Summary
Two-component signal transduction systems are the predominant means by which bacteria sense and respond to environmental stimuli. Bacteria often employ tens or hundreds of these paralogous signaling systems, comprised of histidine kinases (HK) and their cognate response regulators (RR). Faithful transmission of information through these signaling pathways and avoidance of detrimental cross-talk demand exquisite specificity of HK-RR interactions. To identify the determinants of two-component signaling specificity, we examined patterns of amino acid coevolution in large multiple sequence alignments of cognate kinase-regulator pairs. Guided by these results, we demonstrate that a subset of the coevolving residues is sufficient, when mutated, to completely switch the substrate specificity of the kinase EnvZ. Our results shed light on the basis of molecular discrimination in two-component signaling pathways, provide a general approach for the rational rewiring of these pathways, and suggest that analyses of coevolution may facilitate the reprogramming of other signaling systems and protein-protein interactions.
Genome sequencing projects have revealed that most organisms contain large expansions of a relatively small number of signaling families. For instance, the human genome contains large paralogous families of MAP kinases, receptor tyrosine kinases, and TGF-β receptors (Manning et al., 2002). These expansions have enabled organisms to rapidly diversify their information-processing capabilities without having to invent new signaling modalities. However, the expansion of a family of signaling proteins comes at a cost – cells must somehow maintain the specificity of distinct pathways and avoid unwanted cross-talk. Cells use a variety of mechanisms to enforce the specificity of related, but distinct signaling pathways (Schwartz and Madhani, 2004; Ubersax and Ferrell, 2007). In some cases, spatial mechanisms such as scaffolds and sub-cellular localization, or tissue-specific expression in multicellular organisms, can help to impose specificity. Differential timing of expression can also prevent cross-talk. In many cases though, the primary means of ensuring specificity resides at the level of molecular recognition (Newman and Keating, 2003; Skerker et al., 2005; Stiffler et al., 2007; Zarrinpar et al., 2003). However, identifying the amino acids responsible for such molecular-level discrimination is difficult. For signaling pathways, the structure of a kinase-substrate complex can help, but it is often insufficient in pinpointing the specificity determinants. Moreover, structures of signaling complexes are often not available or are difficult to produce given the transient nature of the interaction. Alanine-scanning mutagenesis can identify residues important for a given kinase-substrate interaction, but this approach does not differentiate between residues necessary for catalysis and those determining substrate selectivity. In short, better methods and approaches for mapping specificity determinants in signaling proteins are needed.
A complete understanding of kinase-substrate interaction specificity ultimately demands the successful, rational redesign of a kinase’s specificity. Previous efforts in this direction have mainly involved domain-swapping (Levskaya et al., 2005; Perraud et al., 1998; Utsumi et al., 1989), a relatively gross perturbation that does not yield detailed insight into kinase specificity and substrate selectivity. The rational redesign of a protein kinase thus remains a major challenge. In fact, there are still only a few cases in which the specificity of any protein-protein interaction has been completely redesigned in a systematic, generalizable way (reviewed in (Kortemme and Baker, 2004)).
In bacteria, the predominant family of signaling proteins is the two-component signal transduction system (Stock et al., 2000). These signaling pathways typically consist of a sensor histidine kinase (HK) that autophosphorylates and then transfers the phosphoryl group to a cognate response regulator (RR) that can effect changes in cellular physiology or behavior, often by changing gene expression (Figure 1A). These signaling systems are found in nearly all bacteria, with most species containing 20–30 HK-RR pairs and some containing as many as 200–300. Most histidine kinases have one or two cognate response regulators, and there appears to be minimal cross-talk between different pathways (reviewed in (Laub and Goulian, 2007)). Understanding how a single bacterial cell coordinates many highly related signaling pathways and prevents cross-talk remains a major challenge.
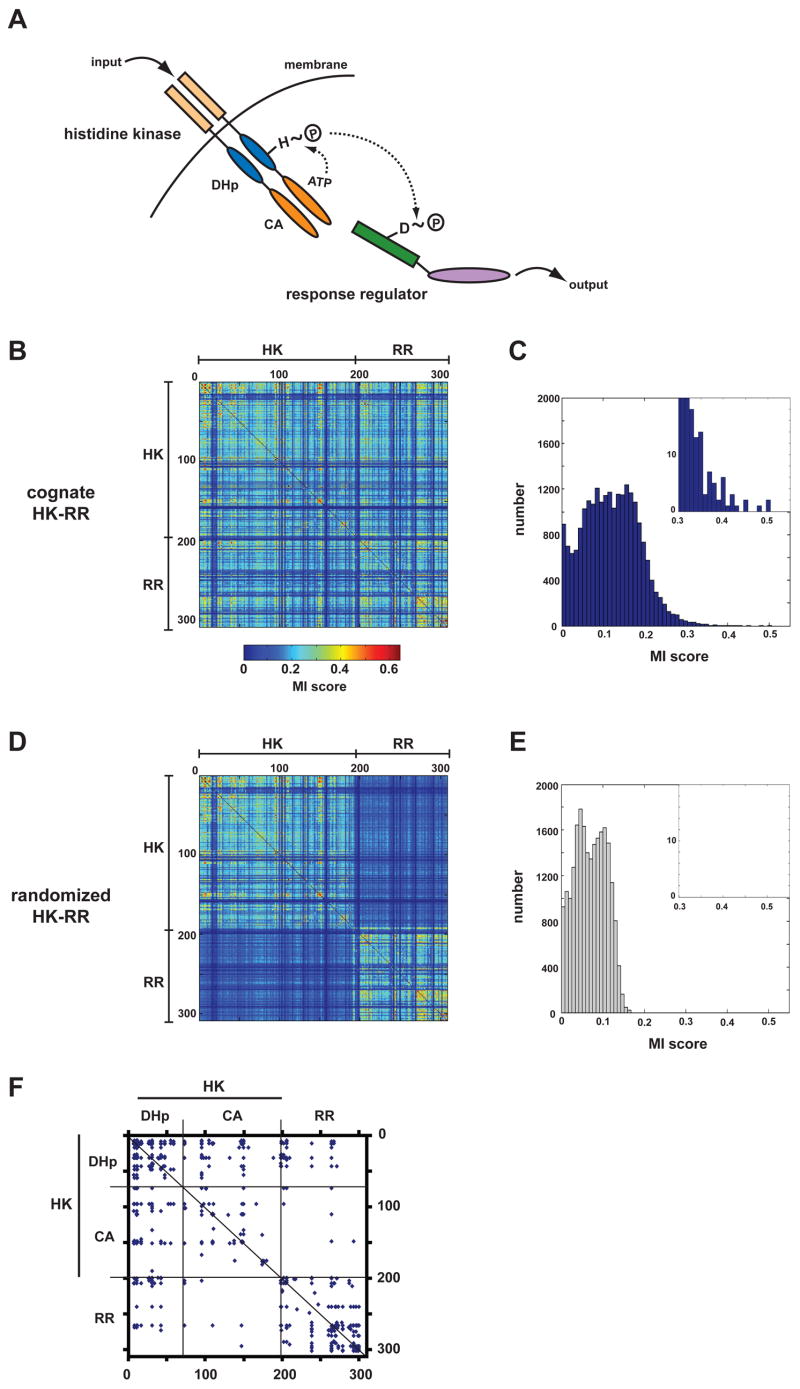
Figure 1. Mutual information analysis of two-component signal transduction proteins.
(A) Schematic of the two-component signaling paradigm. Receipt of a signal stimulates autophosphorylation of a histidine kinase (HK). The phosphoryl group is subsequently passed to a cognate response regulator (RR), which can trigger changes in gene expression or other physiological processes. (B) Identification of covarying residues in histidine kinases and response regulators. Cognate, operon pairs of histidine kinases and response regulators were concatenated and aligned. Using this multiple sequence alignment, every pair of positions was examined for covariation using an analysis of mutual information. The matrix of scores for each pair of positions in the alignment is plotted as a heatmap, according to the color legend shown. Positions within the alignment corresponding to the histidine kinase and the response regulator are indicated. (C) Histogram of interprotein (HK-RR) mutual information scores from panel B. (D) The pairings of histidine kinases and response regulators from panel B were randomized and the mutual information analysis repeated. The heatmap of scores shows that the randomization process reduces interprotein, but not intraprotein (HK-HK, RR-RR), covariation. (E) Histogram of interprotein mutual information scores for the alignment in which HK-RR pairings were scrambled. (F) For the multiple sequence alignment containing cognate HK-RR pairs, the graph indicates the pairs of amino acids with scores >0.35. Regions of the alignment corresponding to the HK and RR are indicated as well as the two HK domains, the dimerization and histidine phosphotransfer (DHp) domain and the catalytic and ATP-binding (CA) domain (see panel A).
Two-component signaling represents an excellent system for probing the molecular basis of specificity in a large, paralogous signaling family. First, both histidine kinases and response regulators are easily identified in sequenced bacterial genomes, in contrast to eukaryotes where the identification of kinase substrates is difficult and incomplete. Second, with respect to phosphotransfer, histidine kinases are known to exhibit a large kinetic preference in vitro for their in vivo cognate response regulator(s) relative to all other response regulators (Fisher et al., 1996; Grimshaw et al., 1998; Skerker et al., 2005). That is, a histidine kinase has an intrinsic ability to recognize its cognate substrate, to the exclusion of all other response regulators. This finding indicates that cellular context is not crucial and the basis of specificity, molecular recognition, can be dissected in vitro.
Structural and mutagenesis studies have helped identify residues necessary for catalyzing phosphotransfer and for the binding of cognate two-component signaling proteins (Janiak-Spens and West, 2000; Jiang et al., 1999; Qin et al., 2003), but the amino acids that dictate the specificity of interaction have remained elusive. It remains a challenge to rationally “rewire” the specificity of a histidine kinase by introducing mutations that change substrate selection without disrupting function. Computational approaches to specificity in two-component pathways have been described (Li et al., 2003; White et al., 2007), but these methods have identified different sets of residues, and experimental verification of their effect on specificity is lacking. Here, to identify specificity determinants, we have examined patterns of aminoacid coevolution between histidine kinases and their cognate response regulators. Using chimeric and mutant proteins, we provide evidence that a subset of the coevolving amino acids determine the substrate specificity of a histidine kinase enabling a rational rewiring of two-component signaling pathways both in vitro and in vivo.
Results
Computational identification of putative specificity-determining residues
As specificity in two-component signaling systems relies on the precise molecular recognition between cognate pairs, a set of amino acids must exist that confer specificity on the interaction. To identify these specificity-determining residues, we searched for amino acids in cognate HK-RR pairs whose identities covary. This approach was based on the notion that, during the course of evolution, any mutation in a specificity-determining residue of one molecule presumably must either revert or be compensated for by a secondary mutation in the cognate molecule that maintains the interaction. This type of approach has been used previously to identify amino acids that interact within proteins (Atchley et al., 2000; Buck and Atchley, 2005; Socolich et al., 2005). To identify covarying amino acids between two-component proteins, we took advantage of the fact that histidine kinases and response regulators encoded in the same operon typically interact with one another in an exclusive, one-to-one fashion (Skerker et al., 2005). Using nearly 200 sequenced bacterial genomes, we identified nearly 1300 HK-RR pairs. These cognate pairs were concatenated and treated as a single sequence and then aligned. To identify positions in this multiple sequence alignment that covary, we calculated the mutual information between each pair of sites (Atchley et al., 2000; Fodor and Aldrich, 2004) (Figures 1B-1C). Mutual information at two sites X and Y in an alignment is defined as: , where column X has n different residues, column Y has m different residues, pj is the probability of residue j in column X, qk is the probability of residue k in column Y, and pjk is the number of sequences with residue j in column X and residue k in column Y divided by the total number of sequences (Atchley et al., 2000; Fodor and Aldrich, 2004). To estimate the background noise of the mutual information values due to sampling bias, we randomized the HK-RR pairings and again measured mutual information between each pair of sites. This randomization did not affect the scores of intramolecular residue pairs, but caused the scores for all intermolecular pairs to fall well below 0.35 (Figures 1D-1E). By contrast, in the original alignment that retains natural HK-RR pairings, there were 43 pairs of residues with intermolecular mutual information scores greater than 0.35 (Figures 1C, 1E). As residues in one molecule often covaried with multiple residues in the other molecule, these 43 pairs involve only 28 residues in total, 16 within the HK and 12 within the RR (Figure 1F).
Histidine kinases always include two domains, a dimerization and histidine phosphotransfer (DHp) domain that contains the phosphorylatable histidine, and a catalytic and ATP-binding (CA) domain that catalyzes autophosphorylation (Stock et al., 2000). Of the 43 highest scoring intermolecular residue pairs in our covariation analysis, 36 were between the RR and the DHp domain of the histidine kinase with only 7 between the RR and the CA domain (Figure 1F). This bias toward the DHp domain was even more pronounced at higher score thresholds (Figure S1); all 33 of the intermolecular residue pairs with scores greater than 0.36 involved residues of the DHp domain of the kinase and the RR. These analyses suggest that the DHp domain dictates the interaction between a histidine kinase and its cognate substrate, as also suggested by yeast two-hybrid and NMR studies (Ohta and Newton, 2003; Tomomori et al., 1999).
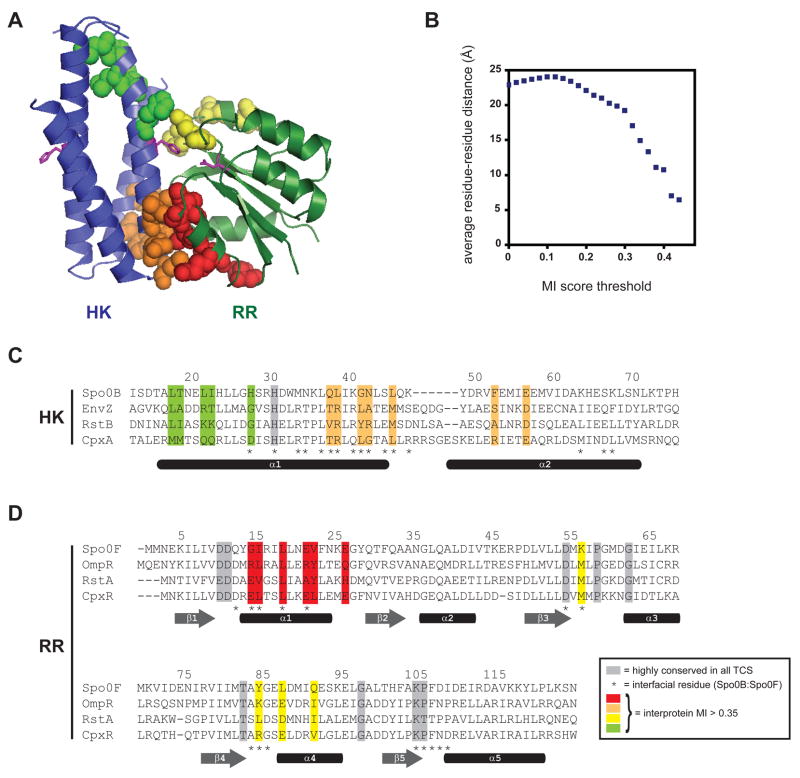
No high-resolution crystal structure of a histidine kinase in complex with a response regulator has been solved. However, the structure of a histidine phosphotransferase, Spo0B, in complex with a response regulator, Spo0F, is considered a reasonable proxy (Zapf et al., 2000) as Spo0B forms a four-helix bundle similar to the DHp domain in histidine kinases (Marina et al., 2005). Figure 2A shows the putative specificity-determining residues mapped onto the Spo0B:Spo0F crystal structure. Nearly all of these residues are solvent-exposed in the individual molecules, but are buried in the interface of the protein complex. To quantify the enrichment for spatially close residue pairs, we calculated the average distance between residue pairs as a function of score threshold (Figure 2B). For all possible residue pairs, the average distance was approximately 23Å. By contrast, at a score threshold of 0.35, there is clear enrichment for residues in close proximity, with an average distance of 10Å. In summary, the mutual information analysis identifies a set of covarying residues that are located at, or very near, the molecular interface of a HK-RR complex. These high-scoring residues do not, however, correlate precisely with the contact residues in the Spo0B:Spo0F cocrystal structure. Some contact residues do not show significant covariation and vice versa (Figures 2C–2D).
Figure 2. Specificity determining residues map to the interface of a two-component signal transduction complex.
(A) Structure of the histidine phosphotransferase Spo0B in complex with the response regulator Spo0F (PDB: 1F51). No HK-RR cocrystal structure has yet been solved. However, the Spo0B:Spo0F serves as a suitable proxy because the four-helix bundle of Spo0B is similar to that formed by the DHp domain of histidine kinases. Residues with the highest intermolecular mutual information scores (> 0.35) are shown by spacefilling. Spo0B is a dimer, but for clarity, only one set of residues is shown. Each molecule contains two regions of high-scoring residues; those in the histidine kinase are colored green and orange and those in the response regulator are colored red and yellow. Additional views are shown in Figure S1. (B) Plot of average residue-residue distances as a function of covariation score for all pairs of positions from the multiple sequence alignment (see Figure 1B and Supplemental File 1) that include one site within the DHp domains of the kinase and one site within the response regulators. (C) Primary sequence alignment of the histidine kinases EnvZ, RstB, and CpxA with the histidine phosphotransferase Spo0B. Residues showing strongest covariation with residues in the response regulator are highlighted in green and orange, as in panel A. (D) Primary sequence alignment of the response regulators Spo0F, OmpR, RstA, and CpxR. Residues showing strongest covariation with residues in the histidine kinase are highlighted in red and yellow, as in panel A. Highly conserved residues are shaded in grey. Interprotein contact residues are marked by asterisks below the sequence alignments along with approximate locations of secondary structure elements.
There are two apparent clusters of covarying residues in each molecule (Figures 2A, 2C-2D). In the histidine kinase, one cluster lies below the phosphotransfer active site, near the base of the four-helix bundle, and the second lies just above the active site histidine. In the response regulator, one cluster lies within alpha-helix 1 while the second lies within the loops connecting beta-strand 3 with alpha-helix 3 and beta-strand 4 with alpha-helix 4.
Analysis of domain-level chimeric histidine kinases
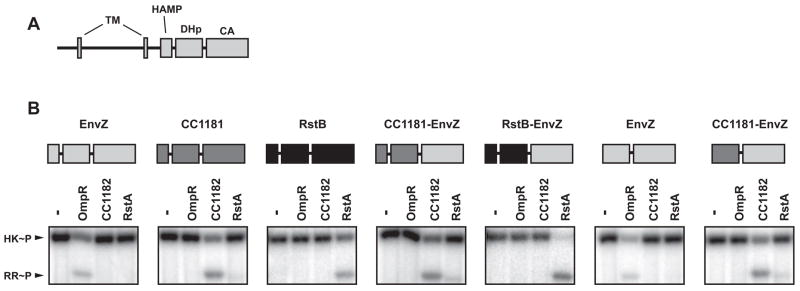
To test whether the highest scoring residues confer specificity to the HK-RR interaction, we constructed a series of chimeric and mutant histidine kinases. First, because our computational analysis demonstrated that nearly all of the significant interprotein covariation involves the DHp domain of the histidine kinase, we created chimeras in which the DHp domains of heterologous kinases were fused to the CA domain of the E. coli kinase EnvZ. We fused the DHp domains of CC1181 (from C. crescentus) and RstB (from E. coli) to the CA domain of E. coli EnvZ (Figure 3). As these kinases are normally anchored in the inner membrane, we removed their transmembrane regions and purified soluble, cytoplasmic portions. The native kinases, EnvZ, CC1181, and RstB, each show a strong kinetic preference in vitro for phosphotransfer to their in vivo cognate substrate, OmpR, CC1182, and RstA, respectively (Figure S3) (Skerker et al., 2005). Both the CC1181-EnvZ and RstB-EnvZ chimeric histidine kinases were capable of autophosphorylation indicating that histidine kinases are, to some extent, modular. With respect to phosphotransfer specificity, each chimera behaved according to the identity of its DHp domain; the CC1181-EnvZ and RstB-EnvZ chimeras were indistinguishable from native CC1181 and RstB, respectively (Figure 3B). These chimeras included the HAMP domains of CC1181 and RstB, a domain often found immediately N-terminal to the DHp domain of histidine kinases (Zhu and Inouye, 2004). To ensure that this domain was not contributing to the change in specificity, we constructed a chimera of CC1181 and EnvZ lacking the HAMP domain. This shorter chimera, containing only the DHp domain of CC1181, was also indistinguishable from native CC1181 and had a strong kinetic preference for phosphotransfer to CC1182. These results indicate that the phosphotransfer specificity of a histidine kinase is dictated almost exclusively by its DHp domain, consistent with the mutual information analysis.
Figure 3. Specificity of phosphotransfer resides in the DHp domain of a histidine kinase.
(A) Schematic of a prototypical histidine kinase. Two transmembrane (TM) regions flank a periplasmic sensor domain followed by the cytoplasmic portion containing HAMP, DHp, and CA domains. (B) Phosphotransfer specificity of domain-swapped chimeric histidine kinases. Each chimera was autophosphorylated and incubated alone or tested for phosphotransfer to the response regulators OmpR, CC1182 and RstA at a 10 second time-point. Schematics above each gel indicate the identity of each domain in the chimera tested. For complete protein sequences, see Supplemental File 2.
Analysis of sub-domain chimeric histidine kinases
Our covariation analysis identified a cluster of seven residues below the histidine active site of the DHp domain (Figure 2A). These residues all lie within a region of the DHp domain that NMR studies implicated in response regulator-binding (Tomomori et al., 1999). To test whether this cluster (colored orange in Figure 2A, 2C) is important for specificity, we made sub-domain chimeras in which only a short segment of the EnvZ DHp domain was replaced with the corresponding sequence of five E. coli histidine kinases, RstB, CpxA, PhoR, AtoS, and PhoQ, which share varying levels of homology with EnvZ (Figures 4A, S2). The transplanted segment included seven of the putative specificity-determining residues, located in the C-terminal portion of helix 1, the N-terminal portion of helix 2, and the loop connecting these helices. The chimeras retained high solubility and showed robust autophosphorylation in vitro (Figures 4B–4F), consistent with normal folding and dimerization. The substrate specificity of each chimeric kinase was assessed by comparing its phosphotransfer activity toward OmpR (cognate substrate of EnvZ) and the new, desired target: RstA, CpxR, PhoB, AtoC, or PhoP. In parallel, we tested the phosphotransfer specificity of the wild-type cognate kinases for these response regulators. Each wild-type histidine kinase exclusively phosphorylated its cognate response regulator (Figures 4B–4F). For example, EnvZ phosphotransfers to OmpR, but not to RstA, whereas RstB phosphotransfers to RstA, but not to OmpR. Strikingly, the sub-domain chimera Chim1, in which a short region of EnvZ was replaced with the corresponding region of RstB, showed robust phosphorylation of RstA and no detectable phosphorylation of OmpR. The pattern of phosphotransfer for Chim1 was indistinguishable from wild-type RstB, consistent with a complete switch in substrate specificity (Figure 4B). Similarly, for each of the other four chimeras, Chim2-Chim5, we observed strong phosphorylation of the new response regulator and no residual phosphorylation of OmpR (Figures 4C–4F). These data indicate that the amino acids at the base of the DHp domain, which includes seven of the highest scoring residues from our computational analysis, are sufficient to determine the phosphotransfer specificity of a histidine kinase.
Figure 4. The base of the DHp domain dictates phosphotransfer specificity of histidine kinases.
(A) Sequences of the DHp domain in the sub-domain chimeras. A small region of the EnvZ DHp domain was replaced with the corresponding portion of five other kinases indicated in parentheses. Amino acids mutated, relative to wild-type EnvZ, are indicated in bold. Colored bars indicate residues with high interprotein (HK-RR) covariation scores. For complete protein sequences, see Supplemental File 2. A schematic of the histidine kinase domain structure and the secondary structure of the DHp domain are shown above the sequences. (B) Rewiring the phosphotransfer specificity of EnvZ to match that of RstB. Autophosphorylated histidine kinases were tested for phosphotransfer to RstA or OmpR and as a control, were incubated without regulator (indicated by a minus sign). The three gel images show the phosphotransfer behavior of (in order from top to bottom): EnvZ, RstB, and the sub-domain chimera Chim1. Similar sets of gel images are shown for rewiring of EnvZ to match the specificity of CpxA (C), PhoR (D), AtoS (E), and PhoQ (F). Phosphotransfer reactions were incubated for 10 seconds (B, C) or 5 minutes (D-F). Black arrowheads indicate the position of the band corresponding to OmpR (top gel in each panel) or the alternative response regulator (bottom two gels in each panel). Kinetic analysis of phosphotransfer specificity of EnvZ (G), RstB (H), CpxA (I), Chim1 (J), and Chim2 (K). In each case an autophosphorylated kinase, indicated above each graph, was examined for phosphotransfer to OmpR, RstA, and CpxR at 0, 10, 30, 60, and 300 seconds. Normalized levels of phosphorylated RR are plotted as fraction maximal versus reaction time in seconds. Data points and error bars represent the mean and standard deviation (n=3). Representative gel images are shown in Figure S4. The slow decrease in phosphorylated RR after reaching a maximal value is due to hydrolysis and phosphatase activity of the kinase. (L) Phosphotransfer profiles for Chim1 and Chim2 versus all 32 E. coli response regulators. Both chimeras phosphorylate only the intended, target response regulator, indicated by black arrowheads.
To quantify the change in substrate specificity for the chimeras, we measured the initial rate of phosphotransfer, allowing estimation of kcat/KM (Fersht, 1985), from Chim1, Chim2, EnvZ, RstB, and CpxA kinases to the OmpR, RstA, and CpxR response regulators (Figures 4G–4K, Table 1). For a given histidine kinase, the ratio of specificity constants (kcat/KM) for two different response regulators is a measure of its kinetic preference for transfer to one substrate over the other. Note, for cognate pairs the phosphotransfer reactions often reached more than 70% of the maximal value within 10 seconds, our first time-point. The initial rates for cognate pairs are thus lower-bound estimates and are used only to assess the order of magnitude change in substrate specificity of mutant kinases. For wild-type EnvZ, we observed a kinetic preference of ~325-fold for phosphotransfer to its cognate substrate OmpR relative to the non-cognate RstA. Conversely, wild-type RstB showed a preference of ~2600-fold for phosphotransfer to RstA over OmpR. The relative kinetic preference of the two kinases is thus ~8 × 105. In comparison, the chimera Chim1 exhibited a ~1400-fold preference for RstA relative to OmpR. Hence, the total change in kinetic preference between Chim1 and EnvZ is ~5 × 105, a value comparable to the difference between wild-type EnvZ and RstB (Table 1). Similarly, we found that the ratio of kinetic preference for EnvZ and CpxA (each with respect to OmpR and CpxR) is ~2 × 105, whereas the ratio for EnvZ and Chim2 is ~5 × 105 (Table 1). These data indicate that the sub-domain chimeras have a nearly complete switch in substrate specificity relative to EnvZ.
Table 1.
Summary of kinetic preference for wild-type, chimeric, and mutant kinases.
| histidine kinase | kinetic preference (RstA v. OmpR)* | kinetic preference relative to EnvZ | kinetic preference (CpxR v. OmpR)# | kinetic preference relative to EnvZ |
|---|---|---|---|---|
| EnvZ | 0.0031 | 1 | 0.0013 | 1 |
| RstB | 2566 | 8 × 105 | - | - |
| Chim1 | 1408 | 5 × 105 | - | - |
| Mut4 | 206 | 7 × 104 | - | - |
| Mut5 | 94 | 3 × 104 | - | - |
| CpxA | - | - | 296 | 2 × 105 |
| Chim2 | - | - | 690 | 5 × 105 |
Specificity constants (kcat/KM) were measured for each kinase relative to the response regulators RstA and OmpR. The kinetic preference value reported is the ratio of these two constants.
Specificity constants (kcat/KM) were measured for each kinase relative to the response regulators CpxR and OmpR. The kinetic preference value reported is the ratio of these two constants.
Next, to ensure that our designed chimeric proteins had been rewired and were not simply promiscuous kinases, we used phosphotransfer profiling, a method for examining the global substrate preference of histidine kinases (Skerker et al., 2005). The sub-domain chimeras Chim1 and Chim2, as well as wild-type EnvZ, CpxA, and RstB, were systematically tested for phosphotransfer to each of the 32 E. coli response regulators. The wild-type kinases specifically phosphorylate their cognate regulators, to the exclusion of all other regulators (Figure S3) (Skerker et al., 2005). In this assay, the chimeras Chim1 (EnvZ-RstB) and Chim2 (EnvZ-CpxA) were specific, on a system-wide level, for RstA and CpxR, respectively (Figure 4L). These comprehensive profiles confirm that the chimeras are not promiscuous but specifically phosphorylate a single response regulator.
Rewiring histidine kinases in vitro by mutating specificity-determining residues
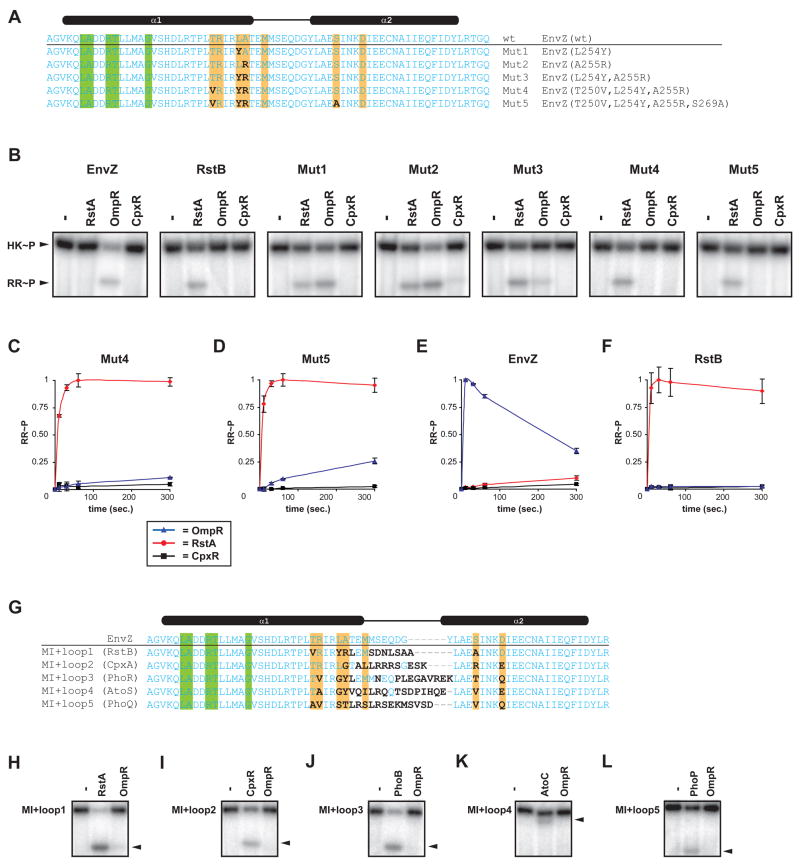
The chimeras described above include seven of the amino acids predicted by our computational analysis to dictate phosphotransfer specificity. We therefore sought to test whether mutating only these residues would be sufficient to change the substrate selectivity of EnvZ. For these experiments, we made mutations in EnvZ in which putative specificity residues were replaced with the corresponding residues from RstB (Figure 5A). Two of the single mutants generated, EnvZ(L254Y) and EnvZ(A255R), showed significant phosphorylation of both RstA and OmpR (Figure 5B). The double mutant EnvZ(L254Y, A255R) preferentially phosphorylated RstA, but still retained some residual activity toward OmpR. Subsequent inclusion of the mutation T250V, however, eliminated the phosphorylation of OmpR but maintained robust phosphotransfer to RstA (Figure 5B). Using phosphotransfer profiling to examine system-wide specificity, we verified that this mutant kinase EnvZ(T250V, L254Y, A255R) exclusively phosphotransfers to RstA (data not shown). Finally, we constructed a quadruple mutant, EnvZ(T250V, L254Y, A255R, S269A), in which each of the putative specificity residues in EnvZ had the identity of the corresponding position in RstB (Figure 5A). This mutant also phosphorylated RstA, but not OmpR (Figure 5B). These data indicate that changing as few as three residues is sufficient to change the substrate preference of EnvZ to that of RstB.
Figure 5. Rewiring two-component signal transduction pathways.
(A) Sequences of point mutants constructed in which amino acids with high interprotein mutual information scores were mutated in EnvZ to match the corresponding amino acid in RstB. Sequences shown correspond only to the DHp domain. Sites differing from the wild-type EnvZ are in bold. Colored bars indicate residues with high interprotein mutual information scores. (B) Phosphotransfer specificity of EnvZ, RstB, and Mut1-Mut5. In each case the kinase was autophosphorylated and then incubated alone or examined for phosphotransfer to RstA, OmpR, and CpxR after 10 second incubations. (C,D) Kinetics of phosphotransfer for Mut4 and Mut5. Normalized RR~P levels are plotted as fraction maximal versus reaction time in seconds. Data points and error bars represent the mean and standard deviation (n=3). Representative gel images are shown in Figure S4. (E, F) Graphs showing the kinetic preference of wild-type EnvZ and RstB (see Figures 4G-4H) are shown for comparison to Mut4 and Mut5. (G) Sequences of the MI+loop (mutual information+loop) mutants. Amino acids that were mutated, relative to wild-type EnvZ, are indicated in bold. Colored bars indicate residues with high interprotein (HK-RR) covariation scores. MI+loop mutant kinases (MI+loop1-5) were tested for phosphotransfer to OmpR and the new intended target: RstA (H), CpxR (I), PhoB (J), AtoC (K), or PhoP (L). Black arrowheads indicate position of the target response regulator band within each gel. Experiments were performed exactly as in Figures 4B-4F and can be compared directly.
To quantify the change in substrate specificity for these variants, we measured approximate kcat/KM ratios for the triple (Mut4: T250V, L254Y, A255R) and quadruple (Mut5: T250V, L254Y, A255R, S269A) mutants (Figures 5C–5D, Table 1). Mut4 exhibited a ~200-fold preference for RstA relative to OmpR while Mut5 had a ~100-fold preference for RstA over OmpR. These mutant kinases thus produce changes in specificity, relative to wild-type EnvZ, of ~7 × 104 and ~3 × 104, respectively (Figures 5C–F). As noted earlier, the ratio of kinetic preference of EnvZ and RstB is ~8 × 105. The point mutants thus produce a nearly complete switch in specificity, supporting the classification of amino acids identified in our computational analysis as bona fide specificity-determining residues. Interestingly, most of these residues lie on one face of alpha-helix 1 in the kinase DHp domain. In the Spo0B:Spo0F cocrystal structure, these residues contact residues from alpha-helix 1 of the response regulator (Figures S1D-S1E). Taken together, our data suggest that the docking of these two helices is a primary means of interaction and substrate discrimination in two-component systems.
We also replaced the predicted specificity residues of EnvZ with the corresponding amino acids from CpxA, PhoR, AtoS, and PhoQ (Figure S2). These mutant kinases showed only partial changes in specificity (not shown), suggesting that the computational analysis missed one or more critical specificity-determining residues. We reasoned that the additional residue(s) would be in close proximity to the seven identified specificity residues (those at the base of the DHp domain), because the chimeras Chim2-5 each showed a nearly complete switch in specificity (Figure 4). Moreover, because our results showed that the C-terminal end of helix-1 plays a key role in substrate discrimination, it seemed plausible that residues in the loop adjacent to helix-1 might contribute to specificity. Our computational analysis may have missed a residue in this region because the loop sequences were difficult to align. To test this hypothesis, we made mutations in EnvZ that (i) changed the identity of the seven specificity-determining residues identified above to match those found in another kinase and (ii) replaced the loop connecting helices 1 and 2 to also match that of another kinase (Figure 5G). We made five “MI+loop” mutants in attempting to switch the specificity of EnvZ to that of RstB, CpxA, PhoR, AtoS, and PhoQ. In each case, the mutant kinase showed robust phosphorylation of the intended, target response regulator and no detectable phosphotransfer to OmpR, indicating a nearly complete switch in substrate specificity (Figure 5H–5L). For RstB and CpxA, we confirmed that the loop alone is insufficient to change substrate specificity (not shown). These data suggest that the MI+loop design strategy offers a general means to rewiring histidine kinase specificity.
Rewiring histidine kinase specificity in vivo
The specificity of phosphotransfer in two-component signaling pathways relies predominantly on molecular recognition and, consequently, the substrate specificity of a histidine kinase in vitro typically mirrors its specificity in vivo. We therefore expected that our strategy for rewiring a histidine kinase in vitro would also lead to the redirection of information flow in vivo. To test this prediction, we examined the ability of our EnvZ MI+loop mutants to phosphorylate CpxR in vivo in E. coli using the cpxP promoter driving gfp as a reporter (Figure 6A). Transcription from the cpxP promoter is stimulated by phosphorylated CpxR (Danese and Silhavy, 1998). Each MI+loop mutant of envZ tested was constructed in the context of a full-length envZ gene on a plasmid and transformed into strain AFS161. This strain harbors a disruption of the chromosomal copy of cpxA, so the expression of GFP depends on the ability of the plasmid-borne kinase to phosphorylate CpxR. As seen in Figure 6B, the CpxA MI+loop mutant led to significant expression of GFP indicating that this kinase could produce high levels of phosphorylated CpxR~P in vivo. By contrast, expression of wild-type EnvZ or the PhoQ and PhoR MI+loop mutants produced levels of GFP close to background. We also tested the ability of each mutant kinase to phosphorylate PhoP, a response regulator normally phosphorylated by the histidine kinase PhoQ. For these experiments we monitored expression of YFP driven by the promoter of mgrB (Figure 6C), a gene directly activated by phosphorylated PhoP (Kato et al., 1999). In this case, the reporter strain bears a deletion of phoQ so that phosphorylation of PhoP depends on the plasmid-borne kinase. Only the PhoQ MI+loop mutant led to high levels of YFP; the wild-type EnvZ and the other MI+loop mutants produced only background levels of YFP (Figure 6D). To ensure that the differences observed in these assays was not due to differences in expression level, we confirmed by Western blotting that each mutant kinase was produced at levels equal to or less than the wild-type EnvZ control, pEnvZ (data not shown). Taken together, these reporter gene studies demonstrate that our strategy for rewiring histidine kinase specificity is effective in vivo and enables the rational redirection of phosphorylation flow inside a living cell.
Figure 6. Rewiring histidine kinase specificity in vivo.
(A) Schematic of the reporter construct used to assess CpxR phosphorylation in vivo. The gfp gene is driven by a cpxP promoter, which depends on CpxR~P for activation. (B) GFP fluorescence normalized by OD600 for the cpxP transcriptional reporter strain AFS161 (envZ− cpxA− PcpxP-gfp) containing the indicated plasmids. (C) Schematic of the reporter construct used to assess PhoP phosphorylation in vivo. The yfp gene is driven by a mgrB promoter, which depends on PhoP~P for activation. The cfp gene is driven by a constitutively active promoter and is used for normalization. (D) YFP/CFP fluorescence ratio of the mgrB transcriptional reporter strain AFS237 (envZ− phoQ− PmgrB-yfp PtetA-cfp) containing the indicated plasmids. In each case, the average of measurements from three independent cultures is shown. Error bars indicate standard deviations. The pEnvZ and p(MI+loop) plasmids express full-length versions of EnvZ and the chimeras (see Figure 5), respectively.
Discussion
The specificity of two-component signaling pathways relies on the intrinsic ability of a histidine kinase to discriminate its cognate response regulator from all other possible substrates. Structural and previous mutagenesis studies have not, however, been able to elucidate the molecular basis of specificity in this protein-protein interaction. Here, we used an analysis of amino-acid covariation to guide the identification of specificity determinants in two-component signal transduction pathways and to produce a general method for rewiring the substrate selectivity of EnvZ, both in vitro and in vivo. Given the high homology between two-component signaling proteins, we anticipate that a similar, rational rewiring of other histidine kinases will be possible using the design approach developed here. Two-component proteins have been engineered and used in synthetic circuits previously, but this work has been limited to fusions of heterologous ligand-binding domains to the entire cytoplasmic portion of EnvZ (Levskaya et al., 2005; Utsumi et al., 1989). The ability to now rewire the substrate selectivity of EnvZ, and other histidine kinases, should significantly enhance their use in synthetic signaling circuits.
Protein-protein interactions are crucial to the operation of nearly every cellular process, but our understanding of interaction specificity remains limited. In particular, there are only a few cases in which protein-protein interaction specificity is understood to a level that allows the rational reprogramming, or engineering, of interaction specificity. There have been some recent successes, such as the redesign of colicin-immunity protein interactions (Kortemme et al., 2004) and coiled-coil dimerization (Havranek and Harbury, 2003), but these studies have relied heavily on structural data. Our results demonstrate that the analysis of amino-acid covariation in large multiple sequence alignments can effectively guide the identification of specificity determinants in protein-protein interactions, even those involved in transient signaling events such as a kinase-substrate interaction. Importantly, the analysis of amino-acid covariation does not require structural data, and in fact, provides complementary information. For instance, the Spo0B:Spo0F structure clearly revealed the molecular surfaces and amino acids in direct contact during phosphotransfer, but the covariation analysis presented here was necessary to pinpoint the subset of contact residues that dictate specificity. The covariation analysis was particularly valuable because no high-resolution structure of a histidine kinase in complex with a response regulator has yet been solved; Spo0B is a histidine phosphotransferase and considered a suitable proxy for histidine kinases, but may not be suitable for structure-based redesign efforts. As sequence databases continue their rapid expansion, covariation analyses should become increasingly useful for mapping the specificity determinants of other protein-protein interactions. Analyses of coevolution should also significantly aid efforts to design or engineer protein specificity, either as a complement to other computational approaches or by focusing the region of interest for mutagenesis or directed evolution (Bloom et al., 2005; Kortemme and Baker, 2004). Our analysis of histidine kinase-response regulator covariation leveraged the fact that cognate pairs are often encoded in the same operon. The use of covariation analysis in eukaryotes will likely depend on protein-protein interaction data for paralogous protein families of interest, such as that generated recently for human bZIP transcription factors and PDZ-peptide interactions (Newman and Keating, 2003; Stiffler et al., 2007).
Our covariation analyses identified two primary clusters of amino acids in histidine kinases that covary with amino acids in response regulators. These clusters are located above and below the active site histidine (colored green and orange in Figure 2A, respectively). Based on previous NMR studies (Tomomori et al., 1999), we focused on the amino acids below the histidine and demonstrated that these residues are crucial determinants of a histidine kinase’s substrate specificity. The small cluster of residues above the histidine thus do not appear to play a significant role in substrate specificity. These amino acids may have been spuriously identified as amino-acid covariation can result from common ancestry rather than functional constraint. The elimination of sequences with greater than 90% identity in our analyses helps to minimize, but does not eliminate, the confounding effects of phylogeny. A number of approaches have recently been developed to further minimize the influence of common ancestry. Application of one such approach (Dunn et al., 2008) to the multiple sequence alignments used here did not substantially change the top-scoring amino acids, although notably, two amino acids (L230 and A231 in EnvZ) in the cluster above the active site histidine were eliminated (data not shown).
Most of the amino acids showing strong covariation between histidine kinases and response regulators are located at or near the presumed molecular interface formed during phosphotransfer. There is not, however, a perfect correlation between the covarying and interfacial residues. It is only a subset of interfacial residues, mostly at the C-terminal end of alpha helix 1, that play the dominant role in determining substrate selectivity. Changing the specificity of EnvZ to match that of RstB was achieved by mutating only the specificity residues within this helix. Changing the specificity of EnvZ to match that of other kinases, however, also required swapping the loop between alpha helices 1 and 2 (Figure 5). As noted earlier, residues within the loop are difficult to align and hence more refractory to covariation analysis. In this respect, structures of histidine kinases provided useful, complementary insight to the covariation analysis. A similar combination of approaches will likely be helpful in reprogramming other protein-protein interactions.
Finally, the identification of specificity determinants in two-component signaling proteins may also shed light on the system-level properties and evolution of paralogous signal transduction families. The exquisite substrate selectivity exhibited by histidine kinases suggests they have been selected during evolution both for recognition of their cognate substrates and against recognition of non-cognate substrates. Evidence for a similar balance of selective forces shaping the specificity of paralogous gene families has also recently been reported for PDZ-peptide and SH3-peptide interactions in eukaryotes (Stiffler et al., 2007; Zarrinpar et al., 2003). For two-component signal transduction systems, the identification of specificity-determining residues will facilitate study of the selective forces that influence the evolution of these signaling proteins. This understanding, in turn, may help to reveal how cells coordinate multiple, paralogous signaling pathways to maintain information flow while preventing unwanted cross-talk.
Experimental Procedures
Computational analyses
Putative cognate two-component proteins were identified in sequenced bacterial genomes by selecting adjacent genes predicted to encode a histidine kinase (HK) and a response regulator (RR), using custom PERL scripts. For HKs the sensor and transmembrane domains were eliminated and for RRs the output domains were removed. This procedure retained the dimerization and histidine phosphotransfer (DHp) and the catalytic and ATP-binding (CA) domains of the HKs and the receiver domains (RD) of the RRs. For each cognate HK-RR pair the DHp, CA, and RD domains were concatenated into a single sequence and aligned using PCMA (Pei et al., 2003) with some manual adjustment (see Supplemental File 1). Analysis of mutual information was performed using published software (Fodor and Aldrich, 2004). Columns in the alignment containing more than 10% gaps were eliminated from consideration. We also ensured that no two sequences in the alignment had greater than 90% identity to one another. This latter step helped to minimize the detection of amino acids that covary due to phylogenetic relationships rather than functional relationships.
Specificity-determining residues were mapped onto the Spo0B:Spo0F crystal structure (PDB: 1F51) using PyMOL (Delano Scientific). The asymmetric unit contains four Spo0B and four Spo0F molecules. For clarity, only the four-helix bundle of one Spo0B dimer in complex with one Spo0F molecule is shown in Figure 2. Distances between residues were measured as the shortest distance between any non-hydrogen atoms.
Cloning and protein purification
Cloning and protein purification was done as described previously (Skerker et al., 2005) with additional details in Supplemental Experimental Procedures.
Phosphorylation and phosphotransfer assays
In vitro analyses of phosphorylation and phosphotransfer were performed as previously described (Skerker et al., 2005). Briefly, histidine kinases in 10 mM HEPES-KOH [pH 8.0], 50 mM KCl, 10% glycerol, 0.1 mM EDTA, 2 mM DTT, 5 mM MgCl2 were autophosphorylated using 500 μM ATP and 0.5 μCi/μl [γ32P]-ATP (from a stock at ~6000 Ci/mmol, Amersham Biosciences) and then subsequently incubated with a response regulator. Kinase and regulator were present at 2.5 μM each. Reactions were incubated at room temperature and then products separated by 10% SDS-PAGE, exposed to a phosphor screen, and quantified using a Typhoon 9400 Scanner (GE Healthcare) with ImageQuant 5.2. For analysis of phosphotransfer kinetics, autophosphorylated kinases were purified by ultrafiltration before incubation with response regulators. Initial rates were determined by measuring the rate of phosphorylation of a kinase’s cognate substrate between 0 and 10 sec and of non-cognate substrates between 0 and 300 sec.
In vivo analysis of specificity mutations
pEnvZ (Hsing and Silhavy, 1997) contains full-length envZ under control of the lac promoter. Plasmids expressing three of the chimeras, p(MI+loop2), p(MI+loop3), and p(MI+loop5), were constructed by replacing the EnvZ DHp domain in pEnvZ with the corresponding chimeric sequences using the restriction sites NdeI and RsrII. pEB5, an envZ-deleted version of pEnvZ, was used as a control (Batchelor and Goulian, 2003). CpxR-regulated transcription was measured with strain AFS161, an MG1655 derivative (Blattner et al., 1997) that contains a chromosomal copy of gfp under control of the cpxP promoter: MG1655 ΔenvZ::cat cpxA::kan ΔlacIZYA::FRT attλ::[PcpxP-gfp]. PhoP-regulated transcription was measured with strain AFS237, an envZ− phoQ− version of TIM177 (Miyashiro and Goulian, 2007) that contains the mgrB promoter driving yfp and a constitutive tetA promoter driving cfp: MG1655 ΔenvZ::cat ΔphoQ::FRT ΔlacZYA::FRT attλ::[PmgrB-yfp] attHK::[PtetA-cfp]). Strains were grown in minimal A medium (Miller, 1992) containing 0.2% glycerol, 50 μg/ml ampicillin, and 3 or 10 μM IPTG for AFS161- or AFS237-derived strains, respectively. Overnight cultures were diluted 1:1000 into fresh media and grown at 37°C with aeration to an OD600 of 0.1–0.2. GFP fluorescence of AFS161 cultures was measured by a spectrofluorometer (Batchelor et al., 2005). YFP fluorescence of AFS237 cultures was measured by fluorescence microscopy and normalized by the CFP fluorescence (Miyashiro and Goulian, 2007). For each culture, the average YFP/CFP fluorescence ratio was computed for ~150 cells.
Supplementary Material
Acknowledgments
We thank A. Murray for helpful discussions throughout the project and A.M., R. Sauer, and A. Keating for comments on the manuscript. This work was supported by the U.S. Department of Energy (MTL), an NIH grant to the Center for Systems Biology at Harvard where the work was initiated (MTL), and an NIH NIGMS grant (MG).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Atchley WR, Wollenberg KR, Fitch WM, Terhalle W, Dress AW. Correlations among amino acid sites in bHLH protein domains: an information theoretic analysis. Mol Biol Evol. 2000;17:164–178. doi: 10.1093/oxfordjournals.molbev.a026229. [DOI] [PubMed] [Google Scholar]
- Batchelor E, Goulian M. Robustness and the cycle of phosphorylation and dephosphorylation in a two-component regulatory system. Proc Natl Acad Sci USA. 2003;100:691–696. doi: 10.1073/pnas.0234782100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batchelor E, Walthers D, Kenney LJ, Goulian M. The Escherichia coli CpxA-CpxR envelope stress response system regulates expression of the porins ompF and ompC. J Bacteriol. 2005;187:5723–5731. doi: 10.1128/JB.187.16.5723-5731.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom JD, Meyer MM, Meinhold P, Otey CR, MacMillan D, Arnold FH. Evolving strategies for enzyme engineering. Curr Opin Struct Biol. 2005;15:447–452. doi: 10.1016/j.sbi.2005.06.004. [DOI] [PubMed] [Google Scholar]
- Buck MJ, Atchley WR. Networks of coevolving sites in structural and functional domains of serpin proteins. Mol Biol Evol. 2005;22:1627–1634. doi: 10.1093/molbev/msi157. [DOI] [PubMed] [Google Scholar]
- Danese PN, Silhavy TJ. CpxP, a stress-combative member of the Cpx regulon. J Bacteriol. 1998;180:831–839. doi: 10.1128/jb.180.4.831-839.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn SD, Wahl LM, Gloor GB. Mutual information without the influence of phylogeny or entropy dramatically improves residue contact prediction. Bioinformatics. 2008;24:333–340. doi: 10.1093/bioinformatics/btm604. [DOI] [PubMed] [Google Scholar]
- Fersht A. Enzyme Structure and Mechanism. 2. New York, NY: W.H. Freeman and Company; 1985. [Google Scholar]
- Fisher SL, Kim SK, Wanner BL, Walsh CT. Kinetic comparison of the specificity of the vancomycin resistance VanS for two response regulators, VanR and PhoB. Biochemistry. 1996;35:4732–4740. doi: 10.1021/bi9525435. [DOI] [PubMed] [Google Scholar]
- Fodor AA, Aldrich RW. Influence of conservation on calculations of amino acid covariance in multiple sequence alignments. Proteins. 2004;56:211–221. doi: 10.1002/prot.20098. [DOI] [PubMed] [Google Scholar]
- Grimshaw CE, Huang S, Hanstein CG, Strauch MA, Burbulys D, Wang L, Hoch JA, Whiteley JM. Synergistic kinetic interactions between components of the phosphorelay controlling sporulation in Bacillus subtilis. Biochemistry. 1998;37:1365–1375. doi: 10.1021/bi971917m. [DOI] [PubMed] [Google Scholar]
- Havranek JJ, Harbury PB. Automated design of specificity in molecular recognition. Nat Struct Biol. 2003;10:45–52. doi: 10.1038/nsb877. [DOI] [PubMed] [Google Scholar]
- Hsing W, Silhavy TJ. Function of conserved histidine-243 in phosphatase activity of EnvZ, the sensor for porin osmoregulation in Escherichia coli. J Bacteriol. 1997;179:3729–3735. doi: 10.1128/jb.179.11.3729-3735.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janiak-Spens F, West AH. Functional roles of conserved amino acid residues surrounding the phosphorylatable histidine of the yeast phosphorelay protein YPD1. Mol Microbiol. 2000;37:136–144. doi: 10.1046/j.1365-2958.2000.01973.x. [DOI] [PubMed] [Google Scholar]
- Jiang M, Tzeng YL, Feher VA, Perego M, Hoch JA. Alanine mutants of the Spo0F response regulator modifying specificity for sensor kinases in sporulation initiation. Mol Microbiol. 1999;33:389–395. doi: 10.1046/j.1365-2958.1999.01481.x. [DOI] [PubMed] [Google Scholar]
- Kato A, Tanabe H, Utsumi R. Molecular characterization of the PhoP-PhoQ two-component system in Escherichia coli K-12: identification of extracellular Mg2+-responsive promoters. J Bacteriol. 1999;181:5516–5520. doi: 10.1128/jb.181.17.5516-5520.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortemme T, Baker D. Computational design of protein-protein interactions. Curr Opin Chem Biol. 2004;8:91–97. doi: 10.1016/j.cbpa.2003.12.008. [DOI] [PubMed] [Google Scholar]
- Kortemme T, Joachimiak LA, Bullock AN, Schuler AD, Stoddard BL, Baker D. Computational redesign of protein-protein interaction specificity. Nat Struct Mol Biol. 2004;11:371–379. doi: 10.1038/nsmb749. [DOI] [PubMed] [Google Scholar]
- Laub MT, Goulian M. Specificity in Two-Component Signal Transduction Pathways. Annu Rev Genetics. 2007;41:121–145. doi: 10.1146/annurev.genet.41.042007.170548. [DOI] [PubMed] [Google Scholar]
- Levskaya A, Chevalier AA, Tabor JJ, Simpson ZB, Lavery LA, Levy M, Davidson EA, Scouras A, Ellington AD, Marcotte EM, et al. Synthetic biology: engineering Escherichia coli to see light. Nature. 2005;438:441–442. doi: 10.1038/nature04405. [DOI] [PubMed] [Google Scholar]
- Li L, Shakhnovich EI, Mirny LA. Amino acids determining enzyme-substrate specificity in prokaryotic and eukaryotic protein kinases. Proc Natl Acad Sci USA. 2003;100:4463–4468. doi: 10.1073/pnas.0737647100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- Marina A, Waldburger CD, Hendrickson WA. Structure of the entire cytoplasmic portion of a sensor histidine-kinase protein. EMBO J. 2005;24:4247–4259. doi: 10.1038/sj.emboj.7600886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Plainview, NY: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- Miyashiro T, Goulian M. Stimulus-dependent differential regulation in the Escherichia coli PhoQ PhoP system. Proc Natl Acad Sci USA. 2007;104:16305–16310. doi: 10.1073/pnas.0700025104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman JR, Keating AE. Comprehensive identification of human bZIP interactions with coiled-coil arrays. Science. 2003;300:2097–2101. doi: 10.1126/science.1084648. [DOI] [PubMed] [Google Scholar]
- Ohta N, Newton A. The core dimerization domains of histidine kinases contain recognition specificity for the cognate response regulator. J Bacteriol. 2003;185:4424–4431. doi: 10.1128/JB.185.15.4424-4431.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei J, Sadreyev R, Grishin NV. PCMA: fast and accurate multiple sequence alignment based on profile consistency. Bioinformatics. 2003;19:427–428. doi: 10.1093/bioinformatics/btg008. [DOI] [PubMed] [Google Scholar]
- Perraud AL, Kimmel B, Weiss V, Gross R. Specificity of the BvgAS and EvgAS phosphorelay is mediated by the C-terminal HPt domains of the sensor proteins. Mol Microbiol. 1998;27:875–887. doi: 10.1046/j.1365-2958.1998.00716.x. [DOI] [PubMed] [Google Scholar]
- Qin L, Cai S, Zhu Y, Inouye M. Cysteine-scanning analysis of the dimerization domain of EnvZ, an osmosensing histidine kinase. J Bacteriol. 2003;185:3429–3435. doi: 10.1128/JB.185.11.3429-3435.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz MA, Madhani HD. Principles of MAP kinase signaling specificity in Saccharomyces cerevisiae. Annu Rev Genetics. 2004;38:725–748. doi: 10.1146/annurev.genet.39.073003.112634. [DOI] [PubMed] [Google Scholar]
- Skerker JM, Prasol MS, Perchuk BS, Biondi EG, Laub MT. Two-component signal transduction pathways regulating growth and cell cycle progression in a bacterium: a system-level analysis. PLoS Biol. 2005;3:e334. doi: 10.1371/journal.pbio.0030334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Socolich M, Lockless SW, Russ WP, Lee H, Gardner KH, Ranganathan R. Evolutionary information for specifying a protein fold. Nature. 2005;437:512–518. doi: 10.1038/nature03991. [DOI] [PubMed] [Google Scholar]
- Stiffler MA, Chen JR, Grantcharova VP, Lei Y, Fuchs D, Allen JE, Zaslavskaia LA, MacBeath G. PDZ domain binding selectivity is optimized across the mouse proteome. Science. 2007;317:364–369. doi: 10.1126/science.1144592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- Tomomori C, Tanaka T, Dutta R, Park H, Saha SK, Zhu Y, Ishima R, Liu D, Tong KI, Kurokawa H, et al. Solution structure of the homodimeric core domain of Escherichia coli histidine kinase EnvZ. Nat Struct Biol. 1999;6:729–734. doi: 10.1038/11495. [DOI] [PubMed] [Google Scholar]
- Ubersax JA, Ferrell JE., Jr Mechanisms of specificity in protein phosphorylation. Nat Rev Mol Cell Biol. 2007;8:530–541. doi: 10.1038/nrm2203. [DOI] [PubMed] [Google Scholar]
- Utsumi R, Brissette RE, Rampersaud A, Forst SA, Oosawa K, Inouye M. Activation of bacterial porin gene expression by a chimeric signal transducer in response to aspartate. Science. 1989;245:1246–1249. doi: 10.1126/science.2476847. [DOI] [PubMed] [Google Scholar]
- White RA, Szurmant H, Hoch JA, Hwa T. Features of protein-protein interactions in two-component signaling deduced from genomic libraries. Meth Enzymol. 2007;422:75–101. doi: 10.1016/S0076-6879(06)22004-4. [DOI] [PubMed] [Google Scholar]
- Zapf J, Sen U, Madhusudan Hoch JA, Varughese KI. A transient interaction between two phosphorelay proteins trapped in a crystal lattice reveals the mechanism of molecular recognition and phosphotransfer in signal transduction. Structure. 2000;8:851–862. doi: 10.1016/s0969-2126(00)00174-x. [DOI] [PubMed] [Google Scholar]
- Zarrinpar A, Park SH, Lim WA. Optimization of specificity in a cellular protein interaction network by negative selection. Nature. 2003;426:676–680. doi: 10.1038/nature02178. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Inouye M. The HAMP linker in histidine kinase dimeric receptors is critical for symmetric transmembrane signal transduction. J Biol Chem. 2004;279:48152–48158. doi: 10.1074/jbc.M401024200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.