Abstract
The extent to which polypeptide conformation depends on side-chain composition and sequence has been widely studied, but less is known about the importance of maintaining an α-amino acid backbone. Here, we examine a series of peptides with backbones that feature different repeating patterns of α- and β-amino acid residues but an invariant side-chain sequence. In the pure α-backbone, this sequence corresponds to the previously studied peptide GCN4-pLI, which forms a very stable four-helix bundle quaternary structure. Physical characterization in solution and crystallographic structure determination show that a variety of α/β-peptide backbones can adopt sequence-encoded quaternary structures similar to that of the α prototype. There is a loss in helix bundle stability upon β-residue incorporation; however, stability of the quaternary structure is not a simple function of β-residue content. We find that cyclically constrained β-amino acid residues can stabilize the folds of α/β-peptide GCN4-pLI analogues and restore quaternary structure formation to backbones that are predominantly unfolded in the absence of cyclic residues. Our results show a surprising degree of plasticity in terms of the backbone compositions that can manifest the structural information encoded in a sequence of amino acid side chains. These findings offer a framework for the design of nonnatural oligomers that mimic the structural and functional properties of proteins.
Keywords: α/β-peptides, foldamers, protein folding
The remarkable relationships among subunit sequence, folding pattern, and function in proteins and nucleic acids are fundamental to the existence of life, and the development of a detailed understanding of the interplay among these molecular properties remains a significant challenge (1, 2). The complex behavior of proteins and nucleic acids has motivated a wide range of research, which has broadened in recent years to encompass unnatural oligomers that display biopolymer-like conformational behavior and even biopolymer-like functions. Such efforts have included the study of RNA analogues based on pyranose rather than furanose (3), peptides based on β- or γ-amino acids rather than α-amino acids (4, 5), and systems that diverge more profoundly from biological precedents (6, 7). As the folding propensities of new backbone elements have been elucidated, the first steps have been taken to blend natural and unnatural subunits and thereby create folded oligomers with heterogeneous backbones and distinctive behavior (8–10).
The design of synthetic oligomers that display specific structures and functions requires an understanding of sequence → structure → function relationships among unnatural backbones. Past work on the folding behavior of peptides and proteins indicates that limited backbone modification in a given sequence, including replacement of one or two α-residues with β-residues, can be accomplished without significantly altering native structure or function (11–13). More recently, we have shown that a particular type of nonnatural backbone, composed of a mixture of α- and β-amino acid residues in an ααβαααβ pattern, can partially mimic the folding and self-assembly behavior of parent α-peptide side-chain sequences (14, 15). This observation raises an interesting question: How general is the ability of nonnatural peptide backbones to manifest the higher-order structure encoded in a particular sequence of amino acid side chains? This question has previously been explored conceptually. In pioneering work on the development of the simple exact lattice (“beads on a string”) model of protein folding, Dill et al. (16) hypothesized that nonprotein oligomers with appropriately designed sequences should be able to manifest protein-like properties.
Here, we probe the relationship between backbone composition and sequence-encoded folding in the context of different α/β amino acid combinations. We compare the folding and self-assembly behavior, in solution and crystals, of several 33-residue peptides that contain a side-chain sequence known to induce four-helix bundle formation when projected from a purely α-residue backbone. These “α/β-peptides” contain various regular combinations of α- and β-amino acid residues. Our results show that the ability of heterogeneous α/β backbones to form a helix bundle quaternary structure, as dictated by a specific sequence of side chains, is a general feature of diverse α/β backbone patterns, with intriguing variations in assembly mode and propensity among particular backbones. In addition, we find that quaternary structure stability can be enhanced based on fundamental understanding of β-residue conformational propensities, by taking advantage of useful avenues for conformational preorganization that are available for these unnatural subunits.
Results and Discussion
Experimental Design.
α-Helical coiled-coil assemblies constitute one of the most intensely studied families of protein quaternary structure and have proven excellent model systems for recapitulating the folding complexity of proteins in synthetically tractable peptides (17, 18). We selected a well studied sequence as the basis for our studies. GCN4-pLI (1, Fig. 1A) is a 33-residue α-peptide derived from the dimerization domain of the yeast transcriptional regulator GCN4 (19–21). GCN4-pLI forms a very stable four-helix bundle (16 kDa) and features a canonical heptad residue repeat pattern (abcdefg), with hydrophobic Leu and Ile residues occupying most a and d positions, respectively (Fig. 1B). The a and d residue positions align along one face of the α-helix and define the hydrophobic core of the helix bundle assembly. We previously described α/β-peptide 2, an analogue of GCN4-pLI with an ααβαααβ residue repeat (Fig. 1A) (14). Each α → β substitution in 2 consists of a β3-amino acid bearing the side chain of the replaced α-residue in the GCN4-pLI sequence (Fig. 1C). This substitution strategy leads to an oligomer with the same sequence of side chains as 1 but a different backbone. α/β-Peptide 2 adopts a parallel four-helix bundle fold in the crystalline state, analogous to that of GCN4-pLI α-peptide 1; however, the behaviors of 1 and 2 differ in solution, with the α-peptide self-assembling to a tetramer and the α/β-peptide forming a trimer (14). These variations in stoichiometry complicate the interpretation of observed differences in helix bundle stability and led us to wonder whether the loss in specificity for the sequence-encoded oligomerization state was a general consequence of extensive backbone modification. We therefore prepared and tested a series of derivatives with other regular α → β3 replacement patterns that maintain the GCN4-pLI side-chain sequence.
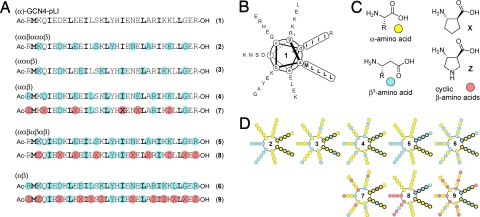
Fig. 1.
Chemical structures and helical wheel diagrams of 1–9. (A) Primary sequences of α-peptide 1 and α/β-peptides 2–9, sorted according to α/β backbone pattern. Bold letters indicate hydrophobic core residues in the GCN4-pLI sequence. Colored circles indicate sequence positions occupied by β-residues, cyan for β3-residues and orange for cyclic β-residues. (B) Helical wheel diagram of 1 with hydrophobic core residues indicated. (C) Structures of an α-amino acid, a β3-amino acid, and the cyclic β-amino acids ACPC (X) and APC (Z). (D) Helical wheel diagrams of the α/β residue patterns of 2–9 based on a heptad repeat. Each circle represents a residue and is colored by residue type, yellow for α-residues, cyan for β3-residues, and orange for cyclic β-residues. Bold circles indicate hydrophobic core positions.
Previously uncharacterized α/β-peptides 3–6 each bear the side-chain sequence of GCN4-pLI with different patterns of α → β3 replacement (Fig. 1): αααβ, ααβ, ααβαβαβ, and αβ, respectively. Among these oligomers, only 5 features a β substitution pattern (ααβαβαβ) that follows the heptad repeat of the α-helix. α/β-Peptide 2 also has a substitution pattern that follows the heptad repeat, which in this case confines the β-residues to the periphery of the helix bundle quaternary structure (14). For 3, 4, and 6, formation of an α-helix-like conformation, presumably a prerequisite for self-association, would cause the β-residues to spiral around the helix axis (Fig. 1D). α/β-Peptides 3–6 all have multiple β-residue substitutions at hydrophobic core positions in the GCN4-pLI sequence in contrast to α/β-peptide 2, which has no core substitutions.
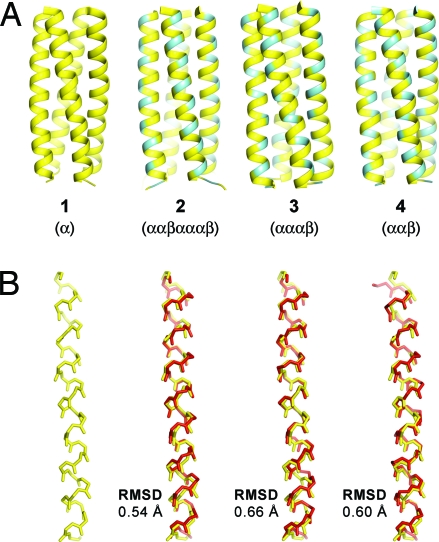
We obtained crystals of 3 and 4 by hanging-drop vapor diffusion, and diffraction data were collected and solved to 2.0 Å and 1.8 Å, respectively [supporting information (SI) Table S1]. Each α/β-peptide adopts a parallel four-helix bundle quaternary structure that is quite similar to those previously observed for GCN4-pLI (1) and α/β derivative 2 (Fig. 2A). Individual helices formed by the different α/β backbones of 3 and 4 show significant homology to an α-helix (Fig. 2B, Table S2). Helical folds adopted by both the αααβ backbone of 3 and the ααβ backbone of 4 display the i → i+4 hydrogen bonding pattern of an α-helix, and Cα atoms of α-residues in each of the two new structures overlay well with the corresponding α-residues in GCN4-pLI (1). The local conformations adopted by α- and β-residues in the helices do not vary over different α/β backbone compositions; backbone dihedral angles for α-residues in 2, 3, and 4 are tightly clustered in the α-helical region of the Ramachandran plot, and the β3-residues show significant internal consistency in their backbone torsional preferences (Table S3). Side-chain rotamer preferences among β-residues in 2–4 do not appear to be significantly affected by the local change in backbone chemical composition (Fig. S1).
Fig. 2.
Comparison of the GCN4-pLI side chain sequence on four different backbone patterns. (A) Helix bundle quaternary structures of each derivative with yellow and blue indicating α- and β3-residues, respectively. (B) Helical secondary structures of each α/β-peptide 2–4 (red) compared with that of α-peptide 1 (yellow); the overlays and accompanying RMSD values are based on Cα atoms in shared α-residues. The coordinates for 1 (PDB: 1GCL) (21) and 2 (PDB: 2OXK) (14) were obtained from previously published structures.
The packing of hydrophobic core residues in GCN4-pLI is a key component in specifying the structure, oligomerization state, and stability of the helix bundle (21, 22). The occurrence of β3-residues in the hydrophobic cores of tetramers formed by 3 and 4 does not substantially alter the helix bundle quaternary structure relative to α-peptide prototype 1. For some core β-residues (β3-hLeu23 of 3 and β3-hLeu16 of 4), the packing is comparable with that of the α-residue at the equivalent position in GCN4-pLI (Fig. S2). At other positions (β3-hIle19 of both 3 and 4), the packing is less efficient, leading to a small cavity in the core of the helix bundle (Fig. S2). This imperfect core packing may contribute to the reduced stability of 3 and 4 relative to α-peptide 1 and α/β-peptide 2 as determined by the measurements detailed below.
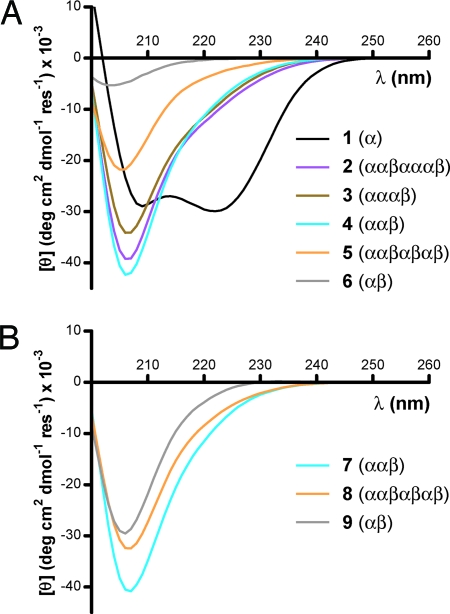
We compared the folding/assembly behavior of 1–6 in aqueous solution (Fig. 3A and Table 1). Analytical ultracentrifugation (AU) suggests that 3 and 4 form discrete tetrameric assemblies (200 μM; 25°C), which is consistent with the self-association stoichiometry of each α/β-peptide in the crystalline state. In contrast, α/β-peptide 5, which has higher β-residue content than 3 or 4, does not appear to self-associate in a well defined stoichiometry at 200 μM. In light of the self-association established for 2–4 by AU, CD data suggest that α-helix-like secondary structure in these α/β-peptides gives rise to an intense CD minimum at ≈206 nm (Fig. 3A). This CD signature is similar to reported spectra for helical α/β-peptides with a 1:1 backbone alternation of α- and β-residues (23). α/β-Peptide 6 appears to be poorly folded, as indicated by a low-intensity CD signature that is independent of concentration and temperature. CD spectra of 6, along with high-temperature CD data obtained during the course of thermal denaturations of 2–5, suggest that unfolded α/β-peptides are generally characterized by a less-intense CD minimum ([θ] ∼6,000 to -10,000 deg cm2 dmol−1 residue−1) that is slightly blue shifted (λmin ≈ 202 nm) with respect to the folded state. α/β-Peptides 3 and 4 both display cooperative thermal unfolding transitions (Fig. S3) with midpoints (Tm) at ≈77° and 82°C, respectively, at 100 μM concentration. The Tm values vary with α/β-peptide concentration, as expected if the helicity detected by CD develops concomitantly with self-assembly (Table 1). Coupling of helix formation and self-assembly is well known for 1 and other α-peptides that form helix bundles (19, 21).
Fig. 3.
CD spectra of 1–9. (A) CD spectra for GCN4-pLI α-peptide 1 and α/β-peptides 2–6 generated from simple α → β3 substitution. (B) CD spectra for GCN4-pLI α/β-peptide derivatives bearing cyclic β-residues. The colors of the spectra in B match the corresponding acyclic β-residue derivatives in A. All spectra were acquired for 100 μM α-peptide or α/β-peptide in 10 mM NaOAc (pH 4.6).
Table 1.
Summary of CD and AU data for 1-9
| Peptide | Backbone pattern | Cyclic β-residues | [θ]min, deg cm2 dmol−1 res−1* |
Tm, °C† |
Nassoc‡ | |
|---|---|---|---|---|---|---|
| 100 μM | 25 μM | |||||
| 1 | (α) | − | −30,000 | >100 | n.d. | 4 |
| 2 | (ααβαααβ) | − | −39,000 | >100 | >100 | 3 |
| 3 | (αααβ) | − | −34,000 | 77 | 63 | 4 |
| 4 | (ααβ) | − | −42,000 | 82 | 73 | 4 |
| 5 | (ααβαβαβ) | − | −22,000 | 44 | – | (1,3) |
| 6 | (αβ) | − | −5,000 | – | – | n.d. |
| 7 | (ααβ) | + | −41,000 | >100 | >100 | 4 |
| 8 | (ααβαβαβ) | + | −32,000 | >100 | >100 | 4§ |
| 9 | (αβ) | + | −30,000 | 80 | 70 | (1,5) |
*Minimum molar ellipticity observed in the CD spectrum of a 100 μM sample in 10 mM NaOAc (pH 4.6) at 25°C.
†Thermal unfolding transitions observed by CD of peptide at the indicated concentration in 10 mM NaOAc (pH 4.6).
‡Apparent association state determined by AU of a 200 μM sample in 10 mM NaOAc (pH 4.6), 150 mM NaCl; numbers in parentheses indicate a mixture of two species.
§Data acquired for a 100 μM sample in 10 mM NaOAc (pH 4.6); higher concentration and the addition of 150 mM NaCl led to aggregation.
Overall, the crystal structures of 2–4 and the solution data for 2–6 demonstrate that a variety of different α/β backbone patterns can manifest the helix bundle quaternary structure encoded in the GCN4-pLI side-chain sequence. The ααβαααβ, αααβ, and ααβ patterns of 2, 3 and 4, respectively, each exhibit a folded structure reminiscent of that displayed by α-peptide 1. Comparisons among 1–6 in solution show a general decline in quaternary structure stability upon β-residue incorporation. Quaternary structure stability is not, however, a simple function of β-residue content: 4, with 11 β-residues, forms a four-helix bundle that has a higher Tm than does the four-helix bundle formed by 3, with 8 β-residues. The peptides containing the largest number of β3-residues, 5 and 6, failed to adopt stable folds in solution. One possible explanation for the poor conformational stability of the peptides with high β-residue content is that the helices formed by these backbones are too different from an α-helix to exhibit the quaternary structure encoded by the GCN4-pLI side-chain sequence in a pure α-residue context. Given the remarkable structural homology observed among 1–4, however, we favor an alternative hypothesis. We propose that the differences in quaternary structural stability among 1–6 are strongly influenced by an entropic penalty associated with the larger number of torsional restrictions necessary for helix formation by a β3-residue relative to an α-residue. This proposal is consistent with differences noted for formation of isolated helices by pure α and pure β3 backbones (24). We tested the residue flexibility hypothesis with the experiments described below.
β3 → Cyclic β-Residue Substitutions.
If the loss of quaternary structural stability that accompanies α → β3-residue replacements among GCN4-pLI analogues 2–6 arises at least in part from increased torsional freedom in the backbone, then it should be possible to recover stability by incorporation of appropriately rigidified β-residues. Previous work with β-peptides (25, 26) and 1:1 α/β-peptides (27, 28) has shown that cyclic β-residues can enhance helicity. The cyclic constraint (e.g., residues X and Z, Fig. 1C) restricts rotation about the Cα–Cβ backbone bond of a β-residue. Theoretical studies of protein-folding energetics suggest that decreasing residue configurational entropy should be an effective general strategy for improving conformational stability in any natural or nonnatural oligomer backbone (29). With the above considerations in mind, we prepared and characterized α/β-peptides 7, 8, and 9, derivatives of 4, 5, and 6 in which a subset of β3-residues is replaced with cyclically constrained analogues (Fig. 1).
We selected β-amino acids with a five-membered ring constraint, such as (S,S)-trans-2-aminocyclopentane carboxylic acid (ACPC; X in Fig. 1), for incorporation into the GCN4-pLI α/β-peptides. This particular cyclic constraint has been shown to promote helical folding in peptides with a 1:1 α/β pattern (27, 28, 30). The backbone dihedral angles adopted by ACPC residues in β- and α/β-peptide crystal structures (26, 27, 30, 31) are similar to those seen for the β3-residues in the crystal structures of 2, 3, and 4. A pyrrolidine-based analogue of ACPC (Z in Fig. 1) was incorporated into 7–9 at sequence positions that originally (i.e., in 4–6) contained β3-residues with a cationic side chain in an effort to minimize deviation from the charge distribution of the original side-chain sequence. Visual inspection of the structures of 3 and 4 suggested that core packing of the four-helix bundle may be improved by replacement of β3-hIle19 with ACPC.
The data obtained for 7, 8, and 9 in solution support our hypothesis that replacing β3-residues with cyclic β-residues can improve helix bundle stability (Table 1). Each peptide containing cyclic residues displays an intense CD minimum near 206 nm similar to those observed for the well folded α/β-peptides containing only acyclic (β3) residues (Fig. 3B). AU analysis indicates that 7–9 all self-associate in aqueous solution, and variable-temperature CD analysis suggests that the assemblies formed by 7–9 are significantly more stable than the assemblies formed by more flexible counterparts 4–6 (Table 1). α/β-Peptide 7 forms a discrete tetramer in solution, which parallels the assembly behavior of flexible analogue 4. α/β-Peptide 8 forms a tetramer, in contrast to the less-specific self-assembly behavior of more flexible analogue 5; however, 8 shows nonspecific high-order aggregation under certain experimental conditions (SI Text). The AU data for 9 can be fit to an equilibrium between monomer and 5-mer. Although this result constitutes a departure from the tetrameric assembly of α prototype 1, the folding behavior of 9 nevertheless represents an improvement relative to flexible analogue 6, for which CD data indicate little or no helical structure.
The use of cyclic β-residues in 7–9 necessarily involves departure from the side-chain sequence that is common among 1–6. Having established that cyclic residue incorporation can improve fold stability in α/β-peptide analogues of GCN4-pLI, we sought to determine the structural consequences of the sequence alterations. We collected diffraction data on crystals of 7 and solved the structure to 2.2 Å resolution (Fig. 4 and Table S1). α/β-Peptide 7 has the same ααβ backbone pattern as 4, and these two oligomers differ only in six substitutions of a β3-residue for a cyclic β-residue. The unit cell dimensions and crystal symmetry suggested that crystals of 7 were isomorphic with those of 4; however, the refined structure of 7 showed some subtle but important differences. The overlay of the helix bundles of 4 and 7 is rather poor (backbone RMSD = 1.15 Å for residues 2–30), considering the high sequence homology between these oligomers (Fig. 4B). The divergence between the structures is localized to the C-terminal half of the sequence; the N-terminal segment shows excellent overlap (backbone RMSD = 0.28 Å for residues 2–17).
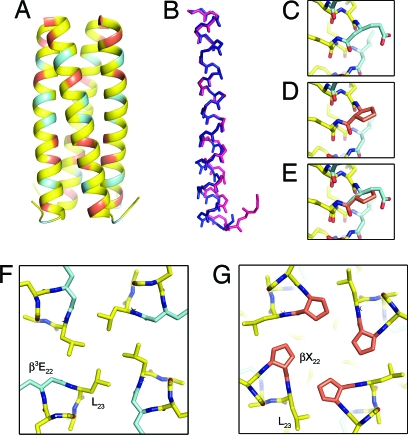
Fig. 4.
Comparison of the crystal structures of α/β-peptides 4 and 7. (A) Four-helix bundle quaternary structure of α/β-peptide 7. (B) Overlay of the helical backbone of 7 (pink) with that of 4 (blue). (C–E) Comparison of an acyclic β3-residue in 4 to the cyclic β-residue at the same position in 7: β3-hGlu10 in 4 (C); β-ACPC10 in 7 (D); overlay of C and D (E). (F and G) Top view of the hydrophobic core packing at residues 22 and 23 in 4 (F) and 7 (G).
Inspection of the backbone conformations of the three cyclic β-residues in the N-terminal segment of 7 indicates that the rigidified subunits effectively mimic the conformation of the flexible β3-residues in 4 (Fig. 4 C–E). The structural differences between 4 and 7 arise not from changes in helical secondary structure but, rather, from a tighter superhelical winding in the C-terminal region of the helix bundle. The source of this tighter winding appears to be the substitution at residue 22, β3-hGlu → ACPC. Glu22 occupies an interface g position in GCN4-pLI (1), and β3-hGlu22 in 4 adopts a similar position (Fig. 4F). In contrast, ACPC22 in α/β-peptide 7 (Fig. 4G) occupies a position in the hydrophobic core that is occupied by Leu23 in 4 and GCN4-pLI oligomers 1–3. This change leads to a break in the heptad repeat of hydrophobic core residues along the GCN4-pLI sequence (SI Text and Fig. S4). Such discontinuities are well documented in natural coiled-coil structures, and the type seen in the structure of 7 is an example of a “helical stammer.” (32) Here, the stammer appears to result, at least in part, from replacing a negatively charged Glu side chain in the parent GCN4-pLI sequence with a hydrophobic ACPC side chain (SI Text).
Overall, the significant and consistent improvements in fold stability observed for oligomers 7–9 relative to more flexible analogues 4–6 show that proper choice and placement of constrained β-residues can result in helix bundle quaternary structures with stabilities and architectures approaching those of an α-peptide prototype. This trend supports the hypothesis that the increased number of low-energy torsions in a β3-residue relative to an α-residue can lead to diminished conformational stability in α/β-peptides relative to analogous α-peptides but that the rigidification strategies possible with β-residues can allow recovery of that lost stability. Subtle differences between the crystal structures of 7 and 4 as well as deviations from tetrameric association of 8 (under some conditions) and 9, however, underscore the importance of primary sequence in dictating folded structure. Cyclic β-residues can rigidify secondary structure and stabilize quaternary structure, but the loss of sequence information resulting from side-chain modification may affect the ability of an unnatural peptide backbone to reproduce sequence-encoded folding behavior displayed by an α-peptide prototype.
Implications for Protein Folding.
The results here provide direct experimental evidence bearing on hypotheses arising from two distinct theoretical treatments of protein folding. A key tenet of the beads on a string protein folding model described by Dill et al. (16) is that the relationship between side chain sequence and folding is independent of backbone composition, and this model predicts that nonprotein backbones should be capable of manifesting protein-like structure. The results for our α/β-peptide analogues of GCN4-pLI bear out this hypothesis to some extent, but an energetic penalty (presumably entropic) for β-residue incorporation on quaternary structure stability is apparent. Interestingly, the loss of quaternary structure stability accompanying α → β-residue substitution is predicted from a key principle of protein folding, the relationship between fold stability and backbone configurational entropy of the unfolded state. The spin glass theory of Wolynes suggests that per-monomer configurational entropy plays an important role in folding and predicts that backbone rigidification should promote folding and higher-order structure in both natural and nonnatural oligomers (29, 33). The theoretical role of backbone rigidity in protein stability is supported by experimental examples such as a study by Matthews et al. (34) that demonstrated that a single Ala → Pro substitution at a solvent-exposed position in T4 lysozyme improves stability without impacting structure or enzymatic activity. Our findings regarding the consequences of cyclic β-residue incorporation on the fold stability of GCN4-pLI α/β-peptides suggest that, analogous to the case for proteins, controlling backbone configurational entropy may prove to be a general strategy for promoting tertiary and quaternary structure in oligomers with mixed α/β backbones. It is noteworthy that cyclic backbone constraint of the Cα–Cβ torsion of β-residues, unlike Xxx → Pro substitution, does not preclude the participation of amide protons in hydrogen bonding.
Conclusions.
We have demonstrated folding and self-assembly in diverse α/β-peptide backbones displaying the primary side-chain sequence of an α-peptide prototype. In three of the oligomer backbones, ααβαααβ, αααβ, and ααβ, systematic α → β3 substitution can lead to faithful mimicry of the quaternary structure encoded by the prototype sequence. In backbone patterns with high β-residue content, ααβ, ααβαβαβ, and αβ, coupled folding and self-assembly can be dramatically enhanced by replacing a subset of acyclic β3-residues with cyclically constrained analogues. Overall, these results reveal a surprising degree of plasticity with respect to the backbone compositions that can recapitulate the quaternary structure encoded by the GCN4-pLI side-chain sequence; however, it is important to note that α → β substitutions can impact both conformational preferences and conformational stability. The relationship between fold stability and backbone composition is complex, and we show here only what is possible to achieve by relatively straightforward modification of a prototype sequence. Our results indicate that the particular side chains selected for substitution with cyclically constrained β-residues are critical in determining how effectively those modifications stabilize a target folded structure. Recent function-oriented studies with α/β-peptides based on a BH3 domain have shown that the properties of α/β-peptides prepared by simple sequence-based design, of the type illustrated here, depend on the frame shift of the backbone repeat within a given sequence (15). Continuing exploration of sequence-based backbone modification will lead to a better understanding of the relationships among backbone composition, side chain sequence, and folding, which should ultimately enable the design of nonnatural oligomers that mimic the structural and functional complexity of proteins. Recent advances in the ribosomal synthesis of peptides with significant unnatural amino acid content are intriguing in this regard (35).
Materials and Methods
Synthesis of α- and α/β-Peptides.
α-Peptide 1 and α/β-Peptides 2–5 were prepared by standard Fmoc automated solid phase synthesis. α/β-Peptides 6–9 were prepared by microwave-assisted Fmoc solid-phase peptide synthesis (36, 37). Detailed synthetic methods can be found in SI Text. Crude peptides were purified to >95% purity by preparative reverse-phase HPLC on a C18 column by using gradients between 0.1% vol/vol TFA in water and 0.1% vol/vol TFA in acetonitrile. The identity and purity of 1–9 were confirmed by MALDI-TOF-MS and analytical HPLC, respectively. Stock solutions for physical characterization were quantified by UV absorbance assuming ε276 = 1450 M−1 cm−1 for the Tyr or β3-hTyr residue in each sequence (38).
Crystallization and Data Collection.
Crystallizations were performed by the hanging-drop vapor-diffusion method. Peptide stock solutions were ≈15 mg/ml in water and mixed 1:1 with the crystallization buffers indicated before equilibration. α/β-Peptide 3 was crystallized from 0.2 M (NH4)2SO4, 0.1 M Tris (pH 8.5), 30% vol/vol PEG 550 monomethyl ether, and the crystal was frozen after cryoprotection in the above buffer supplemented with 5% vol/vol glycerol. α/β-Peptide 4 was crystallized from 0.1 M Hepes (pH 7.5), 4.3 M NaCl, and the crystal was frozen after cryoprotection in the above buffer supplemented with 25% vol/vol glycerol. α/β-Peptide 7 was crystallized from 0.7 M diammonium tartrate, 0.1 M Tris (pH 8.5) and frozen after cryoprotection in 1 M diammonium tartrate, 0.1 M Tris (pH 8.5), 30% vol/vol glycerol. Crystals were frozen in liquid nitrogen, and diffraction data were collected on a Bruker X8 Proteum Diffractometer by using Cu Kα radiation. Data were indexed, integrated, and scaled with the Bruker Proteum2 software package. The crystal of 3 diffracted to 2.0 Å resolution with P21 symmetry and unit cell dimensions of a = 35.2 Å, b = 35.2 Å, c = 47.3 Å, β = 90.9°. Attempts to integrate and scale the data for 3 in higher-symmetry crystal systems led to significantly worse statistics, supporting the assignment of the monoclinic space group. The crystal of 4 diffracted to 1.8 Å resolution with P4212 symmetry and unit cell dimensions a = b = 38.4 Å, c = 46.5 Å. The crystal of 7 diffracted to 2.0 Å resolution with P4212 symmetry and unit cell dimensions a = b = 35.5 Å, c = 45.8 Å.
Structure Determination.
Structure solution was carried out by using the CCP4 software suite (39). Molecular replacement was carried out with Phaser (40). The search model for 3 was derived form the published structure of α-peptide 1 (21), the model for 4 was derived from the published structure of α/β-peptide 2 (14), and the model for 7 was derived from the refined structure of 3. The same Rfree sets were used for 3 and 7 to avoid contamination of the test set used during the refinement of 7. Refinement was accomplished by a combination of Refmac (41) for automated refinement, Coot (42) for manual model building, and ARP/wARP (43) for map improvement by free atom density modification and water building. A Refmac library containing geometric restraints for the β-amino acid residues as well as the α → β and β → α amide linkages was included in the refinement. Helical and superhelical parameters (Table S2) were calculated by using the program TWISTER (44).
CD.
CD measurements were carried out on an Aviv 202SF Circular Dichroism Spectrophotometer with a 1-nm step and 5-sec. averaging time. Thermal melts were carried out in 5°C intervals with an equilibration time of 10 min. after each temperature change. In cases where cooperative folding transitions were observed, Tm values were determined by fitting the data to a simple two-state folding model by using GraphPad Prism. The unfolded baselines for samples that did not show complete unfolding were estimated based on baselines observed for samples that did unfold. Although such estimates may compromise the accurate determination of Tm values, qualitative trends in stability are evident from the raw thermal denaturation data (Fig. S3) and support the relative stability within the series as determined from Tm values.
AU.
AU measurements were carried out on a Beckman–Coulter XL-A Analytical Ultracentrifuge equipped with an An-60 Ti Rotor (Beckman-Coulter part number 361964, Fullerton, CA). All samples were prepared at a final concentration of 200 μM in 10 mM NaOAc (pH 4.6), 150 mM NaCl except for α/β-peptide 8, which was measured at 100 μM in 10 mM NaOAc (pH 4.6). Samples were measured at three or more speeds ranging from 12,000 rpm to 45,000 rpm and monitored at 275 nm (Figs. S5 and S6). Apparent molecular mass for each peptide was determined by global nonlinear fitting of the equilibrium radial absorbance data at all speeds by using the Igor software package (WaveMetrics). Details regarding the fitting procedure can be found in SI Text.
Supplementary Material
Acknowledgments.
We thank Prof. James Keck for providing access to x-ray instrumentation and for helpful comments on the manuscript. We thank PepTech for providing protected β3-amino acid derivatives at a discount. This work was supported by National Institutes of Health (NIH) Grant GM61238 and CA119875 Fellowship (to W.S.H.). CD and AU data were obtained at the University of Wisconsin Biophysics Instrumentation Facility, which was established with support from the University of Wisconsin and National Science Foundation Grant BIR-9512577 and NIH Grant S10 RR13790. The microwave reactor was purchased with support from the University of Wisconsin, Madison Nanoscale Science and Engineering Center (DMR-0425880).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition footnote: Atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3C3F for 3, 3C3G for 4, and 3C3H for 7).
This article contains supporting information online at www.pnas.org/cgi/content/full/0801135105/DCSupplemental.
References
- 1.Schueler-Furman O, Wang C, Bradley P, Misura K, Baker D. Progress on modeling of protein structures and interactions. Science. 2005;310:638–642. doi: 10.1126/science.1112160. [DOI] [PubMed] [Google Scholar]
- 2.Shapiro BA, Yingling YG, Kasprzak W, Bindewald E. Bridging the gap in RNA structure prediction. Curr Opin Struct Biol. 2007;17:157–165. doi: 10.1016/j.sbi.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 3.Eschenmoser A. Chemical etiology of nucleic acid structure. Science. 1999;284:2118–2124. doi: 10.1126/science.284.5423.2118. [DOI] [PubMed] [Google Scholar]
- 4.Cheng RP, Gellman SH, DeGrado WF. β-Peptides: From structure to function. Chem Rev. 2001;101:3219–3232. doi: 10.1021/cr000045i. [DOI] [PubMed] [Google Scholar]
- 5.Seebach D, Beck AK, Bierbaum DJ. The world of β- and γ-peptides comprised of homologated proteinogenic amino acids and other components. Chem Biodivers. 2004;1:1111–1239. doi: 10.1002/cbdv.200490087. [DOI] [PubMed] [Google Scholar]
- 6.Hill DJ, Mio MJ, Prince RB, Hughes TS, Moore JS. A field guide to foldamers. Chem Rev. 2001;101:3893–4011. doi: 10.1021/cr990120t. [DOI] [PubMed] [Google Scholar]
- 7.Hecht S, Huc I, editors. Foldamers: Structure, Properties, and Applications. Weinheim, Germany: Wiley-VCH; 2007. [Google Scholar]
- 8.Chatterjee S, Roy RS, Balaram P. Expanding the polypeptide backbone: Hydrogen-bonded conformations in hybrid polypeptides containing the higher homologues of α-amino acids. J R Soc Interface. 2007;4:587–606. doi: 10.1098/rsif.2006.0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Pol S, Zorn C, Klein CD, Zerbe O, Reiser O. Surprisingly stable helical conformations in α/β-peptides by incorporation of cis-β-aminocyclopropane carboxylic acids. Angew Chem Int Ed. 2004;43:511–514. doi: 10.1002/anie.200352267. [DOI] [PubMed] [Google Scholar]
- 10.Hayen A, Schmitt MA, Ngassa FN, Thomasson KA, Gellman SH. Two helical conformations from a single foldamer backbone: “Split personality” in short α/β-peptides. Angew Chem Int Ed. 2004;43:505–510. doi: 10.1002/anie.200352125. [DOI] [PubMed] [Google Scholar]
- 11.Arnold U, et al. Protein prosthesis: A semisynthetic enzyme with a β-peptide reverse turn. J Am Chem Soc. 2002;124:8522–8523. doi: 10.1021/ja026114n. [DOI] [PubMed] [Google Scholar]
- 12.Roy RS, Karle IL, Raghothama S, Balaram P. α,β hybrid peptides: A polypeptide helix with a central segment containing two consecutive β-amino acid residues. Proc Natl Acad Sci USA. 2004;101:16478–16482. doi: 10.1073/pnas.0407557101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Powers ET, Deechongkit S, Kelly JW. Backbone-backbone H-bonds make context-dependent contributions to protein folding kinetics and thermodynamics: Lessons from amide-to-ester mutations. Adv Protein Chem. 2006;72:39–78. doi: 10.1016/S0065-3233(05)72002-7. [DOI] [PubMed] [Google Scholar]
- 14.Horne WS, Price JL, Keck JL, Gellman SH. Helix bundle quaternary structure from α/β-peptide foldamers. J Am Chem Soc. 2007;129:4178–4180. doi: 10.1021/ja070396f. [DOI] [PubMed] [Google Scholar]
- 15.Horne WS, Boersma MD, Windsor MA, Gellman SH. Sequence-based design of α/β-peptide foldamers that mimic BH3 domains. Angew Chem Int Ed. 2008;47:2853–2856. doi: 10.1002/anie.200705315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dill KA, et al. Principles of protein folding: A perspective from simple exact models. Protein Sci. 1995;4:561–602. doi: 10.1002/pro.5560040401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lupas AN, Gruber M. The structure of α-helical coiled coils. Adv Protein Chem. 2005;70:37–78. doi: 10.1016/S0065-3233(05)70003-6. [DOI] [PubMed] [Google Scholar]
- 18.Woolfson DN. The design of coiled-coil structures and assemblies. Adv Protein Chem. 2005;70:79–112. doi: 10.1016/S0065-3233(05)70004-8. [DOI] [PubMed] [Google Scholar]
- 19.O'Shea EK, Rutkowski R, Kim PS. Evidence that the leucine zipper is a coiled coil. Science. 1989;243:538–542. doi: 10.1126/science.2911757. [DOI] [PubMed] [Google Scholar]
- 20.O'Shea EK, Klemm JD, Kim PS, Alber T. X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil. Science. 1991;254:539–544. doi: 10.1126/science.1948029. [DOI] [PubMed] [Google Scholar]
- 21.Harbury PB, Zhang T, Kim PS, Alber T. A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine-zipper mutants. Science. 1993;262:1401–1407. doi: 10.1126/science.8248779. [DOI] [PubMed] [Google Scholar]
- 22.Richmond TJ, Richards FM. Packing of α-helices: Geometrical constraints and contact areas. J Mol Biol. 1978;119:537–555. doi: 10.1016/0022-2836(78)90201-2. [DOI] [PubMed] [Google Scholar]
- 23.Schmitt MA, Weisblum B, Gellman SH. Interplay among folding, sequence, and lipophilicity in the antibacterial and hemolytic activities of α/β-peptides. J Am Chem Soc. 2007;129:417–428. doi: 10.1021/ja0666553. [DOI] [PubMed] [Google Scholar]
- 24.Cheng RP, DeGrado WF. De novo design of a monomeric helical β-peptide stabilized by electrostatic interactions. J Am Chem Soc. 2001;123:5162–5163. doi: 10.1021/ja010438e. [DOI] [PubMed] [Google Scholar]
- 25.LePlae PR, Fisk JD, Porter EA, Weisblum B, Gellman SH. Tolerance of acyclic residues in the β-peptide 12-helix: Access to diverse side-chain arrays for biological applications. J Am Chem Soc. 2002;124:6820–6821. doi: 10.1021/ja017869h. [DOI] [PubMed] [Google Scholar]
- 26.Appella DH, et al. Residue-based control of helix shape in β-peptide oligomers. Nature. 1997;387:381–384. doi: 10.1038/387381a0. [DOI] [PubMed] [Google Scholar]
- 27.Schmitt MA, Choi SH, Guzei IA, Gellman SH. Residue requirements for helical folding in short α/β-peptides: Crystallographic characterization of the 11-helix in an optimized sequence. J Am Chem Soc. 2005;127:13130–13131. doi: 10.1021/ja0536163. [DOI] [PubMed] [Google Scholar]
- 28.Price JL, Horne WS, Gellman SH. Discrete heterogeneous quaternary structure formed by α/β-peptide foldamers and α-peptides. J Am Chem Soc. 2007;129:6376–6377. doi: 10.1021/ja071203r. [DOI] [PubMed] [Google Scholar]
- 29.Wolynes PG. Protein folding and beyond: Energy landscapes and the organization of living matter in time and space. In: Zewail A, editor. Physical Biology from Atoms to Medicine. London: Imperial College Press; 2008. pp. 267–288. [Google Scholar]
- 30.Choi SH, Guzei IA, Gellman SH. Crystallographic characterization of the α/β-peptide 14/15-helix. J Am Chem Soc. 2007;129:13780–13781. doi: 10.1021/ja0753344. [DOI] [PubMed] [Google Scholar]
- 31.Appella DH, et al. Synthesis and structural characterization of helix-forming β-peptides: trans-2-Aminocyclopentanecarboxylic acid oligomers. J Am Chem Soc. 1999;121:7574–7581. [Google Scholar]
- 32.Brown JH, Cohen C, Parry DAD. Heptad breaks in α-helical coiled coils: Stutters and stammers. Proteins. 1996;26:134–145. doi: 10.1002/(SICI)1097-0134(199610)26:2<134::AID-PROT3>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 33.Bryngelson JD, Wolynes PG. Spin glasses and the statistical mechanics of protein folding. Proc Natl Acad Sci USA. 1987;84:7524–7528. doi: 10.1073/pnas.84.21.7524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matthews BW, Nicholson H, Becktel WJ. Enhanced protein thermostability from site-directed mutations that decrease the entropy of unfolding. Proc Natl Acad Sci USA. 1987;84:6663–6667. doi: 10.1073/pnas.84.19.6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hartman MCT, Josephson K, Lin C-W, Szostak JW. An expanded set of amino acid analogs for the ribosomal translation of unnatural peptides. PLoS ONE. 2007;2:e972. doi: 10.1371/journal.pone.0000972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murray JK, Gellman SH. Parallel synthesis of peptide libraries using microwave irradiation. Nat Protocols. 2007;2:624. doi: 10.1038/nprot.2007.23. [DOI] [PubMed] [Google Scholar]
- 37.Murray JK, Gellman SH. Application of microwave irradiation to the synthesis of 14-helical β-peptides. Org Lett. 2005;7:1517–1520. doi: 10.1021/ol0501727. [DOI] [PubMed] [Google Scholar]
- 38.Gill SC, Von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 39.Collaborative Computational Project Number 4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 40.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr D. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 41.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 42.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 43.Lamzin VS, Wilson KS. Automated refinement of protein models. Acta Crystallogr D. 1993;49:129–147. doi: 10.1107/S0907444992008886. [DOI] [PubMed] [Google Scholar]
- 44.Strelkov SV, Burkhard P. Analysis of α-helical coiled coils with the program TWISTER reveals a structural mechanism for stutter compensation. J Struct Biol. 2002;137:54–64. doi: 10.1006/jsbi.2002.4454. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.