucocorticoid receptor (GR) mediates virtually all actions of glucocorticoids, and the nature and magnitude of a cell’s response to these steroids are determined primarily by hormone concentration and GR signaling capacity. DAX-1 (dosagesensitive sex reversal-adrenal hypoplasia congenita critical region on the X-chromosome, gene 1) is an orphan nuclear receptor that functions as a corepressor, and deletion or mutation of DAX-1 causes a decrease in glucocorticoid production. However it is unclear whether DAX-1 also alters GR function as a transcription factor. Here, we demonstrate that DAX-1 acts as a novel selective GR modulator. It specifically inhibits ligand-dependent GR transactivation with little effect on GR-mediated transrepression. As demonstrated by coimmunoprecipitation and glutathione- S-transferase pull-down assays, DAX-1 physically interacts with GR, but this interaction does not influence either ligand-induced GR nuclear translocation or subsequent GR association with glucocorticoid-responsive elements. Instead, DAX-1 competes with coactivators such as GR-interacting protein 1 for binding to the receptor. Specifically, suppression of GR transactivation is mediated by the N-terminal half of DAX-1, and in particular the LXXLL motifs. Thus we demonstrate that DAX-1 directly modulates GR signaling in addition to affecting glucocorticoid hormone levels.
GLUCOCORTICOIDS ARE a class of stress- induced steroid hormones that are endogenously synthesized in the adrenal cortex. Most, if not all, glucocorticoid-induced actions are mediated via the glucocorticoid receptor (GR), which is expressed in virtually every tissue and cell type. The GR, a member of the nuclear receptor superfamily of proteins, resides in the cytoplasm as a multiprotein complex and translocates to the nucleus upon ligand binding. Ligand-bound GR homodimers then interact with either cognate glucocorticoid-responsive elements (GREs) or other transcription factors such as nuclear factor-κB (NF-κB), activator protein 1 (AP-1), Sma and Mad related protein, or signal transducer and activator of transcription, to mediate transactivation or transrepression of target genes.
Clinically, glucocorticoids are extensively prescribed for their antiinflammatory or immune-suppressive effects in the treatment of asthma, dermatitis, rheumatoid arthritis, and autoimmune diseases or prevention of graft rejection (1,2,3,4). However, long-term use of steroids is often associated with a wide range of adverse effects including weight gain, glaucoma, myopathy, hypertension, and osteoporosis (5,6,7,8,9,10). It is widely accepted that GR-mediated transrepression of target genes, particularly proinflammatory cytokines and cytokine receptors, accounts for the beneficial effects of glucocorticoids in combating inflammation and autoimmune diseases, whereas the side effects are associated with GR-mediated transactivation. Therefore, selective GR modulators that enhance GR-mediated transrepression but reduce transactivation are expected to have great therapeutic value due to improved benefit to risk ratio (5,11).
In contrast to GR, DAX-1 (dosage-sensitive sex reversal-adrenal hypoplasia congenita critical region on the X-chromosome, gene 1) is an atypical nuclear receptor in that it does not contain the zinc-finger DNA-binding domain that is highly conserved among most members of the receptor superfamily. Instead, the protein harbors 3.5 repeats of 65–67 amino acids at the N terminus, each of which contains a LXXLL motif that was previously identified in nuclear receptor coactivators. Functionally, DAX-1 has been characterized as a corepressor. It suppresses transcription of its own gene by binding to the promoter region (12). Alternatively, DAX-1 associates with several other nuclear receptors and blocks gene regulation mediated by those factors (13,14,15,16,17). In addition, DAX-1 has been shown to recruit other corepressors such as nuclear receptor corepressor and Alien to form a super complex (13,18).
In humans, DAX-1 gives rise to male-to-female sex reversal when the corresponding genomic region XP21 is duplicated (19,20,21,22) and adrenal hypoplasia congenita (AHC) and hypogonadotropic hypogonadism (HH) when mutated or deleted (19,23,24). AHC typically presents as adrenal insufficiency with decreased levels of circulating glucocorticoids and mineralocorticoids. The intracellular levels of steroid and receptor largely determine the nature and magnitude of the cellular response to glucocorticoids (25,26,27,28). Thus, it is reasonable to assume that DAX-1 may ultimately disturb GR signaling by reducing glucocorticoid production, particularly in AHC patients.
Based on the fact that DAX-1 functions as a corepressor and that the protein indirectly impedes GR signaling, we hypothesized that DAX-1 may also affect the transcriptional activity of GR, but this issue remains unclear. In this report, we investigated the effect of DAX-1 expression on GR signaling. Our data indicate that DAX-1 is a selective regulator of GR actions by preferentially inhibiting transactivation, but not transrepression, as demonstrated by both transfected reporter constructs and endogenous genes. Furthermore, we demonstrate that DAX-1 physically interacts with GR, and functional LXXLL motifs in the N terminus of DAX-1 are critical to suppression of GR transactivation.
RESULTS
DAX-1 Inhibits GR-Mediated Reporter Gene Transactivation
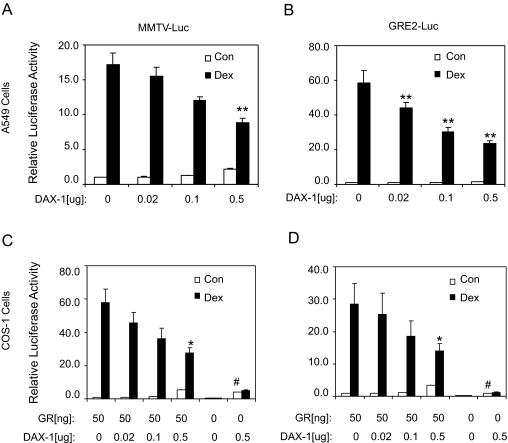
Our initial goal in these investigations was to determine whether DAX-1 altered the ability of the GR to signal. Using A549 human lung carcinoma cells, we show that DAX-1 inhibited dexamethasone (dex)-stimulated luciferase activity on both mouse mammary tumor virus (MMTV)-Luc and GRE2-Luc constructs in a concentration-dependent manner (Fig. 1, A and B). At the highest concentration, it suppressed the luciferase activity by approximately 50%. To confirm that this effect was mediated by GR, GR-deficient COS-1 cells were transfected with the same constructs in the presence or absence of GR (Fig. 1, C and D). In the presence of GR only, dex led to a dramatic enhancement of reporter activity, and this increase was suppressed in a dose-dependent manner when DAX-1 was also transfected. In the absence of GR, no enhancement of reporter activity was observed in response to dex, and the presence of DAX-1 had little effect in reducing the constitutive activity of the reporter genes. These data suggest that DAX-1 inhibits ligand-induced GR-mediated transactivation of MMTV-Luc and GRE2-Luc.
Figure 1.
DAX-1 Inhibits Ligand-Induced GR Transactivation in Both A549 (A and B) and COS-1 (C and D) Cells
A549 cells were transfected with either MMTV-Luc (A) or GRE2-Luc (B), pGL3-hRL, and increasing concentrations of DAX-1. COS-1 cells were transfected with either MMTV-Luc (C) or GRE2-Luc (D) and pGL3-hRL as well as increasing concentrations of DAX-1 in the presence or absence of GR. The transfection mixture was supplemented with mock plasmid to obtain the same amount of total DNA. After transfection, cells were treated with vehicle or 100 nm dex in low serum OPTI-MEM media for 20 h. Reporter luciferase activity was determined and normalized to the Renilla luciferase. The mean ± se from at least three independent experiments is shown. *, P < 0.05; and **, P < 0.01 represent significant differences between samples transfected with mock plasmid and DAX-1 when treated with dex. #, P < 0.05 represents significant differences between samples transfected with mock plasmid and DAX-1 in the absence of dex. Con, Control.
Interestingly, expression of DAX-1 also caused a small increase in constitutive luciferase activity on both reporter constructs and in both A549 and COS-1 cells, independent of dex and functional GR levels. The mechanism by which DAX-1 enhances the constitutive luciferase activity is unclear, but it probably reflects nonspecific activation of the basic transcription machinery because DAX-1 is known to interact with other corepressors and quench their inhibitory effect (13,18).
DAX-1 Does Not Affect GR-Mediated Transrepression
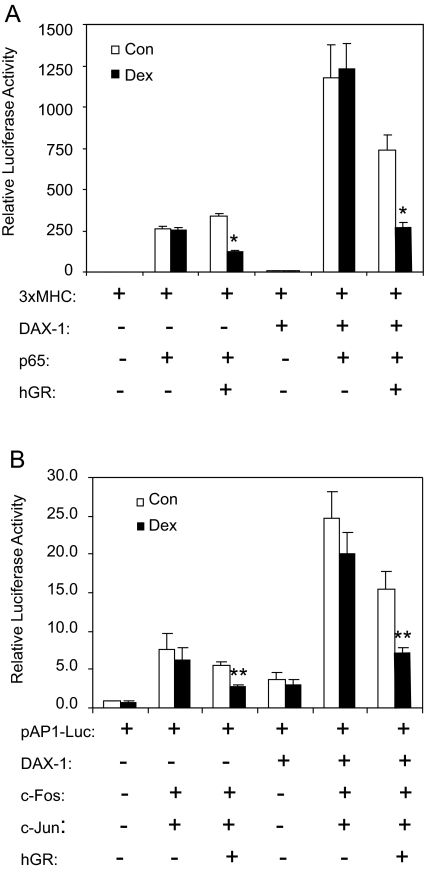
To determine whether DAX-1 also affects GR-mediated transrepression, COS-1 cells were transfected with 3xMHC-Luc and p65 (a NF-κB-responsive reporter and the NF-κB subunit, respectively) in the presence or absence of GR and/or DAX-1. As shown in Fig. 2A, expression of p65 enhanced luciferase activity by approximately 250-fold, and this was not affected by GR in the absence of ligand. After treatment with dex, this NF-κB activity was inhibited significantly by 50–60%, consistent with previous publications (29). In the presence of DAX-1 alone, it promoted the constitutive 3xMHC-Luc activity by approximately 5-fold, similar to that shown in Fig. 1. Cotransfection of DAX-1 and p65 resulted in a synergistic increase (1200-fold) in luciferase activity that was not influenced by dex. When cells were cotransfected with all three plasmids (p65, GR, and DAX-1), a GR-dependent suppression of 3xMHC luciferase activity was observed even in the absence of dex. This suppression likely occurred due to blockage by GR of nonspecific gene activation elicited by DAX-1. When the cells were subsequently treated with dex, an additional 50–60% suppression in luciferase activity was detected. This reduction was quantitatively similar to that obtained in cells transfected with GR and p65 only, which suggests that the presence of DAX-1 had no effect on the transrepression potential by GR.
Figure 2.
DAX-1 Does Not Affect Ligand-Induced GR Transrepression
A, COS-1 cells were transfected with a NF-κB-responsive reporter 3xMHC-Luc (200 ng) and pGL3-hRL (50 ng) in the presence or absence of NF-κB subunit p65 (25 ng), hGRα (250 ng), and DAX-1 (2500 ng). B, COS-1 cells were transfected with AP-1-responsive reporter pAP1-Luc (200 ng) and pGL3-hRL in the presence or absence of the AP-1 subunits c-Fos (12.5 ng) and c-Jun (12.5 ng), hGRα (250 ng), and DAX-1 (2500 ng). After transfection, the cells were treated with vehicle or 100 nm dex in low serum OPTI-MEM media for 20 h. Reporter luciferase activity was determined and normalized to the Renilla luciferase internal control. The mean ± se from at least three independent experiments is shown. *, P < 0.05; and **, P < 0.01 represent significant differences between vehicle and dex in samples transfected with the same constructs. Con, Control.
To confirm the effect of DAX-1 on GR transrepression, COS-1 cells were transfected with an AP-1 responsive construct (pAP-1-Luc), and AP-1 (c-Jun and c-Fos) in the presence or absence of GR and/or DAX-1 (Fig. 2B). Similar to NF-κB, dex treatment suppressed 50% of AP-1 transactivation in cells cotransfected with GR and AP-1. Expression of DAX-1 slightly enhanced constitutive AP-1 reporter activity, and cotransfection of DAX-1 and AP-1 caused a further synergistic increase. When cells were transfected with GR, DAX-1, and AP-1, dex induced a 50% decrease in AP-1-mediated activity. This reduction is quantitatively similar to that obtained in cells transfected with AP-1 and GR alone. Together these data demonstrate that DAX-1 has little effect on GR-mediated transrepression.
It is interesting that DAX-1 had a synergistic effect with p65 and/or c-Fos/Jun on their corresponding reporter genes. However, this synergy is probably due to the nonspecific activation of the basal transcription machinery resulting from corepressor sequestration by DAX-1 because no physical interaction between DAX-1 and p65 was detected by coimmunoprecipitation (data not shown).
DAX-1 Has Selective Effects on Endogenous GR-Regulated Genes
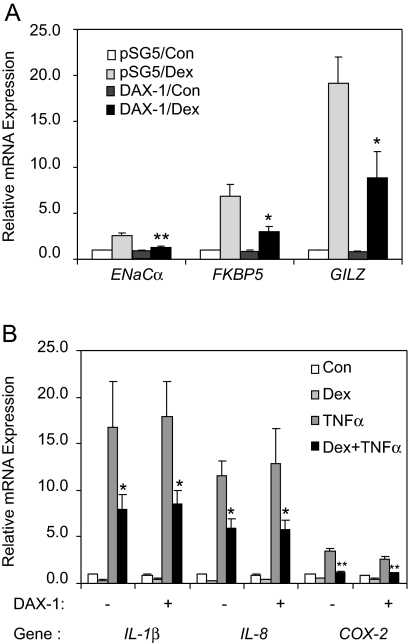
To examine the effect of DAX-1 on endogenous GR-transactivated target genes, glucocorticoid induced leucine zipper (GILZ), FK506 binding protein 51 (FKBP5), and epithelial sodium channel α (ENaCα) were selected as bona fide glucocorticoid targets (30,31,32,33,34,35). After transfection with DAX-1 or mock plasmid pSG5, A549 cells were treated with 100 nm dex or vehicle. Quantification of target mRNAs was determined by real-time PCR and normalized to cyclophilin B, a gene shown to be unresponsive to glucocorticoids (36). As shown in Fig. 3A, dex increased expression of ENaCα, FKBP5, and GILZ approximately 2.6-, 6.9-, and 19.2-fold, respectively. In cells cotransfected with DAX-1, the magnitude of dex-induced up-regulation was significantly lower, indicating that DAX-1 inhibits the ability of endogenous GR to transactivate target genes in the context of normal chromatin.
Figure 3.
DAX-1 Has Selective Effects on Endogenous GR Target Genes
A, Genes transactivated by GR. A549 cells were transfected with 0.5 μg pSG5 or DAX-1, and then treated with or without 100 nm dex in low serum OPTI-MEM media for 3 h. B, Genes transrepressed by GR. A549 cells were transfected with 0.5 μg pSG5 or DAX-1 and then treated with either vehicle, 10 ng/ml TNFα, 100 nm dex, or both for 2 h. Total RNA was extracted and target gene expression was determined using real-time quantitative PCR, after normalization to the internal control, cyclophilin B. The mean ± se from at least three independent experiments is shown. *, P < 0.05; and **, P < 0.01 represent significant differences between cells treated with TNFα and cells treated with TNFα and dex. Con, Control.
Next, we investigated the effect of DAX-1 on GR transrepression of the endogenous targets IL-1β, IL-8, and COX-2. Induction of these genes by TNFα-mediated activation of NF-κB has been shown previously to be repressed by glucocorticoids (37,38,39). DAX-1 or mock vector-transfected A549 cells were treated with vehicle, 10 ng/ml TNFα, 100 nm dex, or both for 2 h. This time period was found to be the point of maximum gene expression during preliminary time-course experiments (data not shown). TNFα dramatically activated expression of IL-1β (15- to 20-fold), IL-8 (10- to 15-fold), and COX-2 (3- to 5-fold) (Fig. 3B). In contrast, dex treatment inhibited transcription of each of these target genes under both constitutive and TNFα-induced conditions. More importantly, the same extent of suppression was observed on all three genes regardless of the presence or absence of DAX-1, suggesting that DAX-1 has no effect on ligand-activated GR transrepression of endogenous target genes. Furthermore, expression of DAX-1 did not affect levels of the endogenous GR target genes, unlike the transiently transfected reporter constructs.
DAX-1 Physically Interacts with GR
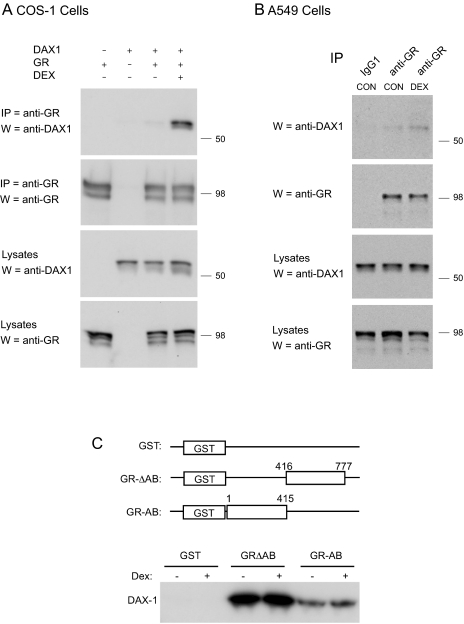
To better understand the molecular basis for DAX-1-mediated suppression of GR transactivation, we investigated whether DAX-1 physically associates with GR. As shown in Fig. 4A, DAX-1 was coimmunoprecipitated by a GR-specific antibody, demonstrating that DAX-1 and GR do interact when expressed in transfected COS-1 cells. Moreover, glucocorticoid treatment promoted an increase in the association because more DAX-1 was coimmunoprecipitated from cells treated with dex than with vehicle, even though similar levels of GR were immunoprecipitated and DAX-1 was expressed at similar levels in the lysates (compare outer two lanes, Fig. 4A). To investigate whether endogenous DAX-1 and GR can form a complex, we performed coimmunoprecipitation experiments in A549 cells that express both native proteins. As shown in Fig. 4B, DAX-1 was coimmunoprecipitated by the GR-specific antibody, and dex treatment promoted an increase in the association consistent with our observations in COS-1 cells.
Figure 4.
DAX-1 Physically Interacts with GR
A, Coimmunoprecipitation in COS-1 cells. COS-1 cells were transfected with GR, DAX-1, or both GR and DAX-1. Cells were treated with vehicle (CON) or 100 nm dex for 1 h as indicated. Lysates were prepared and proteins immunoprecipitated (IP) with a mouse anti-GR antibody. Recovered proteins were resolved by SDS-PAGE and probed by Western blotting (W) using a rabbit anti-DAX-1 antibody. The membrane was then washed and reprobed with the mouse anti-GR antibody. Input levels of DAX-1 and GR are shown in lower two blots. B, Coimmunoprecipitation in A549 cells. A549 cells expressing endogenous GR and DAX-1 were treated with vehicle (CON) or 100 nm dex for 15 min. Lysates were prepared and proteins immunoprecipitated with a mouse anti-GR antibody or an equivalent amount of a mouse IgG1 antibody that does not recognize GR. Recovered proteins were analyzed as described above. C, GST pull-down assay. GST-GR deletion constructs were expressed in bacteria, and partially purified lysates were incubated in the presence or absence of dex with DAX-1, which was synthesized by in vitro transcription and translation reaction. The complexes were pulled down by glutathione-couple agarose beads, washed and resolved by Western blotting, and probed with antibodies against DAX-1. Blots shown are representative of two to four independent experiments. CON, control.
To further confirm this observation as well as to define the regions of GR responsible for the interaction, glutathione-S-transferase (GST) pull-down assays were performed. DAX-1 was highly enriched by the GST-GR fusion proteins as opposed to the GST alone (Fig. 4C), even though the amount of the GST protein used in the experiment was greater than the amount of the GST-GR chimeras (data not shown). Both of the GST-GR fusions were expressed at similar levels (data not shown), yet the C-terminal half of GR displayed a significantly greater binding of DAX-1 (Fig. 4C). This observation is consistent with previous reports that the C terminus of nuclear hormone receptors is mainly responsible for interaction with DAX-1 (15,16,40). In contrast to the coimmunoprecipitation studies discussed above, dex treatment did not affect the level of DAX-1 bound to the C-terminal half of GR. This most likely reflects conformational differences between the full-length GR expressed in cells and a fragment of GR expressed as a GST-fusion protein in bacteria.
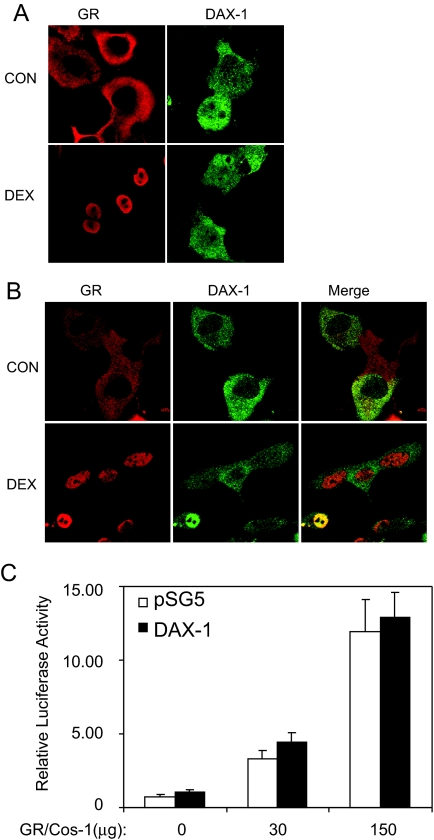
DAX-1 Does Not Block Ligand-Induced GR Nuclear Translocation and DNA Binding
Next, we investigated whether DAX-1 affects ligand-induced GR nuclear localization. COS-1 cells expressing GR alone, DAX-1 alone, or both GR and DAX-1 were treated with or without 100 nm dex, and the distribution of the proteins was assessed by immunocytochemistry. In cells expressing GR alone (Fig. 5A, left panels), the receptor was predominantly located in the cytoplasm of cells in the absence of hormone. After a 1-h treatment with dex, GR was detected exclusively in the nuclei of the transfected cells. In cells expressing DAX-1 alone (Fig. 5A, right panels), a diverse distribution pattern was observed for DAX-1 that was unaffected by dex treatment. DAX-1 was found either primarily in the cytoplasm or equally distributed between the cytoplasm and nucleus in most of the cells. In some cells, however, DAX-1 resided predominantly in the nucleus. This diversity of DAX-1 localization is in agreement with previous observations (15).
Figure 5.
DAX-1 Does Not Affect GR Nuclear Translocation and Interaction with GREs
A, COS-1 cells were transfected separately with GR or DAX-1. The cells were then treated with vehicle (CON) or 100 nm dex for 1 h and processed for immunocytochemistry. Shown are representative confocal microscopic images of the distribution of GR (red) and DAX-1 (green). B, COS-1 cells were cotransfected with both GR and DAX-1. The cells were then treated with vehicle (CON) or 100 nm dex for 1 h and processed for immunocytochemistry. Shown are representative confocal microscopic images of the distribution of GR (red) and DAX-1 (green). As indicated by the yellow color in the merged image, the distribution of GR and DAX-1 overlay one another in both the cytoplasm and nucleus. C, GR interaction with GREs. The effect of DAX-1 on GR interaction with GREs was determined using cytosolic ELISA as described in Materials and Methods. GR-containing COS-1 cell lysate (0, 30, and 150 μg) was supplemented with control COS-1 lysate (without GR) to achieve the same amount of total protein (150 μg) in each sample. Samples were then added to 96-well plates previously coated with biotinylated TAT-GRE, and the amount of GR bound to TAT-GRE was immunodetected using anti-GR primary antibody with the SuperSignal ELISA Fento Maximum Sensitivity Substrate and represented by luciferase activity. The data are plotted as mean ± se from three independent experiments.
In cells coexpressing both proteins, the presence of DAX-1 did not appear to alter the distribution of GR (Fig. 5B). Ligand-free GR was still contained predominantly in the cytoplasm (Fig. 5B, upper left panel), and ligand-bound GR was still detected only in the nuclei (Fig. 5B, lower left panel). In a similar set of experiments, expression of DAX-1 did not alter the cellular distribution of a yellow fluorescent protein (YFP)-GR fusion protein (data not shown). These data suggest that DAX-1 does not affect gross ligand-induced GR nuclear translocation, which is consistent with its selective effect on GR transactivation. Furthermore, expression of GR did not appear to alter the cellular distribution of DAX-1. DAX-1 still showed a diverse staining pattern independent of hormone treatment (Fig. 5B, middle panels). Finally, as indicated by the yellow color in the merged images (Fig. 5B, right panels), the distributions of the two proteins overlay one another in both the cytoplasm and nucleus consistent with GR and DAX-1 forming a complex.
To determine whether DAX-1 inhibits GR interaction with GREs, cytosolic ELISA was conducted. As shown in Fig. 5C, luciferase activity was detected in a concentration-dependent manner in response to the total input of dex-bound GR. The presence of DAX-1 in the complex had no effect on the amount of GR bound to GRE at either concentration, suggesting that DAX-1 does not inhibit GR binding to GREs.
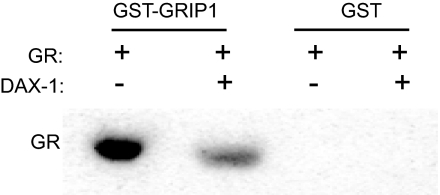
DAX-1 Competes with GR-Interacting Protein 1 (GRIP1) for Binding to GR
We hypothesized that inhibition of GR transactivation by DAX-1 is due to the latter’s ability to compete with coactivators that bind GR such as GRIP1. To test this hypothesis, GST-pull-down assays were performed with GST-GRIP1 [amino acids (aa) 563-1121] and ligand-activated GR in the presence or absence of DAX-1. As shown in Fig. 6, GR protein was highly enriched by GST-GRIP1 fusion protein as opposed to GST alone, confirming the interaction between GRIP1 and GR (41,42,43). In the presence of DAX-1, the amount of GR precipitated by the GST-GRIP1 was significantly decreased, suggesting that DAX-1 competes with GRIP1 for binding to GR.
Figure 6.
DAX-1 Competes with GRIP1 for Binding to GR
GR was expressed in COS-1 cells by transient transfection. After transfection, the receptor was activated with 100 nm dex for 30 min, and cell lysates were collected. Equal amounts of lysate were subsequently incubated with either GST-GRIP1 or GST and in the presence or absence of in vitro translated DAX-1. Western blot analysis was performed to determine the level of GR protein. The blot is a representative of two independent experiments.
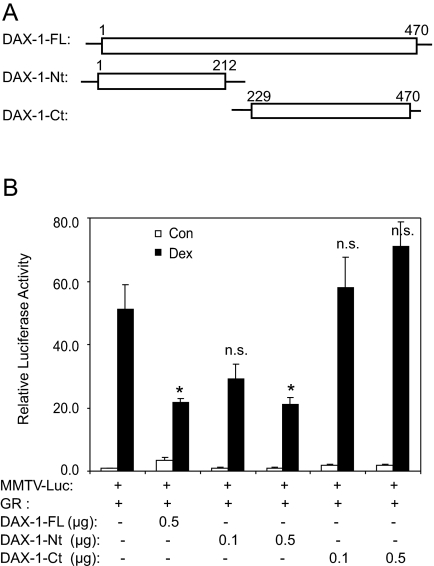
The N Terminus of DAX-1 Is Responsible for Suppression of GR Transcriptional Activity
To investigate the molecular basis by which DAX-1 represses GR, deletion constructs containing either the N terminus (aa 1–212, DAX-1-Nt) or C terminus (aa 229–470, DAX-1-Ct) were generated and assessed for their ability to modulate GR transactivation in COS-1 cells (Fig. 7). As shown previously, full-length DAX-1 inhibited GR activity by approximately 50%. The N-terminal half caused similar suppression whereas the C terminus did not, indicating that the former region of DAX-1 is required for the repression of GR signals. It should be noted that expression of the C-terminal half of DAX1 could not be determined at the protein level because available antibodies only recognize the N terminus of DAX-1. However, at the mRNA level, quantitation by real-time PCR shows the C terminus to be expressed at the same level as full-length DAX-1 (data not shown). At higher concentrations, the C terminus slightly enhanced GR transactivation. However, it is likely that this increase resulted from nonspecific enhancement because DAX-1 is known to interact with other corepressors via its C terminus and quench their inhibitory effect (13,18).
Figure 7.
The N Terminus of DAX-1 Is Responsible for Its Suppressor Activity on GR Transactivation
DAX-1 deletion constructs were generated using site-directed mutagenesis (A) and assessed for their activity in repressing GR transactivation in COS-1 cells (B). COS-1 cells were transfected overnight using TransIT transfection reagents mixed with MMTV-Luc, pRL, GR, DAX-1-FL (the full-length DAX-1), DAX-1-Ct, or DAX-1-Nt plasmids. The cells were then fed with fresh OPTI-MEM for 8 h followed by incubation with 100 nm dex or vehicle for an additional 20 h. Luciferase activity was determined, and firefly luciferase activity was normalized to Renilla luciferase activity. The data are plotted as mean ± se from at least three independent experiments. *, P < 0.05 represents significant differences between samples transfected with pSG5 and those transfected with DAX-1. P > 0.05. n.s., No significant difference; Con, control.
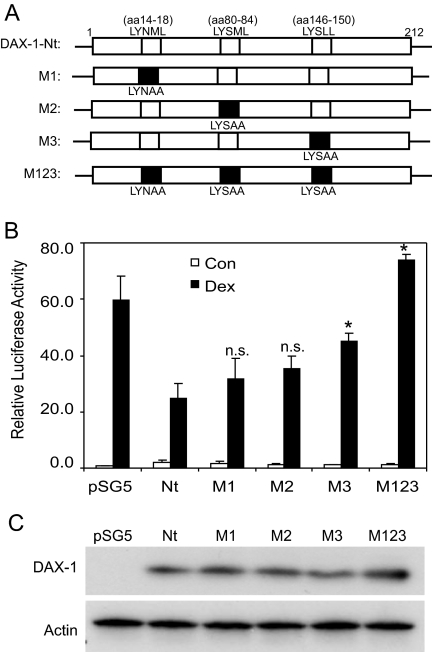
Three LXXLL Motifs Are Required for Repression of GR Transactivation as Well as Interaction with GR
DAX-1 possesses three repetitive regions in its N terminus, each of which contains a leucine-rich motif (LXXLL sequence) resembling the NR-box of nuclear receptor coactivators (17). To investigate whether these motifs are accountable for its repressor activity on GR, site-directed mutagenesis was used to convert each motif to an inactive form (LXXAA) either individually or in combination (Fig. 8A). The resultant mutants were assessed for their ability to modulate GR-mediated transcription in COS-1 cells. As shown earlier in Fig. 7B, the wild-type DAX-1 N terminus repressed GR transactivation by about 50% (Fig. 8B). Disruption of the individual LXXLL motifs resulted in less repression. Among these three motifs, the third one (aa 146–150) appears to play a more critical role than the other two, because mutation of this fragment caused a significant loss of repressor activity. In accordance, mutation of all three motifs M123 completely abolished the inhibitory effect, indicating that all three motifs are required to achieve full suppression of GR. The possibility that the loss of repressor activity was due to variations in protein expression of the mutants was ruled out because Western analysis demonstrated comparable levels of DAX-1 protein among the various mutants (Fig. 8C).
Figure 8.
The LXXLL Motifs Are Required for Suppression of GR by DAX-1
A, Schematics of LXXLL mutants by site-directed mutagenesis. B, Analysis of inhibitory effects of the LXXLL mutants on GR transactivation was assessed in COS-1 cells. COS-1 cells were transfected overnight using TransIT transfection reagents mixed with MMTV-Luc, pRL, GR, DAX-1-Nt, and various N-terminal mutants. The cells were then fed with fresh OPTI-MEM for 8 h followed by incubation with 100 nm dex or vehicle for an additional 20 h. Luciferase activity was determined and firefly luciferase activity was normalized to Renilla luciferase activity. The data are plotted as mean ± se from at least three independent experiments. *, P < 0.05 represents significant differences between dex-treated samples transfected with DAX-1-Nt and those transfected with LXXLL motif mutants. P > 0.05. n.s., No significant difference. C, Levels of protein expression for various DAX-1 mutants by Western blotting. A representative blot from three independent experiments is shown. Con, Control.
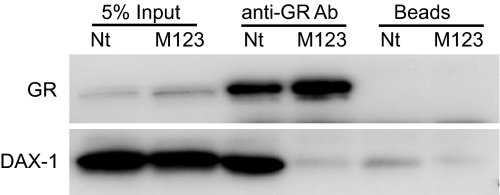
The loss of repressor activity for M123 may be due to its inability to interact with GR. To evaluate this option, coimmunoprecipitations were performed, and results are shown in Fig. 9. It is apparent that the GR-specific antibody precipitated DAX-1-Nt, but not M123 under the same conditions. These data indicate that the LXXLL motifs are necessary for physical interaction between DAX-1 and GR.
Figure 9.
The LXXLL Motifs Are Required for Interaction of DAX-1 with GR
Coimmunoprecipitation of GR with DAX-1-Nt or DAX-1-Nt-M123. COS-1 cells were transfected with GR (2 μg) together with either DAX-1-Nt (8 μg) or DAX-1-Nt-M123 (8 μg overnight). Cleared cell lysate was processed for coimmunoprecipitation in the presence (labeled as “anti-GR Ab”) or absence (labeled as “Beads”) of anti-GR antibody. Protein A beads were added, and the complexes were further incubated for an additional hour. The beads were precipitated and washed, and proteins were eluted and resolved on SDS-PAGE. Total cell lysate (5%) used for immunoprecipitation was loaded on the gel to serve as a positive control to demonstrate the presence of GR and DAX-1-Nt or M123. The presence of GR and DAX-1 was detected by Western blotting with the corresponding antibodies. The blot is a representative of three experiments. Ab, Antibody.
DAX-1 Antagonizes Coactivation of GR by GRIP1
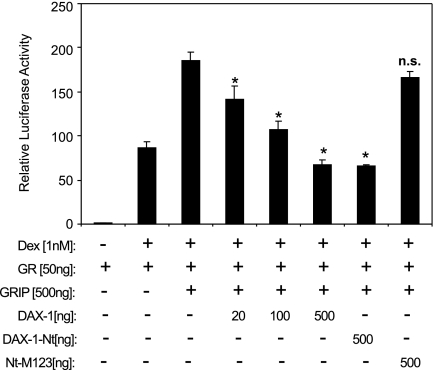
We hypothesized that competition of DAX-1 with coactivators such as GRIP1 for binding to GR should result in antagonism of GRIP1-elicited coactivation of GR. To investigate this question, COS-1 cells were transiently transfected with GRE2-Luc, GR, and GRIP1 in the presence or absence of increasing concentrations of DAX-1 or its mutants. As expected, GRIP1 enhanced GR transactivation dramatically (Fig. 10). This enhancement is blocked by DAX-1 in a concentration-dependent manner. Furthermore, we demonstrate that this blockage is mediated by the DAX-1-Nt, and in particular the LXXLL-motifs, because mutation of all three motifs completely abolished the inhibitory effect of DAX-1 on GRIP1-stimulated GR transactivation.
Figure 10.
DAX-1 Antagonizes GRIP1-Elicited Coactivation of GR
COS-1 cells were transfected overnight using TransIT transfection reagents mixed with 100 ng pRL, 400 ng MMTV-Luc, 50 ng GR, 500 ng GRIP, and 20, 100, and 500 ng DAX-1, 500 ng DAX-1-Nt, or 500 ng DAX-1-M123 plasmid. The cells were fed with fresh OPTI-MEM for 8 h followed by incubation with 100 nm dex or vehicle for an additional 20 h. Luciferase activity was determined, and firefly luciferase activity was normalized to the Renilla luciferase activity. Data are plotted as mean ± se from at least three independent experiments. *, P < 0.05 represents significant differences between extracted samples transfected with mock plasmid and those transfected with DAX-1 constructs. P > 0.05. n.s., No significant difference.
DISCUSSION
In the current study, we demonstrate that DAX-1 is a selective GR-modulating protein that specifically inhibits ligand-induced GR transactivation but not transrepression by competing for coactivator binding to GR. These findings, together with the known effect of DAX-1 on steroid production, suggest that DAX-1-mediated perturbation of GR signaling occurs at both the hormonal and the receptor levels.
DAX-1 is generally characterized as a corepressor protein that has been proposed to inhibit the actions of several nuclear receptors via diverse mechanisms along receptor signaling cascades. For example, DAX-1 blocks transcription of its own gene and the steroidogenic acute regulatory protein (StAR) by associating with the stem loop regions of their respective promoter sequences (12). In contrast, blockage of nuclear translocation accounts for the inhibitory effect of DAX-1 on the androgen receptor (15). DAX-1 is also a dominant-negative regulator of retinoic acid receptor (RAR)-mediated transcription by competing with RAR for binding to DNA-responsive elements (23). Here, we demonstrate that DAX-1 inhibits GR actions by inhibiting coactivator recruitment, and not via blocking GR nuclear translocation or interaction with GREs (Fig. 5). By this mechanism, DAX may have the opportunity to more finely regulate GR transcriptional activity because the antagonism will be a function of the expression and relative levels of coregulators.
Suppression of nuclear receptor activity by DAX-1 could involve either displacement of coactivators (16,17), recruitment of corepressors (13), or both (17). Unlike most other members of the nuclear receptor family, DAX-1 possesses a unique N terminus that contains 3.5 repeats of 65–67 amino acids, each of which harbors a LXXLL motif (17). Our study demonstrates that these N-terminal motifs of DAX-1 are responsible for inhibition of GR activity. These data indicate that the interaction mediated by the LXXLL motifs between these two nuclear receptors is a prerequisite for the suppression of GR by DAX-1, indicating that displacement of coactivators by the LXXLL motifs is the likely mechanism. Although displacement of coactivators has also been suggested as a mechanism for DAX-1 inhibition of Nur77 (16,17), it is conceivable that the mechanism of DAX-1 inhibition of GR is distinct from that of Nur77. In the case of GR, the N terminus of DAX-1 is not only necessary but also sufficient in achieving the full inhibition by the wild-type DAX-1, whereas in the case of Nur77 it is the C terminus, but not the N terminus, that is indispensable. Mechanistically, DAX-1 suppression of GR is more in line with that of estrogen receptor where the N-terminal LXXLL motifs of DAX-1 are essential for inhibition of both nuclear receptors (17).
After association with other nuclear receptors via its N terminus, DAX-1 has been shown to actively recruit corepressors such as nuclear receptor corepressor (13) or Alien (18) via its ligand-binding domain, thus achieving its suppressive effect (44). However, our data indicate that inhibition of GR in the current cellular context occurs independent of the C terminus of DAX-1, suggesting that recruitment of other corepressors via this mechanism is not required. However, we cannot rule out that DAX-1 may recruit corepressors to further down-regulate GR activity. This is a possibility in cells where abundant DAX-1-interacting corepressors are present.
Rat DAX-1 has been shown to be transcriptionally up-regulated by dex-bound GR in a complex with steroidogenic factor-1 (45). In addition, our data using human lung carcinoma A549 cells suggested that DAX-1 may be down-regulated by ligand-activated GR (data not shown). Together these data suggest that GR plays an important role in the transcriptional regulation of DAX-1, possibly via a species-, or tissue-specific manner. Considering that DAX-1 is in turn suppressive of GR transactivational potential, these data indicate that a complex feedback loop involving the two nuclear receptors may be present. Further characterization of the feedback loop is necessary to better reveal its potential involvement in modulating steroidogenesis and hormone actions.
DAX-1 inhibits the transactivational potential of GR, but the maximum suppression achieved was only 50%. The exact molecular basis for this is unclear, but it could be accounted for by several possibilities. First, the limited amount of DAX-1 protein expressed as a result of transient transfection may not be sufficient to achieve the full inhibition. Second, a dynamic equilibrium may exist between GR and LXXLL-containing coactivators and/or corepressors. Finally, it is well known that GR contains two transactivation domains, one of which is ligand-independent N-terminal domain 1 and another is ligand-dependent C-terminal domain 2. It appears that DAX-1 affects only the ligand-elicited transcriptional activation.
Deletion or mutation of DAX-1 is known to cause AHC associated with HH, leading to decreased levels of circulating glucocorticoids and mineralocorticoids. Furthermore, our current data demonstrated that DAX-1 could also directly suppress GR transcriptional activity. Together, these data suggest that DAX-1 plays a dual role in modulating glucocorticoid action at both the hormone level and the receptor level. Whereas the pathogenesis of AHC/HH resulting from DAX-1 dysfunction most likely takes place at a critical stage of development of the adrenal gland, the effect of DAX-1 on GR could be more widespread because it simply depends on the specific tissue levels of the coregulator. Numerous mutations or deletions have been identified in the dax-1 gene from patients with AHC. It is conceivable that the mutations in the N terminus, in particular those that impact the integrity of LXXLL motifs or their accessibility to nuclear receptors, may play an important role in modifying GR transactivation potential, resulting in an altered response of GR to endogenous and/or exogenous glucocorticoids.
It is interesting that DAX-1 functions as a selective GR-modulating protein by inhibiting only ligand-induced GR transactivation and not transrepression. It is widely accepted that transrepression activities of GR are responsible for the beneficial effects of glucocorticoids in the treatment of inflammation and tissue rejection, whereas the adverse side effects are associated with transactivation activities of GR during long-term administration of glucocorticoids. Thus, dissociation of GR transactivation from transrepression has been an active field of research for apparent therapeutic value. Our studies showing that DAX-1 is uniquely involved in GR-mediated transactivation represent an undescribed avenue for future research to achieve an improved therapeutic index.
MATERIALS AND METHODS
Reagents and Antibodies
Dexamethasone (dex) (1,4-pregnadien-9α-fluoro-16α-methyl-11β,17,21-triol-3,20-dione) was purchased from Steraloids (Newport, RI). Goat antirabbit antibodies conjugated with horseradish peroxidase were purchased from Jackson ImmunoResearch Laboratories, Inc. (West Grove, PA). The goat antimouse Alexa Fluor 594 and goat antirabbit Alexa Fluor 488 antibodies were from Invitrogen (Carlsbad, CA). The mouse anti-GR antibody was purchased from BD Biosciences. Protease inhibitors were purchased from Roche Molecular Biochemicals (Indianapolis, IN). Oligonucleotide primers for PCR and mutagenesis were synthesized by Integrated DNA Technologies, Inc. (Coralville, IA). The primer probe sets for real-time quantitative RT-PCR were obtained from Applied Biosystems (Foster City, CA). All mutagenesis experiments were conducted using the QuikChange Site-Directed Mutagenesis kit from Stratagene (La Jolla, CA). The peroxidase-labeled antirabbit immunoglobulin G secondary antibody and enhanced chemiluminescence detection kit were purchased from Amersham Pharmacia Biotech (Piscataway, NJ). The quick TNT coupled reticulocyte lysate translation system was purchased from Promega Corp. (Madison, WI).
Plasmids
The following plasmids have been described previously: pCMV5-hGRα, GR-responsive reporter constructs MMTV-Luc and tyrosine aminotransferase (TAT)-GRE2-Luc (46,47), Renilla luciferase construct pGL3-hRL (48), YFP-GR fusion construct pEYFP-hGRα (49), and human NF-κB p65 subunit construct pCMV-p65 and its reporter gene 3xMHC-Luc (29,47,50). The AP-1 expression plasmid (pCMV-cFos and pCMV-cJun) was obtained from Dr. Robert Scheinman (University of Colorado Health Sciences Center, CO), and pAP-1-Luc (the AP-1-responsive reporter construct) was purchased from Stratagene (La Jolla, CA).
The GST-hGR fusion plasmid was constructed by inserting the PCR product of full-length hGRα cDNA into the unique BamHI and XhoI sites of pGEX-4T-1 vector. GST-hGRα-AB containing the N terminus (aa 1–415) of GR was generated by introducing a stop codon at aa 416 of GST-hGRα plasmid by site-directed mutagenesis. The plasmid pGEX-4T-1-hGRαΔAB (R602S) was produced by inserting the PCR-amplified hGRαΔAB fragment (aa 416–777) into the BamHI and XhoI sites of pGEX4T-1 vector; the resultant plasmid was mutated (Arg-Ser) at site 602 to increase protein solubility in Escherichia coli cells.
The plasmid pSG5-hDAX-1 was kindly provided by Dr. Eckardt Treuter (Karolinska Institute, Sweden). DAX-1 deletion constructs were generated by digesting pSG5-DAX-1 with restriction enzymes BglII and BspE1. The longer fragment containing DAX-1 N terminus and intact pSG5 vector was religated and was mutated to introduce a stop codon at aa 212 of hDAX-1. The resultant plasmid is called pSG5-DAX-1-Nt. The shorter fragment containing DAX-1 C terminus was modified by restriction digestion and inserted into the pSG5 vector that was predigested with EcoRI, filled in with Klenow, and subsequently digested with BamHI. The ligation product was mutated to introduce a start codon at aa 229, and the resultant plasmid is referred to as pSG5-DAX-1-Ct. LXXLL mutants (M1, M2, M3, M123) were produced by QuikChange site-directed mutagenesis using pSG5-DAX1-Nt as the template based on standard protocols with the exception that a 65 C annealing temperature was used.
Mammalian Cell Transfections
A549 cells were maintained in DMEM/F12 (Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum and antibiotics at 37 C in 5% CO2 atmosphere. COS-1 cells were maintained in DMEM supplemented with 10% calf serum combined with heat-inactivated fetal bovine serum and l-glutamine as well as antibiotics at 37 C in 5% CO2 atmosphere. A549 or COS-1 cells (1.0 × 105/well) were plated in 12-well plates 1 d before transfection. Cells were transfected using either GenePORTER2 (Gene Therapy Systems, San Diego, CA) or TransIT (Mirus, Madison, WI) transfection reagents with appropriate plasmids in duplicate wells based on the manufacturer’s instructions. The following day, the cells were fed with OPTI-MEM media (Invitrogen) containing reduced serum for 8 h followed by incubation with 100 nm dex or vehicle for an additional 20 h. Total time from transfection to assay was between 44 and 48 h. For luciferase assays, cells were washed twice with PBS and lysed in 150 μl 1× cell lysis buffer (Roche) at room temperature for 20–30 min with gentle shaking. Lysates (15 μl) were assayed for their luciferase activity using Dual-Luciferase Reporter Assay (Promega) by LuminoScan luminometer (Thermo Labsystems, Milford, MA). Firefly luciferase activity was normalized to the internal control Renilla luciferase.
Real-Time PCR
For real-time quantitative RT-PCR, cells were washed with PBS, and total RNA was extracted using RNeasy RNA extraction kit (QIAGEN, Valencia, CA). The abundance of individual mRNAs was determined by the ABI Prism 7900 HT Sequence Detection System (Applied Biosystems) using the absolute quantification method. Amplification of target mRNA and internal control cyclophilin B was performed in separate wells, and the target mRNA was normalized to the internal control before further analysis.
Coimmunoprecipitation
COS-1 cells were cotransfected with GR alone, DAX-1 alone, or both GR and DAX-1. Cells were harvested 24 h later and replated using fully supplemented medium stripped of endogenous steroids. The next day, dishes were treated with either vehicle or 100 nm dex for 1 h. Cells were harvested, resuspended in lysis buffer (20 mm Tris-HCl, pH 7.5; 2 mm EDTA; 150 mm NaCl; 0.5% Triton X-100) containing protease inhibitors, and incubated with rotation for 90 min at 4 C in the absence or presence of 1 μm dex. After centrifugation, the supernatant was removed and protein concentration was determined. Equivalent amounts of protein were precleared for 1 h with protein G agarose beads. The mouse anti-GR antibody was then added to the samples and incubated overnight at 4 C. The next day protein G agarose beads were added for a 90-min incubation at 4 C. The beads were washed five times with cold lysis buffer, resuspended in sample buffer containing 5% 2-mercaptoethanol, and heated for 10 min at 95 C. Recovered proteins were resolved on 8% Tris-glycine SDS-PAGE, transferred to nitrocellulose, and immunoblotted with an epitope-purified rabbit anti-DAX-1 antibody developed in our laboratory using the peptide sequence published previously (51). Blots were washed and reprobed with the mouse anti-GR antibody. For the coimmunoprecipitation of endogenous GR and DAX-1, A549 cells were treated with vehicle or 100 nm dex for 15 min and processed as described above. Proteins were immunoprecipitated with the mouse anti-GR antibody or an equivalent amount of a mouse IgG1 antibody that does not interact with GR. For both COS-1 and A549 experiments, input levels of GR and DAX-1 were assessed by immunoblotting equivalent amounts of protein with the mouse anti-GR and rabbit anti-DAX-1 antibodies.
GST-Protein/Protein Interaction Assay
GST pull-down assays were performed using ProFound Pull-Down GST Protein:Protein Interaction Kit (Pierce Chemical Co., Rockford, IL) according to the manufacturer’s instructions. Briefly, pGEX 4T-1, pGEX-GRΔAB, and pGEX-GRAB expressed in BL-20 Gold E. coli were partially purified with glutathione-coupled Sepharose beads and incubated overnight at 4 C with DAX-1 expressed by in vitro quick-coupled transcription and translation system (TNT) (Promega). The beads were washed and proteins eluted in electrophoresis sample buffer by boiling for 10 min. The denatured proteins were resolved in SDS-PAGE, transferred to a polyvinylidine difluoride membrane, and immunodetected as described above.
Similarly, to study the interaction among GR, GRIP1, and DAX-1, pGEX-4T-1 and pGEX-2TK-GRIP1 were expressed in BL-20 Gold E. coli, and the proteins were partially purified with glutathione-coupled Sepharose beads. The beads were divided and incubated overnight at 4 C with ligand-activated GR that was transiently expressed in COS-1 cells and with or without DAX-1 expressed by TNT reaction. The beads were washed and proteins eluted in electrophoresis sample buffer by boiling for 10 min. The denatured proteins were resolved in SDS-PAGE, transferred to a polyvinylidine difluoride membrane, and immunodetected as described above.
Analysis of Intracellular Localization Using Confocal Microscopy
COS-1 cells were transfected with GR alone, DAX-1 alone, or both GR and DAX-1. Cells were harvested 24 h later and replated in 35-mm glass bottom dishes (MatTek, Ashland, MA) using fully supplemented medium stripped of endogenous steroids. The next day, dishes were treated with either vehicle or 100 nm dex for 1 h. Cells were then washed with cold PBS and fixed for 30 min at room temperature with 4% paraformaldehyde in PBS. The cells were then washed with PBS and permeabilized for 30 min at room temperature in PBS containing 0.1% vol/vol Triton X-100 and 2% wt/vol BSA. To detect GR and DAX-1, the mouse anti-GR antibody (1:1000) and the rabbit anti-DAX-1 antibody (1:1000) were added to the cells and incubated for 1 h at room temperature. The cells were then washed with PBS and incubated for 30 min at room temperature with 5% normal goat serum in PBS. The goat antimouse Alexa Fluor 594 (1:500) and goat antirabbit Alexa Fluor 488 (1:500) secondary antibodies were then added to the cells and incubated for 45 min at room temperature. The cells were washed in PBS and analyzed on a Zeiss laser scanning confocal microscope (LSM 510; Carl Zeiss, Thornwood, NY) Images were collected sequentially using dual excitation (488 nm from argon laser, 543 nm from HeNe laser) and emission filter sets (band pass, 500–530 nm; long pass, 560 nm).
Competitive DNA Binding ELISA
The effect of DAX-1 on GR/GRE binding was evaluated using competitive DNA binding ELISA as published previously with minor modification of the cytosol preparation (46). Specifically, COS-1 cells transfected with the hGR plasmid were washed with ice-cold PBS and scraped into centrifuge tubes placed on ice. Dex was added to the cells at a final concentration of 100 nm for 1 h to transform the receptor to an active state. Whole-cell lysate was then prepared by homogenization in MENG buffer (25 mm 3[N-morpholino]propanesulfonic acid, pH 7.1; 1 mm EDTA; 10% glycerol) supplemented with protease inhibitors. Homogenates were centrifuged at 22,000 × g for 10 min at 4 C, and protein concentrations were determined using a Bio-Rad protein assay kit (Bio-Rad Laboratories, Hercules, CA). Increasing amounts of GR-containing lysate were supplemented with nontransfected COS-1 lysate to obtain an equivalent amount of total protein among all samples. The combined sample was then mixed with either control or DAX-1-containing lysate for 1 h at 25 C. The complex was further incubated for 15 min in 1× DNA ELISA buffer (20 mm HEPES, 80 mm KCl, 5 mm MgCl2, 2 mm dithiothreitol, 10% glycerol) containing 0.1 mg/ml polydeoxyinosinic deoxycytidylic acid for a total of 100 μl. Samples were then added to 96-well plates previously coated with biotinylated TAT-GRE, and the amount of GR bound to TAT-GRE was immunodetected using anti-GR antibody 57 and the SuperSignal ELISA Femto Maximum Sensitivity Substrate (Pierce) as described previously (46).
Statistical Analysis
The data presented are the mean ± se. The statistical significance of differences between treatments was determined by nonparametric Mann-Whitney test. Values of P < 0.05 or less were considered statistically significant.
Acknowledgments
We thank Ms. Grace Kissling (National Institute of Environmental Health Sciences, Research Triangle Park, NC) for the statistical analysis; Dr. Diane Dong for making the plasmid pGEX-4T-1-hGRαΔAB; and Dr. Matt Yudt for pGEX-2TK-GRIP1 (aa 563-1121). We thank Mr. Jeff Reece for technical help on confocal microscopy; members of the Cidlowski laboratory for their critical reviews of the manuscript; and The National Cancer Institute Fellows Editorial Board for its constructive comments on the manuscript.
Footnotes
This work was supported by the Intramural Research Program of the National Institute of Environmental Health Sciences, National Institutes of Health.
Disclosure Statement: The authors have nothing to disclose.
First Published Online April 16, 2008
Abbreviations: aa, Amino acid; AHC, adrenal hypoplasia congenital; DAX-1, dosage-sensitive sex reversal-adrenal hypoplasia congenita critical region on the X-chromosome, gene 1; DAX-1-Ct, DAX-1 C terminus; DAX-1-Nt, DAX-1 N terminus; dex, dexamethasone; ENaCα, epithelial sodium channel α; FKBP5, FK506 binding protein 51; GILZ, glucocorticoid-induced leucine zipper; GR, glucocorticoid receptor; GRE, glucocorticoid-responsive element; GRIP1, GR-interacting protein 1; GST, glutathione-S-transferase; HH, hypogonadotropic hypogonadism; MMTV, mouse mammary tumor virus; NF-κB, nuclear factor-κB; TAT, tyrosine aminotransferase; YFP, yellow fluorescent protein.
References
- Coghlan MJ, Elmore SW, Kym PR, Kort ME 2003 The pursuit of differentiated ligands for the glucocorticoid receptor. Curr Top Med Chem 3:1617–1635 [DOI] [PubMed] [Google Scholar]
- Barnes PJ, Greening AP, Crompton GK 1995 Glucocorticoid resistance in asthma. Am J Respir Crit Care Med 152:S125–S140 [DOI] [PubMed] [Google Scholar]
- Langhoff E, Ladefoged J, Jakobsen BK, Platz P, Ryder LP, Svejgaard A, Thaysen JH 1986 Recipient lymphocyte sensitivity to methylprednisolone affects cadaver kidney graft survival. Lancet 1:1296–1297 [DOI] [PubMed] [Google Scholar]
- Kirkham BW, Corkill MM, Davison SC, Panayi GS 1991 Response to glucocorticoid treatment in rheumatoid arthritis: in vitro cell mediated immune assay predicts in vivo responses. J Rheumatol 18:821–825 [PubMed] [Google Scholar]
- Schacke H, Docke WD, Asadullah K 2002 Mechanisms involved in the side effects of glucocorticoids. Pharmacol Ther 96:23–43 [DOI] [PubMed] [Google Scholar]
- Bond WS 1977 Toxic reactions and side effects of glucocorticoids in man. Am J Hosp Pharm 34:479–485 [PubMed] [Google Scholar]
- Mills CM, Marks R 1993 Side effects of topical glucocorticoids. Curr Probl Dermatol 21:122–131 [DOI] [PubMed] [Google Scholar]
- Tarchalska-Krynska B 1994 [Glucocorticosteroids: mechanism of action, pharmacological effects, pharmacokinetics and adverse effects]. Otolaryngol Pol 48 (Suppl 17):41–48 (Polish) [PubMed] [Google Scholar]
- Davis GF 1986 Adverse effects of corticosteroids. II. Systemic. Clin Dermatol 4:161–169 [DOI] [PubMed] [Google Scholar]
- Coskey RJ 1986 Adverse effects of corticosteroids. I. Topical and intralesional. Clin Dermatol 4:155–160 [DOI] [PubMed] [Google Scholar]
- Buttgereit F, Song IH, Straub RH, Burmester GR 2005 [Current insights into the development of new glucocorticoid receptor ligands]. Z Rheumatol 64:170–176 (German) [DOI] [PubMed] [Google Scholar]
- Zazopoulos E, Lalli E, Stocco DM, Sassone-Corsi P 1997 DNA binding and transcriptional repression by DAX-1 blocks steroidogenesis. Nature 390:311–315 [DOI] [PubMed] [Google Scholar]
- Crawford PA, Dorn C, Sadovsky Y, Milbrandt J 1998 Nuclear receptor DAX-1 recruits nuclear receptor corepressor N-CoR to steroidogenic factor 1. Mol Cell Biol 18:2949–2956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agoulnik IU, Krause WC, Bingman III WE, Rahman HT, Amrikachi M, Ayala GE, Weigel NL 2003 Repressors of androgen and progesterone receptor action. J Biol Chem 278:31136–31148 [DOI] [PubMed] [Google Scholar]
- Holter E, Kotaja N, Makela S, Strauss L, Kietz S, Janne OA, Gustafsson JA, Palvimo JJ, Treuter E 2002 Inhibition of androgen receptor (AR) function by the reproductive orphan nuclear receptor DAX-1. Mol Endocrinol 16:515–528 [DOI] [PubMed] [Google Scholar]
- Song KH, Park YY, Park KC, Hong CY, Park JH, Shong M, Lee K, Choi HS 2004 The atypical orphan nuclear receptor DAX-1 interacts with orphan nuclear receptor Nur77 and represses its transactivation. Mol Endocrinol 18:1929–1940 [DOI] [PubMed] [Google Scholar]
- Zhang H, Thomsen JS, Johansson L, Gustafsson JA, Treuter E 2000 DAX-1 functions as an LXXLL-containing corepressor for activated estrogen receptors. J Biol Chem 275:39855–39859 [DOI] [PubMed] [Google Scholar]
- Altincicek B, Tenbaum SP, Dressel U, Thormeyer D, Renkawitz R, Baniahmad A 2000 Interaction of the corepressor Alien with DAX-1 is abrogated by mutations of DAX-1 involved in adrenal hypoplasia congenita. J Biol Chem 275:7662–7667 [DOI] [PubMed] [Google Scholar]
- Habiby RL, Boepple P, Nachtigall L, Sluss PM, Crowley Jr WF, Jameson JL 1996 Adrenal hypoplasia congenita with hypogonadotropic hypogonadism: evidence that DAX-1 mutations lead to combined hypothalmic and pituitary defects in gonadotropin production. J Clin Invest 98:1055–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanaria E, Bardoni B, Dabovic B, Calvari V, Fraccaro M, Zuffardi O, Camerino G 1995 Xp duplications and sex reversal. Philos Trans R Soc Lond B Biol Sci 350:291–296 [DOI] [PubMed] [Google Scholar]
- Swain A, Zanaria E, Hacker A, Lovell-Badge R, Camerino G 1996 Mouse Dax1 expression is consistent with a role in sex determination as well as in adrenal and hypothalamus function. Nat Genet 12:404–409 [DOI] [PubMed] [Google Scholar]
- Swain A, Narvaez V, Burgoyne P, Camerino G, Lovell-Badge R 1998 Dax1 antagonizes Sry action in mammalian sex determination. Nature 391:761–767 [DOI] [PubMed] [Google Scholar]
- Zanaria E, Muscatelli F, Bardoni B, Strom TM, Guioli S, Guo W, Lalli E, Moser C, Walker AP, McCabe ER, Meitinger T, Monaco AP, Sassone-Corsi P, Camerino G 1994 An unusual member of the nuclear hormone receptor superfamily responsible for X-linked adrenal hypoplasia congenita. Nature 372:635–641 [DOI] [PubMed] [Google Scholar]
- Muscatelli F, Strom TM, Walker AP, Zanaria E, Recan D, Meindl A, Bardoni B, Guioli S, Zehetner G, Rabl W, Schwarz HP, Kaplan J-C, Camerino G, Meitinger T, Monaco AP 1994 Mutations in the DAX-1 gene give rise to both X-linked adrenal hypoplasia congenita and hypogonadotropic hypogonadism. Nature 372:672–676 [DOI] [PubMed] [Google Scholar]
- Vanderbilt JN, Miesfeld R, Maler BA, Yamamoto KR 1987 Intracellular receptor concentration limits glucocorticoid-dependent enhancer activity. Mol Endocrinol 1:68–74 [DOI] [PubMed] [Google Scholar]
- Danielsen M, Stallcup MR 1984 Down-regulation of glucocorticoid receptors in mouse lymphoma cell variants. Mol Cell Biol 4:449–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cidlowski JA, Cidlowski NB 1981 Regulation of glucocorticoid receptors by glucocorticoids in cultured HeLa S3 cells. Endocrinology 109:1975–1982 [DOI] [PubMed] [Google Scholar]
- Silva CM, Powell-Oliver FE, Jewell CM, Sar M, Allgood VE, Cidlowski JA 1994 Regulation of the human glucocorticoid receptor by long-term and chronic treatment with glucocorticoid. Steroids 59:436–442 [DOI] [PubMed] [Google Scholar]
- Yudt MR, Jewell CM, Bienstock RJ, Cidlowski JA 2003 Molecular origins for the dominant negative function of human glucocorticoid receptor β. Mol Cell Biol 23:4319–4330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- U M, Shen L, Oshida T, Miyauchi J, Yamada M, Miyashita T 2004 Identification of novel direct transcriptional targets of glucocorticoid receptor. Leukemia 18:1850–1856 [DOI] [PubMed] [Google Scholar]
- Hubler TR, Scammell JG 2004 Intronic hormone response elements mediate regulation of FKBP5 by progestins and glucocorticoids. Cell Stress Chaperones 9:243–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannarile L, Zollo O, D'Adamio F, Ayroldi E, Marchetti C, Tabilio A, Bruscoli S, Riccardi C 2001 Cloning, chromosomal assignment and tissue distribution of human GILZ, a glucocorticoid hormone-induced gene. Cell Death Differ 8:201–203 [DOI] [PubMed] [Google Scholar]
- Mick VE, Itani OA, Loftus RW, Husted RF, Schmidt TJ, Thomas CP 2001 The α-subunit of the epithelial sodium channel is an aldosterone-induced transcript in mammalian collecting ducts, and this transcriptional response is mediated via distinct cis-elements in the 5′-flanking region of the gene. Mol Endocrinol 15:575–588 [DOI] [PubMed] [Google Scholar]
- Wang JC, Derynck MK, Nonaka DF, Khodabakhsh DB, Ha C, Yamamoto KR 2004 Chromatin immunoprecipitation (ChIP) scanning identifies primary glucocorticoid receptor target genes. Proc Natl Acad Sci USA 101:15603–15608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asselin-Labat ML, Biola-Vidamment A, Kerbrat S, Lombes M, Bertoglio J, Pallardy M 2005 FoxO3 mediates antagonistic effects of glucocorticoids and interleukin-2 on glucocorticoid-induced leucine zipper expression. Mol Endocrinol 19:1752–1764 [DOI] [PubMed] [Google Scholar]
- Hermoso MA, Matsuguchi T, Smoak K, Cidlowski JA 2004 Glucocorticoids and tumor necrosis factor α cooperatively regulate toll-like receptor 2 gene expression. Mol Cell Biol 24:4743–4756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haij S, Woltman AM, Bakker AC, Daha MR, van Kooten C 2002 Production of inflammatory mediators by renal epithelial cells is insensitive to glucocorticoids. Br J Pharmacol 137:197–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chivers JE, Cambridge LM, Catley MC, Mak JC, Donnelly LE, Barnes PJ, Newton R 2004 Differential effects of RU486 reveal distinct mechanisms for glucocorticoid repression of prostaglandin E2 release. Eur J Biochem 271:4042–4052 [DOI] [PubMed] [Google Scholar]
- Croxtall JD, van Hal PT, Choudhury Q, Gilroy DW, Flower RJ 2002 Different glucocorticoids vary in their genomic and non-genomic mechanism of action in A549 cells. Br J Pharmacol 135:511–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Kasahara M, Yoshioka H, Morohashi K, Umesono K 2003 LXXLL-related motifs in Dax-1 have target specificity for the orphan nuclear receptors Ad4BP/SF-1 and LRH-1. Mol Cell Biol 23:238–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong H, Kohli K, Trivedi A, Johnson DL, Stallcup MR 1996 GRIP1, a novel mouse protein that serves as a transcriptional coactivator in yeast for the hormone binding domains of steroid receptors. Proc Natl Acad Sci USA 93:4948–4952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong H, Kohli K, Garabedian MJ, Stallcup MR 1997 GRIP1, a transcriptional coactivator for the AF-2 transactivation domain of steroid, thyroid, retinoid, and vitamin D receptors. Mol Cell Biol 17:2735–2744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding XF, Anderson CM, Ma H, Hong H, Uht RM, Kushner PJ, Stallcup MR 1998 Nuclear receptor-binding sites of coactivators glucocorticoid receptor interacting protein 1 (GRIP1) and steroid receptor coactivator 1 (SRC-1): multiple motifs with different binding specificities. Mol Endocrinol 12:302–313 [DOI] [PubMed] [Google Scholar]
- Ito M, Yu R, Jameson JL 1997 DAX-1 inhibits SF-1-mediated transactivation via a carboxy-terminal domain that is deleted in adrenal hypoplasia congenita. Mol Cell Biol 17:1476–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gummow BM, Scheys JO, Cancelli VR, Hammer GD 2006 Reciprocal regulation of a glucocorticoid receptor-steroidogenic factor-1 transcription complex on the Dax-1 promoter by glucocorticoids and adrenocorticotropic hormone in the adrenal cortex. Mol Endocrinol 20:2711–2723 [DOI] [PubMed] [Google Scholar]
- Necela BM, Cidlowski JA 2004 A single amino acid change in the first zinc finger of the DNA binding domain of the glucocorticoid receptor regulates differential promoter selectivity. J Biol Chem 279:39279–39288 [DOI] [PubMed] [Google Scholar]
- Oakley RH, Sar M, Cidlowski JA 1996 The human glucocorticoid receptor β isoform. Expression, biochemical properties, and putative function. J Biol Chem 271:9550–9559 [DOI] [PubMed] [Google Scholar]
- Krone N, Riepe FG, Dorr HG, Morlot M, Rudorff KH, Drop SL, Weigel J, Pura M, Kreze A, Boronat M, de Luca F, Tiulpakov A, Partsch CJ, Peter M, Sippell WG 2005 Thirteen novel mutations in the NR0B1 (DAX1) gene as cause of adrenal hypoplasia congenita. Hum Mutat 25:502–503 [DOI] [PubMed] [Google Scholar]
- Schaaf MJ, Cidlowski JA 2003 Molecular determinants of glucocorticoid receptor mobility in living cells: the importance of ligand affinity. Mol Cell Biol 23:1922–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel MV, McKay IA, Burrin JM 2001 Transcriptional regulators of steroidogenesis, DAX-1 and SF-1, are expressed in human skin. J Invest Dermatol 117: 1559–1565 [DOI] [PubMed] [Google Scholar]
- Tamai KT, Monaco L, Alastalo TP, Lalli E, Parvinen M, Sassone-Corsi P 1996 Hormonal and developmental regulation of DAX-1 expression in Sertoli cells. Mol Endocrinol 10:1561–1569 [DOI] [PubMed] [Google Scholar]