Abstract
Plants depend on light signals to modulate many aspects of their development and optimize their photosynthetic capacity. Phytochromes (phys), a family of photoreceptors, initiate a signal transduction pathway that alters expression of a large number of genes to induce these responses. Recently, phyA and phyB were shown to bind members of a basic helix–loop–helix family of transcription factors called phy-interacting factors (PIFs). PIF1 negatively regulates chlorophyll biosynthesis and seed germination in the dark, and light-induced degradation of PIF1 relieves this negative regulation to promote photomorphogenesis. Here, we report that PIF1 regulates expression of a discrete set of genes in the dark, including protochlorophyllide oxidoreductase (POR), ferrochelatase (FeChII), and heme oxygenase (HO3), which are involved in controlling the chlorophyll biosynthetic pathway. Using ChIP and DNA gel-shift assays, we demonstrate that PIF1 directly binds to a G-box (CACGTG) DNA sequence element present in the PORC promoter. Moreover, in transient assays, PIF1 activates transcription of PORC in a G-box-dependent manner. These data strongly suggest that PIF1 directly and indirectly regulates key genes involved in chlorophyll biosynthesis to optimize the greening process in Arabidopsis.
Keywords: basic helix–loop–helix transcription factors, photomorphogenesis, phytochrome signaling, transcriptional regulation, G-box
Light has a profound effect on plant growth and development. Plants not only rely on light signals to regulate developmental phases, but also to provide spatial and temporal information about their environment. Within plant cells, an array of photoreceptors detects several light characteristics such as wavelength, direction, duration, and intensity. Photoreceptors such as cryptochromes, phototropins, and an unidentified UV-B receptor perceive and respond to blue light, whereas phytochromes (phys) respond to the red (R) and far-red (FR) region of the spectrum (1, 2).
Phys exist in two spectral forms: a R light-absorbing Pr form and a FR light-absorbing Pfr form. R light induces conformation of phys to the Pfr, or “active” form; FR light coverts phys to the Pr, or “inactive” form. In Arabidopsis, phys are encoded by a small multigene family (PHYA-PHYE). All phys are active in R light; however, phyA is light labile and activated by both R and FR light. Both phyA and phyB are predominantly in the cytosol in the Pr form. The Pfr form is induced to translocate into nucleus upon light activation either by unmasking of Nuclear Localization Signal (NLS) present in the C-terminal domain (for phyB) (3) or through associated proteins (for phyA) (4). Activation of phys by light initiates a signaling cascade, which results in changes in gene expression that drive photomorphogenesis (2, 5, 6).
phyA and phyB interact in a conformer-specific manner with basic helix–loop–helix (bHLH) transcription factors called phy-interacting factors (PIFs) (7, 8). PIFs preferentially bind a G-box (CACGTG) DNA sequence element, which is a subclass of an E-box element (CANNTG) present in many light-regulated promoters (9, 10). Interactions between the Pfr form of phyB with PIF3 bound to a G-box promoter motif are hypothesized to directly regulate transcription of light-responsive genes involved in photomorphogenesis (10, 11). However, recent results show that PIFs are stable in the dark and are degraded in response to R and FR light in a phy-dependent manner (8, 12–17), suggesting that activated phys induce degradation of PIFs to promote photomorphogenesis.
Genetic analysis of PIF1 and PIF3-PIF5 suggests that these proteins function as negative regulators of distinct phy-signaling pathways (7, 8). For example, PIF3–PIF5 predominantly control hypocotyl length under R light (9, 18, 19, 20), whereas PIF1 functions as a negative regulator of chlorophyll biosynthesis in the dark and seed germination in FR light (13, 14, 21). PIF1 directly and indirectly regulates gibberellic acid biosynthesis and sensitivity to control seed germination (22). Compared with WT seedlings in the dark, pif1 seedlings accumulate higher amounts of free protochlorophyllide (Pchlide), a phototoxic intermediate in the chlorophyll biosynthetic pathway. Subsequent light exposure causes photooxidative damage and bleaching of pif1 seedlings (13, 21). PIF1 shows transcriptional activation activity in the dark, which is reduced by light-induced degradation of PIF1 to promote chlorophyll biosynthesis and seed germination in light (13, 14). However, the direct target genes by which PIF1 controls chlorophyll biosynthesis have not been identified. Here, we present evidence that PIF1 directly and indirectly regulates key genes in the chlorophyll biosynthetic pathway in the dark to optimize the greening process in Arabidopsis.
Results
PIF1 Regulates Expression of Tetrapyrrole Pathway Genes in the Dark.
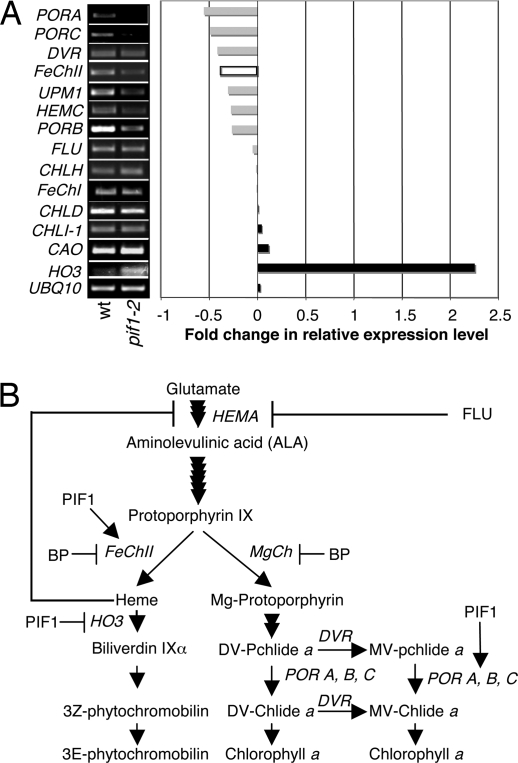
Previously, we have shown that pif1 seedlings have higher levels of Pchlide than WT in the dark (21). Because PIF1 shows strong transcription activation activity in the dark (13, 21), we reasoned that identifying the genes differentially expressed in dark-grown pif1 and WT seedlings may provide further insight into the pif1 phenotype. To this end, we performed whole-genome expression profiling by using Affymatrix Microarray chips on RNA isolated from 4-day-old dark-grown WT and pif1 null mutant seedlings. Using P ≤ 0.05, the Bioconductor microarray analysis software identified only three genes (2.81X, PIF1; 1.96X, At4g17600; 1.91X, At5g44580) differentially expressed between WT and pif1 mutants. One of the three genes is PIF1, which shows a 2.8-fold reduction in expression between WT and the mutant, confirming the validity of our analysis method.
Because the Bioconductor software might be too stringent to detect small expression changes in pif1 seedlings, we used an alternative approach for data analyses as described (23). Using this approach, we identified additional differentially expressed genes (data not shown). Because of PIF1's role in chlorophyll biosynthesis, we focused our analyses on genes involved in the tetrapyrrole pathway [supporting information (SI) Table S1] (24). Interestingly, a few key genes encoding enzymes involved in tetrapyrrole pathway showed expression changes of at least 1.5-fold between the dark grown WT and pif1 samples (Fig. 1 and Table S1). To independently verify our microarray results, a semiquantitative RT-PCR assay was performed. The RT-PCR results largely support the microarray data (Fig. 1A and Table S1). Microarray analysis for ferrochelataseI (FeChI) (At5g26030) and ferrochelataseII (FeChII) (At2g30390), both of which are involved in the conversion of protoporphyrin IX (PPIX) to heme (25, 26), did not show a significant difference between the WT and pif1 samples (Table S1). However, semiquantitative and quantitative RT-PCR (qRT-PCR) analyses showed that FeChII is down-regulated in pif1 seedlings compared with WT (Fig. 1A and Table S1). Taken together, these results suggest that PIF1 is a subtle regulator that controls a small set of key genes involved in chlorophyll biosynthesis.
Fig. 1.
PIF1 regulates key genes involved in the regulation of the tetrapyrrole pathway. (A) (Right) Bar graph shows fold changes of selected genes in pif1 seedlings compared with WT seedlings based on microarray (filled bars) and qRT-PCR (open bars) data. (Left) Independent verification of microarray results using semiquantitative RT-PCR assays of genes involved in tetrapyrrole pathway. RNA was isolated from 4-day-old etiolated seedlings. (B) Tetrapyrrole pathway showing genes directly or indirectly regulated by PIF1. DV-Pchlide, divinylprotochlorophyllide; MV-Pchlide, monovinylprotochlorophyllide; DV-Chlide, divinylchlorophyllide; MV-Chlide, monovinylprotochlorophyllide.
PIF1 Directly Regulates PORC in the Chlorophyll Biosynthesis Pathway.
Because PIFs bind the E/G-box DNA sequence element (CANNTG) (10, 21), we analyzed the upstream promoter region of the differentially expressed genes for the presence of these elements by using the PLACE web site (www.dna.affrc.go.jp/PLACE/signalscan.html). Results show that most of the differentially expressed genes have promoters with two or more E/G boxes (Table S2).
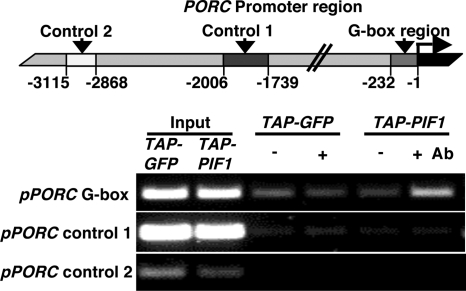
To determine whether these genes are directly regulated by PIF1, we transformed pif1 plants with a construct expressing PIF1 fused to a tandem affinity purification (TAP) tag, 35S:TAP-PIF1 (Fig. S1A) (27). As a control, we expressed a 35S:TAP-GFP construct in the WT background. After confirming that the 35S:TAP-PIF1 transgene complemented pif1 phenotypes (Fig. S1 B–G), we used both transgenic lines in a ChIP assay. After immunoprecipitation of protein–DNA complexes using antibody to the MYC tag, enriched DNA sequences were amplified by using primers to the promoter regions of the candidate genes. ChIP assay results show that the PORC promoter region was amplified from the immunoprecipitation (IP) fraction of 35S:TAP-PIF1 seedlings, but not in the 35S:TAP-GFP or without antibody samples (Fig. 2). Under these conditions, we observed no amplification of the promoter regions of the PORA, DVR, HO3, and FeChII genes. To determine whether these genes were targeted by PIF1 in slightly younger or older seedlings, the ChIP assay was performed on tissue from a range of developmental stages; however, no amplification of these promoters was observed (data not shown). These data suggest that PORC is a direct target of PIF1, whereas PORA, PORB, HO3, and FeChII genes are indirect targets of PIF1.
Fig. 2.
PORC is a direct target of PIF1. (Upper) Illustration of the PORC promoter region. The specific regions amplified by the ChIP assays are shown with nucleotide numbers. (Lower) Gel photographs showing the amplified products from the ChIP assay. The ChIP assay was performed on 3-day-old dark-grown seedlings expressing the TAP-PIF1 or TAP-GFP fusion proteins. Antibody to the MYC tag was used to immunoprecipitate TAP-PIF1/TAP-GFP and associated DNA fragments. DNA was amplified by using primers specific to the region containing the G-box element or control regions in PORC promoter as indicated. +/−, indicates with or without antibody; input, sample before IP.
PIF1 Binds G-Box Motifs Within the PORC and FeChII Promoters.
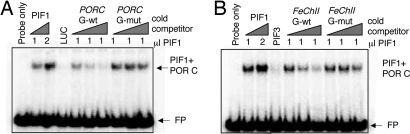
Previously, we have shown that PIF1 binds a synthetic G-box motif by using a gel-shift assay (10, 21). To determine whether PIF1 directly binds the G/E boxes within PORC, a gel-shift assay was performed as described (9, 21). Results show that PIF1 binds the labeled PORC G-box fragment (Fig. 3A). The PORC promoter fragment containing a mutated G-box element did not compete with the WT G-box fragment for PIF1 binding. Because the FeChII promoter has an identical G-box as in the PORC promoter and FeChII expression is regulated by PIF1, we also examined whether PIF1 directly binds to the G-box present in the FeChII promoter. Cold FeChII promoter probe successfully competed with labeled PORC fragments for PIF1 binding (Fig. 3B). Further, mutated G-box FeChII probe did not compete for PIF1 binding with PORC. Control proteins, in vitro expressed-LUC and PIF3, did not bind the PORC G-box sequence in this assay (Fig. 3B). However, a similar PIF3 preparation bound a synthetic G-box originally identified as the PIF3 binding site (data not shown) (10). PIF1 did not bind to PORA and PORB E-box sequences under these experimental conditions (data not shown). These results suggest that PIF1 directly binds to the G-box present in both PORC and FeChII promoters in vitro in a sequence-specific manner.
Fig. 3.
PIF1 binds the G-box motif present in PORC and FeChII native promoters in vitro. (A) Fifteen thousand cpm of 32P-dCTP labeled PORC promoter fragment containing the G-box was incubated with in vitro TNT-expressed PIF1 as indicated. Competition for PIF1 binding was performed with 5×, 25×, or 125× cold PORC probe or mutated G-box (Gm) cold PORC probe. (B) PIF1 binding to PORC-labeled probe was blocked by either WT or G-box-mutated FeChII cold probe. FP, free probe. LUC (A) and PIF3 (B) indicate in vitro-expressed proteins used as controls.
PIF1 Regulates PORC and FeChII Expression in Vivo.
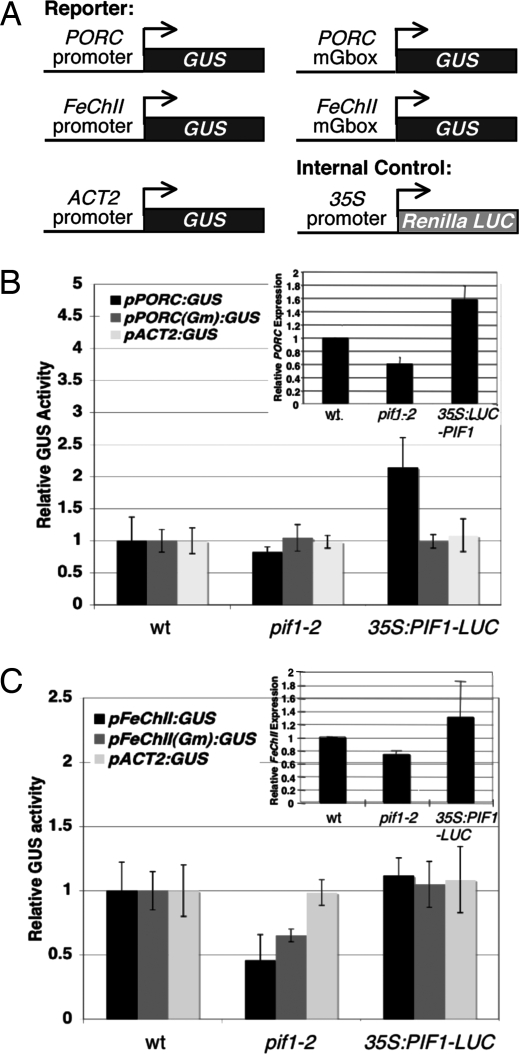
Given that PIF1 is a transcription factor, we wanted to determine whether PIF1 can activate transcription from a native promoter in vivo. As a control, we transiently expressed a non-PIF1 target promoter driving β-glucuronidase (GUS) (pACT2:GUS) in WT, pif1, and PIF1 overexpression (35S:LUC-PIF1) seedlings using the transient assay that we developed (21). GUS assay results show that all three genotypes express the same level of pACT2:GUS, suggesting that PIF1 does not control expression from this promoter (Fig. 4). To determine whether PIF1 can activate transcription from a native promoter, we transiently expressed the native PORC or FeChII promoters driving GUS expression in WT, pif1, and 35S:LUC-PIF1 seedlings (Fig. 4A). Results show that pPORC:GUS activity is significantly higher in 35S:LUC-PIF1 seedlings than in the WT or pif1 seedlings (Fig. 4B). To confirm our results, we measured endogenous PORC expression in these lines by using qRT-PCR assays and found a similar expression pattern as observed for the reporter GUS assays (Fig. 4B Inset). Strikingly, the increased GUS activity in 35S:LUC-PIF1 seedlings expressing pPORC:GUS is eliminated when the G-box within the PORC promoter is mutated (Fig. 4B). These results strongly suggest that PIF1 directly regulates PORC expression in a G-box-dependent manner.
Fig. 4.
PIF1 activates transcription from PORC and FeChII promoters in vivo. (A) Illustration of reporter and internal control constructs used in transient promoter activation assay. (B) Dark-grown WT, pif1, or 35S:LUC-PIF1 seedlings (3.5 days old) were transiently transformed with pACT2:GUS or pPORC:GUS or plasmid containing a mutated promoter G-box motif (pPORCGm:GUS). Relative expression of GUS was measured. WT GUS expression levels are set to 1. n = 3 biological replicates, ± SE. (Inset) qRT-PCR data showing relative expression of PORC in WT, pif1, and 35S:LUC-PIF1 seedlings. WT PORC expression levels are set to 1. n = 5 trials, each with three technical replicates, ± SE. (C) As in B except seedlings were transformed with pFeChII:GUS or pFeChIIGm:GUS. n = three biological replicates, ± SE. (Inset) qRT-PCR data showing relative expression of FeChII in WT, pif1, and 35S:LUC-PIF1 seedlings. WT FeChII expression levels have been set to 1. n = 3 trials, each with three technical replicates, ± SE.
GUS activity in pif1 lines expressing pFeChII:GUS was significantly reduced compared with GUS activity in pFeChII:GUS-expressing WT seedlings (Fig. 4C). Moreover, 35S:LUC-PIF1 lines in the pif1 background showed WT levels of FeChII expression, demonstrating rescue of the pif1 phenotypes in the dark (13). However, the 35S:LUC-PIF1 seedlings did not show overexpression of FeChII. Using qRT-PCR, we found that endogenous FeChII expression levels in WT, pif1, and 35S:LUC-PIF1 seedlings reflect the expression patterns found in the pFeChII:GUS assays (Fig. 4C Inset). In contrast to what was observed in the pPORCGm:GUS assays, the pFeChGm:GUS lines showed no significant change in GUS activity in the WT, pif1, and 35S:LUC-PIF1 backgrounds (Fig. 4C). These results suggest that PIF1 is necessary for activation of FeChII expression in a G-box-independent manner.
Because both PORC and FeChII are modestly induced in light (24), we investigated whether PIF1 plays a role in light regulation of these genes by using the qRT-PCR assays. Results show that the expression of PORC is modestly, but significantly, reduced in pif1 seedlings compared with WT seedlings (Fig. S2). However, pif1 seedlings display a WT FeChII expression level under these light conditions. Because PIF1 is rapidly degraded under light (13), and PORC and FeChII levels are reduced in the dark in the pif1 seedlings compared with WT seedlings (Figs. 1A, 4 B and C, and Fig. S2), these results suggest that PIF1 does not play a significant role in the light-induced expression of these genes.
pif1 Seedlings Have Reduced POR Enzyme Activity.
Microarray and RT-PCR data show that POR genes are down-regulated in pif1 seedlings compared with WT seedlings in the dark (Fig. 1A). To determine whether the transition from Pchlide to chlorophyllide (Chlide) was aberrant in pif1 seedlings, we performed spectrofluorometric analyses on acetone extracts of 4-day-old dark-grown pif1 and WT seedlings with or without a 5-min white light treatment. The results show that although dark-grown pif1 seedlings have a higher relative fluorescence peak at 632 nm, indicative of Pchlide, the relative fluorescence peak at 670 nm, indicative of Chlide, is lower in pif1 seedlings than in WT seedlings after the light treatment (Fig. 5). These in vivo enzyme assay results suggest that pif1 seedlings have reduced levels of POR enzyme activity and, consistent with our microarray data, support our hypothesis that PIF1 regulates expression of the POR genes in the dark (Fig. 1).
Fig. 5.
pif1 seedlings have altered Pchlide and Chlide levels compared with WT seedlings. (A) Relative fluorescence of Pchlide (632 nm) in 4-day-old dark grown WT or pif1 seedlings. (B) Relative fluorescence of Pchlide and Chlide (670 nm) in 4-day-old dark-grown seedlings exposed to 5 min of 80 μmol·m−2·s−1 white light.
PIF1 Regulates Genes Involved in Heme Biosynthesis.
One of the major points of regulation in the chlorophyll pathway is the conversion of PPIX to either Mg-PP, which leads to chlorophyll production, or heme, which leads to phytochromobilin production (Fig. 1B) (25). Heme negatively regulates the chlorophyll pathway by down-regulating δ-aminolevulinic acid (ALA) production (Fig. 1B) (25, 28). Because pif1 seedlings show a reduced level of FeChII and an increased level of HO3 expression in the dark (Table S1 and Figs. 1A and 4C), it is possible that pif1 seedlings have reduced levels of heme compared with WT seedlings. Lower heme levels would result in less feedback inhibition of ALA production and a higher level of Pchlide production (25). Because direct measurement of heme in etiolated Arabidopsis seedlings poses significant technical challenges, we took an indirect approach as described (28). Exogenous application of the iron chelator 2′-2′-bipyridyl (BP) prevents conversion of PPIX to heme and allows accumulation of Mg-PP to detectable levels in seedlings. We measured Mg-PP levels in dark-grown WT and pif1 seedlings incubated with or without BP. Our results show that after BP treatment pif1 seedlings accumulate significantly higher amounts of Mg-PP than WT seedlings (Fig. S3). These data suggest that pif1 seedlings have a reduced amount of heme, possibly resulting from reduced expression of FeChII and an increased expression of HO3 (Figs. 1A and 4C). Alternatively, the higher levels of Mg-PP observed in the pif1 background may be a result of defects in the conversion of ALA to PPIX (Fig. 1B).
To address this notion, we measured PPIX levels in dark-grown seedlings treated with or without 10 mM ALA. Because Pchlide and PPIX fluorescence emission spectra overlap, and given that Pchlide levels are higher in the pif1 background (Fig. 5) (21), absorbance at 503 nm was measured. The results show that pif1 seedlings contain a WT level of PPIX (Fig. S4), suggesting that the elevated levels in Mg-PP found in the pif1 seedlings are a consequence of reduced levels of heme compared with WT seedlings.
Because heme is a negative feedback regulator of the early rate-limiting step in the pathway, reduced levels of heme are expected to increase the rate of ALA biosynthesis (Fig. 1B) (25). We measured the rate of ALA biosynthesis by using a protocol as described (29). The rate of ALA synthesis in pif1 seedlings is ≈2-fold higher than that in WT seedlings (Fig. 6). The modest increase in the rate of ALA synthesis is consistent with the modest increase in Pchlide levels in pif1 seedlings compared with WT seedlings (Fig. 5A). Taken together, these data suggest that PIF1 subtly regulates the level of heme in the dark to fine-tune the tetrapyrrole pathway in Arabidopsis.
Fig. 6.
Increased rate of ALA synthesis in pif1 seedlings compared with WT seedlings. Rate of ALA synthesis measured by absorbance at 553 nm in 3-day-old WT and pif1 seedlings grown in 8-h light/16-h dark cycles. n = 6 biological replicates, ± SE. Samples were harvested at the end of the dark period before the onset of light.
Discussion
Exquisite regulation of the tetrapyyrole pathway in the dark is required to avoid photooxidative damage of seedlings upon illumination. This study provides genetic, molecular, and biochemical evidence that PIF1 directly and indirectly regulates key genes to fine-tune the tetrapyrrole pathway. Several lines of evidence suggest that PORC is a direct target of PIF1. First, microarray and RT-PCR/qRT-PCR assays established that PORC expression is reduced in dark-grown pif1 seedlings compared with WT seedlings (Fig. 1A and Table S1). Second, the ChIP assay shows that PIF1 binds to the promoter of PORC in vivo (Fig. 2). Third, PIF1 directly binds to the G-box element in the PORC promoter (Fig. 3A). Fourth, in transient expression assays PIF1 activates transcription of PORC in a G-box-dependent manner (Fig. 4 A and B). Fifth, regulation of PORC is consistent with our physiological data showing that after initial light exposure Chlide levels in pif1 seedlings are reduced compared with Chlide levels in WT seedlings (Fig. 5). Taken together, these results strongly suggest that PIF1 is a direct regulator of PORC expression.
Expression analyses data suggest that PIF1 regulates all three POR genes, with PORA and PORB displaying the most significant changes in expression (Table S1). However, direct interaction studies show that PORC is the only direct target of PIF1. One distinction between PORA, PORB, and PORC is the cis-elements present in their respective promoters. PORA and PORB promoters have E-boxes, whereas the PORC promoter contains a G-box motif (Table S2). The PIF1 homodimer binds only G-boxes and not E-boxes in in vitro gel-shift assays (Fig. 3 and data not shown). It is probable that PIF1 regulates PORA and PORB expression indirectly and PORC expression directly. Further, POR gene expression is developmentally regulated. PORA and PORB function in young seedlings during the transition from dark to light, and PORC functions in light-grown plants (25). Therefore, PIF1 might control chlorophyll biosynthesis not only during the initial dark-to-light transition, but also during daily light–dark cycles.
The tetrapyrrole pathway is primarily regulated by metabolic intermediates and transcriptional regulation of metabolic enzymes (25). Higher Pchlide content in dark-grown pif1 seedlings suggests that PIF1 either represses genes involved in Pchlide production or activates a repressor that down-regulates Pchlide production. Two well established repressors of the chlorophyll pathway are FLORESCENT (FLU) and heme (25). Both FLU and heme are negative feedback regulators targeting early steps in the chlorophyll pathway to repress production of downstream intermediates (25, 29) (Fig. 1B). Expression analyses confirm that PIF1 does not regulate FLU expression or the expression of other genes involved in conversion of ALA to Pchlide (Fig. 1A, Fig. S4, Table S1, and data not shown). Conversely, PIF1 indirectly activates the expression of FeChII and indirectly represses the expression of HO3 in the dark. FeChII encodes a ferrochelatase enzyme that converts PPIX to heme, and HO3 encodes a heme oxygenase enzyme that converts heme to biliverdin IXα (Table S1 and Figs. 1 and 4C). Although PIF1 regulation of FeChII is subtle (Fig. 1A), the net effect of FeChII and HO3 expression may lead to lower heme content in pif1 seedlings compared with WT seedlings. Reduced heme content relieves the feedback inhibition of ALA synthesis and results in a higher level of Pchlide in pif1 seedlings compared with WT seedlings (Fig. 5A) (21). Increased levels of Mg-PP in pif1 seedlings compared with WT seedlings after BP treatment (Fig. S3A) and the comparable level of PPIX after ALA treatment (Fig. S4A) suggest that pif1 seedlings have less endogenous heme than WT seedlings. Moreover, pif1 seedlings have a modest increase (≈2-fold) in the rate of ALA synthesis compared with WT seedlings (Fig. 6). Interestingly, a reduction in plastidic FeCh in tobacco resulted in an increased rate of ALA synthesis and higher chlorophyll production (25, 30), similar to our results. Combined, our data strongly suggest that PIF1 controls heme levels to optimize Pchlide production in the dark.
Previous work shows that PIF1 functions as a negative regulator of chlorophyll biosynthesis under prolonged light conditions (13, 21). Initially, this finding appears to contradict our conclusion that pif1 seedlings have reduced POR enzyme activity. However, because POR expression is reduced but not eliminated in the pif1 background (Fig. 1A), it is possible that the amount of Pchlide, not the POR enzyme levels, is a limiting factor for chlorophyll biosynthesis under prolonged light conditions. pif1 seedlings have an increased rate of ALA synthesis caused by reduced heme content compared with WT seedlings (Fig. S3 and Fig. 6), resulting in increased Pchlide synthesis in pif1 seedlings (Fig. 5). Therefore, the higher Pchlide level will result in higher chlorophyll synthesis in pif1 seedlings compared with WT seedlings upon prolonged light exposure. Further experiments are necessary to determine whether the POR enzymes or their substrate (Pchlide) is the rate-limiting factor under prolonged light conditions.
PIF1, PIF3, and PIF4 bind a G-box DNA sequence element present in light-regulated promoters, raising questions about how PIFs specify gene targets (Figs. 3 and 4) (9, 10, 21, 31). Our results show that PIF3 does not bind to the G-box present in the PORC and FeChII promoters (Fig. 3B). Both PORC and FeChII promoters contain the G-box sequence, A[CACGTG]T, flanked with an adenine (A) at the 5′ end and a thymine (T) at the 3′ end. Indeed, random DNA binding site selection studies for PIF3 did not isolate any G-box sequence flanked by a 3′ T (10). These results suggest that PIF binding is specified by the sequence flanking the G-box motif in gene promoters, as has been shown for animal bHLH DNA binding (32).
PIFs interact with differential affinities to phys, and PIFs function in distinct phy signaling pathways (8). However, how these interactions result in light regulation of gene expression is still unclear. Our data show that PIF1 constitutively activates gene expression in the dark and does not play a major role in light regulation of these genes (Figs. 1, 4, and S2), which is consistent with the light-induced degradation of PIF1. These results are also consistent with recent reports that both PIF1 and PIF3 constitutively activate gene expression in the dark (22, 33). Therefore, how phys regulate gene expression in response to light remains to be determined.
Although PIF1 regulates key genes in the tetrapyrrole pathway, the effects are subtle. Other bHLH proteins in addition to PIF1 may regulate the expression of PIF1 target genes. The promoters of most of these genes have multiple E/G-boxes within the 500 bp upstream of ATG (Table S2). It is possible that PIF1 binds E-box motifs as heterodimers with other bHLH proteins. The Arabidopsis genome encodes ≈162 bHLH proteins (32), and many of these factors regulate photomorphogenesis (8). It is likely that combinatorial control by multiple factors is necessary to optimize the greening process.
In conclusion, our data show that PIF1 directly and indirectly regulates key genes in the tetrapyrrole pathway in the dark to prepare young etiolated seedlings to respond to light. PIF1 appears to act both positively and negatively to fine-tune the chlorophyll biosynthetic pathway (Fig. 1). Because PIF1 is degraded in light and reaccumulates in the dark (13), PIF1 might provide plants an adaptive advantage under natural light–dark cycles by reducing the daily photooxidative damage at dawn, and thereby ensures robustness and fitness of plants under an ambient light environment.
Materials and Methods
Plant Material and Growth Conditions.
Arabidopsis thaliana ecotype Columbia (Col-0) and the pif1–2 null allele was used for these experiments (13, 21). All seeds were freshly harvested (2–3 months old). Plants were grown on MS media, and seeds were sterilized as in ref. 9.
Microarray Analyses.
Total RNA was isolated from 4-day-old WT and pif1 dark-grown seedlings. Microarray hybridizations and probe synthesis were performed as in ref. 23 on RNA from three independent biological samples. To identify genes that are regulated by PIF1, the data files were also analyzed by using Microsoft Excel as described (23).
RNA Isolation, RT-PCR, and qRT-PCR.
Total RNA was isolated from 4-day-old dark-grown WT, pif1, and 35S:LUC-PIF1 transgenic seedlings by using the RNase Plant Mini Kit (Qiagen) and reverse-transcribed by using SuperScript II (Invitrogen) per the manufacturer's protocol. The qRT-PCR assays used the Power SYBR Green RT-PCR Reagents Kit (Applied Biosystems). Primer sequences used for RT-PCR and qRT-PCR can be found in Table S3, and additional details are available in SI Text.
ChIP Assay.
ChIP assays were performed as in ref. 34, except 3-day-old dark-grown 35S:TAP-PIF1 and 35S:TAP-GFP seedlings were vacuum-infiltrated with 1% formaldehyde for 1 h at 4°C, and cross-linking was quenched by vacuum infiltration with 0.125 M glycine for 3 min. mAb against MYC tag (Calbiochem) was used for IP.
DNA Gel-Shift Assay.
DNA gel-shift assays were performed as described (9, 10). PIF1, PIF3, and LUC were synthesized by using the Rabbit Reticulocyte TNT system (Promega) as described (9). A 70-bp PORC promoter fragment containing a G-box motif was labeled with 32P-dCTP. Cold competitor probe was generated from dimerized oligos of the PORC or FeChII promoter region containing the G-box promoter motif. Probe sequences are shown in Table S3.
Transient Transfection of Promoter-GUS Fusions.
To construct pPORC:GUS, a 1.6-kb promoter region of the PORC gene was cloned into the pENTR vector (Invitrogen), sequenced, and recombined into pBGWFS7 destination vector (35). The G-box element in the PORC promoter was mutated by using a site-directed mutagenesis kit (Stratagene) to produce pPORCGm:GUS. A 1.0-kb promoter region of the FeChII gene was used to construct pFeChII:GUS and pFeChIIGm:GUS as described above. A 1.4-kb promoter region of the ACT2 gene (At3g18780) was used to construct pACT2:GUS as described above. The DNA-coated beads were bombarded into 3.5-day-old WT, pif1, or 35S:LUC-PIF1 transgenic seedlings under dim light as described (21). Seedlings were grown vertically in individually wrapped plates in darkness and opened just before bombardment. Immediately after bombardment, the seedlings were exposed to 15 min of FR light (34 μmol·m−2·s−1) before growing in the dark for 16 h. Total protein was extracted in the darkroom under safe green light, and the protein concentration, Renilla Luciferase, and GUS activity were determined as described (13, 21).
Analysis of Chlorophyll Pathway Intermediates.
Pchlide and Chlide were extracted as in ref. 28 except 4-day-old dark-grown WT and pif1 seedlings were used. Spectrofluorometery (TimeMaster Pro; Photon Technologies International) was performed at an excitation wavelength of 440 nm and an emission wavelength of 600–700 nm, and data were curve-fitted by using PeakFit, version 4.11 (Systat Software). The ALA feeding experiment was carried out as described (28), except ALA or buffer control was vacuum-infiltrated for 5 min at 25 Hg into 4-day-old WT and pif1 seedlings. Measurement of ALA synthesis rate was carried out as in ref. 29 on 3-day-old seedlings grown in 8-h light/16-h dark cycles, and samples were harvested at the end of the dark period.
Additional details are provided in SI Text.
Supplementary Material
Acknowledgments.
We thank Joshua Russell for assistance with the spectrofluorometer, Sharyn Perry for help with ChIP assay, and Phi Luong and Julie Sottilo for technical assistance. This work was supported by National Science Foundation Grant IBN-0418653 and a set-up fund from the University of Texas at Austin (to E.H.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper has been deposited in the GenBank database (accession no. GSE11594).
This article contains supporting information online at www.pnas.org/cgi/content/full/0803611105/DCSupplemental.
References
- 1.Chen M, Chory J, Fankhauser C. Light signal transduction in higher plants. Annu Rev Genet. 2004;38:87–117. doi: 10.1146/annurev.genet.38.072902.092259. [DOI] [PubMed] [Google Scholar]
- 2.Whitelam G, Halliday K. Light and Plant Development. Oxford: Blackwell; 2007. [Google Scholar]
- 3.Chen M, Tao Y, Lim J, Shaw A, Chory J. Regulation of phytochrome B nuclear localization through light-dependent unmasking of nuclear-localization signals. Curr Biol. 2005;15:637–642. doi: 10.1016/j.cub.2005.02.028. [DOI] [PubMed] [Google Scholar]
- 4.Hiltbrunner A, et al. FHY1 and FHL act together to mediate nuclear accumulation of the phytochrome A photoreceptor. Plant Cell Physiol. 2006;47:1023–1034. doi: 10.1093/pcp/pcj087. [DOI] [PubMed] [Google Scholar]
- 5.Rockwell NC, Su Y-S, Lagarias JC. Phytochrome structure and signaling mechanisms. Annu Rev Plant Biol. 2006;57:837–858. doi: 10.1146/annurev.arplant.56.032604.144208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiao Y, Lau OS, Deng XW. Light-regulated transcriptional networks in higher plants. Nat Rev Genet. 2007;8:217–230. doi: 10.1038/nrg2049. [DOI] [PubMed] [Google Scholar]
- 7.Quail PH. Phytochrome interacting factors. In: Whitelam G, Halliday K, editors. Light and Plant Development. Oxford: Blackwell; 2007. pp. 81–105. [Google Scholar]
- 8.Castillon A, Shen H, Huq E. Phytochrome interacting factors: Central players in phytochrome-mediated light signaling networks. Trends Plants Sci. 2007;12:514–521. doi: 10.1016/j.tplants.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 9.Huq E, Quail PH. PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J. 2002;21:2441–2450. doi: 10.1093/emboj/21.10.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martinez-Garcia JF, Huq E, Quail PH. Direct targeting of light signals to a promoter element-bound transcription factor. Science. 2000;288:859–863. doi: 10.1126/science.288.5467.859. [DOI] [PubMed] [Google Scholar]
- 11.Quail PH. Phytochrome photosensory signaling networks. Nat Rev Mol Cell Biol. 2002;3:85–93. doi: 10.1038/nrm728. [DOI] [PubMed] [Google Scholar]
- 12.Bauer D, et al. Constitutive photomorphogenesis 1 and multiple photoreceptors control degradation of phytochrome interacting factor 3, a transcription factor required for light signaling in Arabidopsis. Plant Cell. 2004;16:1433–1445. doi: 10.1105/tpc.021568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shen H, Moon J, Huq E. PIF1 is regulated by light-mediated degradation through the ubiquitin-26S proteasome pathway to optimize photomorphogenesis of seedlings in Arabidopsis. Plant J. 2005;44:1023–1035. doi: 10.1111/j.1365-313X.2005.02606.x. [DOI] [PubMed] [Google Scholar]
- 14.Oh E, et al. Light activates the degradation of PIL5 protein to promote seed germination through gibberellin in Arabidopsis. Plant J. 2006;47:124–139. doi: 10.1111/j.1365-313X.2006.02773.x. [DOI] [PubMed] [Google Scholar]
- 15.Nozue K, et al. Rhythmic growth explained by coincidence between internal and external cues. Nature. 2007;448:358–361. doi: 10.1038/nature05946. [DOI] [PubMed] [Google Scholar]
- 16.Shen Y, Khanna R, Carle CM, Quail PH. Phytochrome induces rapid PIF5 phosphorylation and degradation in response to red-light activation. Plant Physiol. 2007;145:1043–1051. doi: 10.1104/pp.107.105601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Al-Sady B, Ni W, Kircher S, Schafer E, Quail PH. Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol Cell. 2006;23:439–446. doi: 10.1016/j.molcel.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 18.Khanna R, et al. A novel molecular recognition motif necessary for targeting photoactivated phytochrome signaling to specific basic helix-loop-helix transcription factors. Plant Cell. 2004;16:3033–3044. doi: 10.1105/tpc.104.025643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim J, et al. Functional characterization of phytochrome interacting factor 3 in phytochrome-mediated light signal transduction. Plant Cell. 2003;15:2399–2407. doi: 10.1105/tpc.014498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fujimori T, Yamashino T, Kato T, Mizuno T. Circadian-controlled basic helix-loop-helix factor, PIL6, implicated in light-signal transduction in Arabidopsis thaliana. Plant Cell Physiol. 2004;45:1078–1086. doi: 10.1093/pcp/pch124. [DOI] [PubMed] [Google Scholar]
- 21.Huq E, et al. Phytochrome-interacting factor 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science. 2004;305:1937–1941. doi: 10.1126/science.1099728. [DOI] [PubMed] [Google Scholar]
- 22.Oh E, et al. PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by directly binding to the GAI and RGA promoters in Arabidopsis seeds. Plant Cell. 2007;19:1192–1208. doi: 10.1105/tpc.107.050153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hudson ME, Lisch DR, Quail PH. The FHY3 and FAR1 genes encode transposase-related proteins involved in regulation of gene expression by the phytochrome A-signaling pathway. Plant J. 2003;34:453–471. doi: 10.1046/j.1365-313x.2003.01741.x. [DOI] [PubMed] [Google Scholar]
- 24.Matsumoto F, et al. Gene expression profiling of the tetrapyrrole metabolic pathway in Arabidopsis with a mini-array system. Plant Physiol. 2004;135:2379–2391. doi: 10.1104/pp.104.042408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tanaka R, Tanaka A. Tetrapyrrole biosynthesis in higher plants. Annu Rev Plant Biol. 2007;58:321–346. doi: 10.1146/annurev.arplant.57.032905.105448. [DOI] [PubMed] [Google Scholar]
- 26.Singh DV, Cornah JE, Hadingham S, Smith AG. Expression analysis of the two ferrochelatase genes in Arabidopsis in different tissues and under stress conditions reveals their different roles in haem biosynthesis. Plant Mol Biol. 2002;50:773–788. doi: 10.1023/a:1019959224271. [DOI] [PubMed] [Google Scholar]
- 27.Rubio V, et al. An alternative tandem affinity purification strategy applied to Arabidopsis protein complex isolation. Plant J. 2005;41:767–778. doi: 10.1111/j.1365-313X.2004.02328.x. [DOI] [PubMed] [Google Scholar]
- 28.Terry MJ, Kendrick RE. Feedback inhibition of chlorophyll synthesis in the phytochrome chromophore-deficient aurea and yellow-green-2 mutants of tomato. Plant Physiol. 1999;119:143–152. doi: 10.1104/pp.119.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goslings D, et al. Concurrent interactions of heme and FLU with Glu tRNA reductase (HEMA1), the target of metabolic feedback inhibition of tetrapyrrole biosynthesis, in dark- and light-grown Arabidopsis plants. Plant J. 2004;40:957–967. doi: 10.1111/j.1365-313X.2004.02262.x. [DOI] [PubMed] [Google Scholar]
- 30.Papenbrock J, et al. Impaired expression of the plastidic ferrochelatase by antisense RNA synthesis leads to a necrotic phenotype of transformed tobacco plants. Plant J. 2001;28:41–50. doi: 10.1046/j.1365-313x.2001.01126.x. [DOI] [PubMed] [Google Scholar]
- 31.Shin J, Park E, Choi G. PIF3 regulates anthocyanin biosynthesis in an HY5-dependent manner with both factors directly binding anthocyanin biosynthetic gene promoters in Arabidopsis. Plant J. 2007;49:981–994. doi: 10.1111/j.1365-313X.2006.03021.x. [DOI] [PubMed] [Google Scholar]
- 32.Toledo-Ortiz G, Huq E, Quail PH. The Arabidopsis basic helix-loop-helix transcription factor family. Plant Cell. 2003;15:1749–1770. doi: 10.1105/tpc.013839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Al-Sady B, Kikis EA, Monte E, Quail PH. Mechanistic duality of transcription factor function in phytochrome signaling. Proc Natl Acad Sci USA. 2008;105:2232–2237. doi: 10.1073/pnas.0711675105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gendrel AV, Lippman Z, Yordan C, Colot V, Martienssen RA. Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1. Science. 2002;297:1871–1873. doi: 10.1126/science.1074950. [DOI] [PubMed] [Google Scholar]
- 35.Karimi M, De Meyer B, Hilson P. Modular cloning in plant cells. Trends Plants Sci. 2005;10:103–105. doi: 10.1016/j.tplants.2005.01.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.