Abstract
Analysis of phenotypes associated with specific mutants has been instrumental in determining the roles of a bacterial gene in a biological process. However, this technique does not allow one to address whether a specific gene or gene set is necessary to maintain such a process once it has been established. In the study of microbial pathogenesis, it is important but difficult to determine the temporal requirement of essential pathogenic determinants in the entire infection cycle. Here we report a Cre/loxP-based genetic system that allowed inducible deletion of specific bacterial genes after the pathogen had been phagocytosed by host cells. Using this system, we have examined the temporal requirement of the Dot/Icm type IV protein transporter of Legionella pneumophila during infection. We found that deletion of single essential dot/icm genes did not prevent the internalized bacteria from completing one cycle of intracellular replication. Further analyses indicate that the observed phenotypes were due to the high stability of the examined Dot/Icm protein. However, postinfection deletion within 8 h of the gene coding for the Dot/Icm substrate, SdhA, abolishes intracellular bacterial growth. This result indicates that the Dot/Icm transporter is important for intracellular bacterial growth after the initial biogenesis of the vacuole. Our study has provided a technical concept for analyzing the temporal requirement of specific bacterial proteins or protein complexes in infection or development.
Keywords: bacterial pathogenesis, Cre/loxP, Dot/Icm, type IV secretion
Study of the function of a bacterial protein often is achieved by analyzing the distinct phenotypes of mutants lacking the corresponding gene. Due to technical barriers, it is difficult, if not impossible, to analyze whether any of these genes are required for maintaining established development or infection status. For instance, genes critical for the formation of biofilm had been identified in numerous bacteria, but whether or how long these genes are necessary for a biofilm structure to persist is unknown. Similarly, many pathogens use specialized protein secretion systems to deliver panels of virulence factors into host cells to construct a permissive niche necessary for multiplication. In most cases, however, it is not known whether these pathogenic elements are necessary for the entire infection cycle.
Legionella pneumophila, the etiological agent of the Legionnaires' disease, is a Gram-negative bacterium ubiquitously found in freshwater environments. This bacterium uses the Dot/Icm type IV secretion system to transfer a large cohort of effectors to facilitate the biogenesis of a phagosome that supports its growth (1). Pathogens may require a specific gene or gene sets during each stage of infection. Accordingly, bacterial genes specifically induced during late phases of infection have been identified by methods such as recombinase-based in vivo expression technology (RIVET) (2). In contrast to these observations, the current model suggests that a functional Dot/Icm transporter is required only in the very early phase (within minutes) of infection. This conclusion was based on results from two studies. First, repression of dotA, an essential component of the Dot/Icm complex expressed from a regulatable promoter, did not prevent the bacteria from undergoing a cycle of replication (3). Second, Coers et al. (4) showed that in the absence of thymidine, a thy− Dot/Icm+ strain, which itself was unable to replicate intracellularly without exogenous thymidine (5), can rescue growth of a thy+ Dot/Icm− strain coresiding in the same vacuole. Alternative possibilities that could account for these observations have not been addressed. The DotA protein could be stable so that a functional Dot/Icm transporter could continue to translocate effectors even when dotA is no longer expressed or is expressed at a low level. It is necessary therefore to examine whether loss of other components of the Dot/Icm complex could result in a similar phenotype. Moreover, effector translocation by the thy− mutant could continue in the absence of thymidine, thus allowing the coresiding Dot/Icm-deficient bacteria to grow. In support of this notion, recent experiments suggested that in order to successfully establish a replicative vacuole, activities of the Dot/Icm are required for a longer period after bacterial uptake. First, expression of many dot/icm genes is constitutive throughout bacterial growth, suggesting the presence of a transporter during the entire intracellular growth cycle (6). Second, some effectors, such as LidA, SidJ, SdhA, and LepB, are constantly synthesized by the bacterium, and some of these proteins were translocated into the host cytosol in large quantities in the late phase of the infection (7–11). Finally, Dot/Icm-dependent induction of apoptotic cell death, an event important for L. pneumophila growth, does not become apparent until there are >10 bacteria per vacuole (12). To address these discrepancies, we have developed an inducible in vivo gene disruption strategy that is useful for the analysis of temporal requirements for specific genes during bacterial infection or development. Using this system, we have analyzed the temporal requirement of the L. pneumophila Dot/Icm protein transporter during intracellular bacterial growth.
Results
The Development of a Genetic System for Inducible Gene Deletion After Bacterial Uptake.
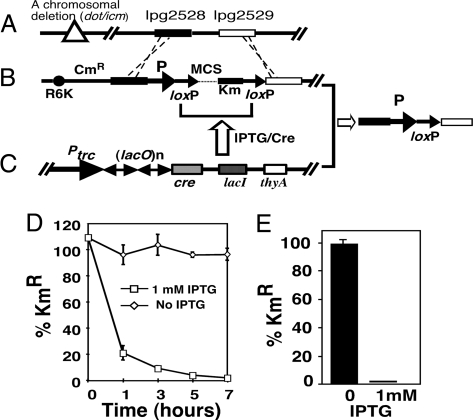
To develop a system that allows the deletion of a specific bacterial gene at any time during infection, we constructed two plasmids suitable for isopropyl β-d-thiogalactoside (IPTG)-inducible bacterial gene knockout by a multistep cloning process (Materials and Methods). The first was a π protein-dependent plasmid used for recombination-based introduction of a gene flanked by two loxP sites (referred to as floxed throughout the text) into a neutral site on the L. pneumophila chromosome. In this construct, a floxed multiple cloning site and a kanamycin resistance gene were flanked by two 1.0-kb DNA fragments from the site of L. pneumophila gene lpg2528 (Fig. 1A). Insertion of the floxed cassette into the 154-bp intergenic region between the convergently transcribed genes lpg2528 and lpg2529 did not interfere with intracellular growth of L. pneumophila (data not shown). To ensure wild-type level gene expression, the construct was designed so that the floxed gene was expressed from its cognate promoter (Fig. 1B).
Fig. 1.
Schematic diagram of a chromosome encoded system for IPTG-inducible deletion of bacterial genes. (A) Structure of the intergenic region between lpg2528 (a hypothetical protein) and lpg2529 (a putative α-amylase), into which the floxed gene is inserted. (B) An oriR6K-based plasmid used for introduction of floxed bacterial genes into corresponding deletion mutants. This plasmid contains a multiple cloning site and a kanamycin resistance gene flanked by two directly repeated loxP sites flanked by two DNA fragments upstream (black bar) and downstream (white bar) of the insertion site. Promoter (arrow P) of the floxed gene was cloned upstream of the first loxP site. The sacB and Cm resistance genes were used to select for recombinant strains that harbor the floxed gene correctly inserted at this chromosomal locus. (C) A plasmid harboring cre that is tightly controlled by LacIQ. Multiple copies of the lac operator sequence (lacO) situated downstream of the Ptrc promoter were used to achieve controlled expression of cre. The thyA gene conferring thymidine autotrophic to the mutant was used to maintain the plasmid in L. pneumophila. (D) Deletion of the floxed icmQ gene in bacteria grown in broth. Bacterial cultures of OD600 = 3.0 were split into two subcultures, and IPTG was added to one of them. At the indicated time points, a fraction of each culture was withdrawn, washed with PBS, and plated onto nonselective media and media containing kanamycin, respectively. (E) IPTG-induced deletion of floxed icmQ in intracellular bacteria. One hour after uptake, infections were induced for 2 h. After removing the inducer, bacterial counts were enumerated to obtain the ratios of kanamycin-resistant cells. Similar results were obtained in several independent experiments.
The second plasmid supplied the Cre recombinase, which was expressed from a promoter tightly regulated by the lac repressor LacI. In this plasmid, multiple copies of the lac operator were placed between the Ptrc promoter and the cre gene (Fig. 1C). In the absence of IPTG, no Cre was synthesized. Thus the floxed gene was expressed to complement the corresponding chromosomal deletion. Addition of IPTG, a compound that can efficiently penetrate some eukaryotic cell membranes, would inactivate the LacIQ repressor. This inactivation would lead to the production of Cre, which would subsequently induce recombination between the two loxP sites, resulting in the deletion of the floxed gene (Fig. 1 B and C). The positioning of the nptII gene between the two loxP sites allowed for easy testing of the efficiency of the deletion based on the sensitivity of the bacteria to kanamycin. Introduction of several dot/icm genes, including icmQ, dotA, dotG, and dotO, into the chromosomal site fully restored virulence of the corresponding mutant (see Results; data not shown). We first examined the time required for IPTG-induced gene deletion in broth-grown bacteria using the icmQ knock-in strain. In the absence of IPTG, all of the cells exhibited resistance to kanamycin, indicating the presence of icmQ in these bacteria (Fig. 1D). However, within 1 h of the addition of 1 mM IPTG, >80% of the bacteria became sensitive to the antibiotics, and the ratio of kanamycin-resistant bacteria dropped to <1% after 7 h of induction (Fig. 1). The relatively low rates of deletion may be due to the less active metabolism in postexponentially grown bacteria, because induction of exponentially grown bacteria led to almost complete deletion within 3 h [supporting information (SI) Fig. S1]. These data indicated that IPTG-induced gene deletion could be achieved in broth-grown L. pneumophila.
To examine the system's effectiveness in deleting the floxed genes in host cells, we determined the extent of IPTG-induced deletion of icmQ after bacterial uptake. Thirty minutes after infection, IPTG was added to infected macrophages and the deletion of icmQ was determined. We found that 2 h of IPTG induction was sufficient to delete the floxed gene in >99% of the bacteria (Fig. 1E). In contrast, nearly 100% of the bacteria from uninduced samples remained kanamycin-resistant (Fig. 1E). Similar deletion efficiency was observed in strains harboring other floxed genes (data not shown). Taken together, our efforts have produced a genetic system that allows inducible deletion of bacterial genes after pathogen phagocytosis.
Deletion of Individual dot/icm Genes Within 1 h After Bacterial Uptake Did Not Abolish L. pneumophila Intracellular Growth.
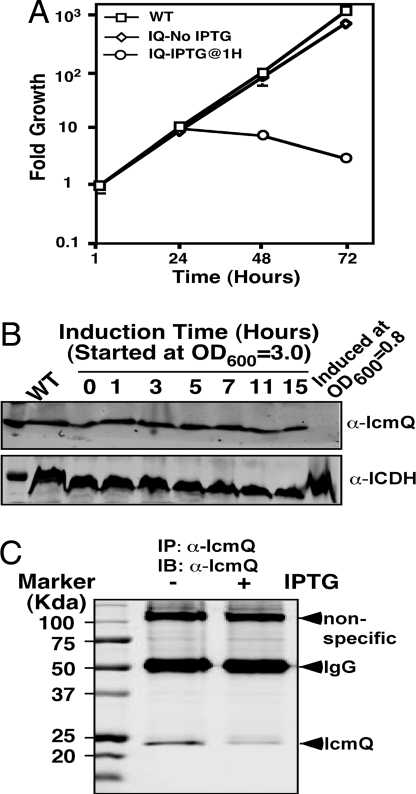
To examine the usefulness of our Cre/loxP-based in vivo gene deletion system, we attempted to determine the temporal requirement of the Dot/Icm type IV secretion system by L. pneumophila during infection. We first tested the icmQ knock-in strain. Thirty minutes after bacterial uptake, we induced a gene deletion by incubating infected cells with 1 mM IPTG for 5 h. After removing the inducer, we determined total bacterial counts at several time points. Twenty-four hours after infection, there was a 10-fold increase in bacterial growth in infections using the wild-type L. pneumophila strain Lp02 (Fig. 2A). A similar rate of multiplication was obtained in uninduced infections by using the icmQ knock-in strain (Fig. 2A). In the same time, however, IPTG-treated samples also exhibited robust growth, increasing by ≈9-fold (Fig. 2A), but no further growth was observed in the IPTG-induced samples beyond the first 24 h of incubation (Fig. 2A). We also examined the growth of strains harboring floxed dotA, dotG, or dotO. Deletion of none of these genes after bacterial internalization led to a significant growth defect in the first 24 h of infection (Fig. S2). These data were reminiscent of earlier observations that repression of dotA expression in internalized L. pneumophila did not affect its first growth cycle (3).
Fig. 2.
Intracellular growth and levels of IcmQ protein after IPTG-induced gene deletion. (A) Deletion of icmQ in intracellular bacteria did not abolish the first cycle of replication. Mouse macrophages were infected with indicated strains at an MOI of 0.05 for 1 h. IPTG was then added for 5 h to a subset of samples infected with the icmQ knock-in strain. Bacterial counts at the indicated time points were determined. Data shown were from a representative of several independent experiments performed in triplicate. (B) IcmQ levels in broth-grown bacteria after gene deletion. IPTG was added to bacterial cultures at the indicated cell densities, and samples withdrawn at the indicated time points were probed for IcmQ. The cytosolic isocitrate dehydrogenase (ICDH) was probed as a loading control. (C) IcmQ levels in bacteria grown in macrophages. Lysates prepared from cells infected for 24 h were subjected to immunoprecipitation with an anti-IcmQ antibody, and SDS/PAGE-resolved samples were probed. Note that the ≈100-kDa band nonspecifically recognized by the antibody was used as a loading control.
To determine whether the observed phenotypes were due to the stability of IcmQ, we examined its protein levels in L. pneumophila after IPTG induction. In broth-grown bacteria, when the inducer was added to cultures at early exponential phase (OD < 1.0), induction for 15 h resulted in bacterial cells that did not contain detectable IcmQ (Fig. 2B, last lane). As expected, these bacteria were unable to replicate intracellularly (data not shown). In contrast, addition of IPTG to bacteria grown to late exponential phase (OD600 = 3.0) did not lead to a detectable decrease in IcmQ (Fig. 2B). In phagocytosed bacteria, 24 h after infection, the level of IcmQ in IPTG-induced samples decreased but still was readily detectable (Fig. 2C). These data suggested that protein stability is responsible for the observed growth after gene deletion.
The IcmQ Protein Is Sufficient to Maintain a Functional Transporter for 20 h After Gene Deletion.
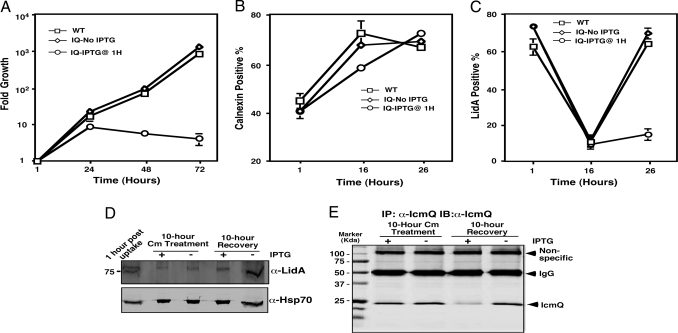
To address whether protein stability indeed accounted for the phenotypes, we further examined the temporal requirement of IcmQ during L. pneumophila infection. We chose to focus on this gene because its expression is driven by a promoter that is the weakest among the examined dot/icm genes and its protein level in bacterial cells is low (6, 13). Moreover, a high-quality, anti-IcmQ antibody is available to monitor its levels under different conditions (13). Earlier studies showed that bacteriostatic antibiotics such as chloramphenicol (Cm) and erythromycin did not detectably alter the route of intracellular trafficking of internalized L. pneumophila (14, 15). We hypothesized that after gene deletion, halting the bacterial growth with inhibitory concentrations of such antibiotics would provide more time for the protein to decay, perhaps leading to more severe growth defects. Thus, after IPTG induction for 5 h, we treated the samples with 5 μg/ml Cm for 10 h. After removing the antibiotic, the bacteria were then allowed to resume growth for an additional 19 h. The drug treatment did not detectably affect intracellular growth of the wild type or the uninduced icmQ knock-in strain (Fig. 3A). Unexpectedly, bacteria in IPTG-treated samples also exhibited robust growth but consistently displayed 2- to 3-fold defect (Fig. 3A). Thus, inhibition of bacterial growth for 10 h after deleting the icmQ gene significantly affected but did not abolish intracellular replication of L. pneumophila. Furthermore, infected cells in Cm-treated samples appeared healthy, probably because the antibiotic inhibits the expression of both pro-death and pro-survival effectors.
Fig. 3.
The Dot/Icm transporter remained active after IPTG-induced icmQ gene deletion. (A) Deletion of icmQ followed by 10 h of Cm treatment did not abolish bacterial intracellular replication. Infections were conducted as in Fig. 2. After removing IPTG, infected cells were incubated with 5 μg/ml Cm for 10 h and the bacterial growth were allowed to continue for 19 h. (B) Association of calnexin with bacterial vacuoles. Postnucleus supernatant was prepared from infected cells at the indicated times and was stained with a calnexin-specific antibody. The 16-h point includes 5 h of IPTG induction followed by 10 h of Cm treatment. The 26-h point includes 10 h of growth after removing Cm. (C and D) Translocation of LidA. Cells infected at an MOI of 5 were stained and scored for LidA (C) or were fractionated with digitonin followed by immunobloting (D). Hsp70 on the same membrane was probed as a loading control. Similar results were obtained in at least three independent experiments, and data shown were from one representative experiment. (E) IcmQ levels in infected cells after Cm treatment and recovery. Lysates of infected cells were subjected to immunoprecipitation with an anti-IcmQ antibody, and the protein was detected by Western blot. Note that the nonspecific band of ≈100 kDa was used as a loading control.
To examine whether the Dot/Icm transporter was still active after the gene deletion, we first examined the association of the endoplasmic reticulum protein calnexin with bacterial vacuoles (7). At all time points examined, similar rates of calnexin-positive vacuoles were detected in wild-type strain infections and in uninduced icmQ knock-in strain infections (Fig. 3B). IPTG induction and Cm treatment caused lower rates of calnexin-positive vacuoles, but such defect was restored after removing the antibiotic (Fig. 3B). These data further suggested that postphagocytic deletion of the icmQ gene followed by Cm treatment for 10 h did not abolish the activity of the Dot/Icm system.
To confirm the activity of the Dot/Icm system, we examined its ability to translocate the effector LidA (6) after these treatments. Without Cm treatment, the LidA-positive vacuoles were ≈65% and the rate was maintained at the same level at all time points examined (data not shown). However, Cm treatment for 10 h markedly decreased the association of LidA with the bacterial vacuoles (Fig. 3C). Furthermore, 10 h of recovery incubation fully restored the rates of LidA-positive vacuoles in samples infected with the wild-type bacterium or with the uninduced icmQ knock-in strain (Fig. 3C). In IPTG-induced samples, however, LidA-positive vacuoles also increased, but only marginally (Fig. 3C).
Because detection of translocated effectors by immunostaining often required higher levels of proteins, we examined the levels of injected LidA by digitonin fractionation of infected cells (16). Before Cm treatment, translocated LidA was readily detectable in infected cells; the drug treatment led to a decrease in its levels, but the protein was still detectable (Fig. 3D). Furthermore, after the removal of Cm and recovery, translocated LidA increased in both induced samples and uninduced samples, with the latter at a higher level (Fig. 3D). These data further indicated that the Dot/Icm system is still functional for substrate transfer after gene deletion followed by 10 h of growth inhibition.
Finally, we examined the IcmQ protein under these conditions. Immediately after Cm treatment, the amount of IcmQ was indistinguishable between induced and uninduced samples (Fig. 3E). However, after the 10-h recovery incubation, IcmQ in induced samples decreased to ≈20% of that of the uninduced samples (Fig. 3E and Fig. S3). Taken together, these results indicated that the IcmQ protein is extremely stable and that its cellular levels may be lowered only by dilution associated with cell division. The latter property is likely responsible for the robust growth of L. pneumophila after gene deletion.
Deletion of the sdhA Gene in Internalized Bacteria Within 8 h After Infection Severely Affected Intracellular Growth of L. pneumophila.
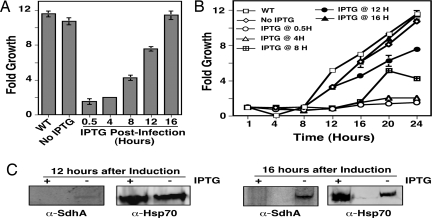
Clearly the Dot/Icm transporter was extremely stable, and disruption of one of its component genes after phagocytosis did not abolish the multiplication of internalized bacteria. Next we considered the possibility that some effectors of the Dot/Icm may have a lower stability, especially in host cell cytosol. Furthermore, continuous translocation of the effector after gene deletion could quickly deplete the protein from the bacterial cytoplasm, which could likely lead to more severe growth defects. Thus, we examined the temporal requirement for SdhA, a Dot/Icm substrate essential for intracellular growth in mouse macrophages (9). In the absence of IPTG, the sdhA knock-in strain underwent an almost 10-fold growth in 24 h, a rate indistinguishable from that of the wild-type strain; such growth continued throughout the entire 3-day experiment (Fig. 4A and Fig. 4S). Importantly, in samples that received IPTG 30 min after infection, no bacterial growth was detected (Fig. 4A). In analyses in which samples were induced at different times after uptake, adding IPTG at 4 h completely abolished bacterial replication (Fig. 4 A and B). Similarly, initiation of the induction at 8 h led to significant disruption of bacterial multiplication. Considerable inhibition of growth also occurred when IPTG was added 12 h after uptake (Fig. 4 A and B). However, when the induction time was extended to 16 h, no disruption of growth was observed (Fig. 4).
Fig. 4.
Deletion of sdhA within 8 h after phagocytosis abolished bacterial intracellular growth. (A) Growth of L. pneumophila after IPTG-induced sdhA deletion. Macrophages were infected at an MOI of 0.05, and the inducer was added at the indicated times. Growth of the bacteria was determined 24 h after uptake. (B) Kinetics of the growth after sdhA deletion. The growth of the bacteria in infections similar to A was determined at 4-h time intervals. Similar results were obtained in three independent experiments each done in triplicate. (C) Detection of SdhA after gene deletion. Protein in the soluble fraction of lysates of infected cells was obtained by TCA precipitation, and SdhA was detected by immunoblotting. The Hsp70 was probed as a loading control.
To correlate the growth phenotypes with the presence of the SdhA protein, we attempted to examine SdhA in infected cells after IPTG-induced gene deletion. We were unable to detect SdhA in cells infected with any of the testing strains in the first 8 h of incubation (data not shown). However, this protein became detectable when infections had proceeded for >12 h, but only in uninduced samples (Fig. 4C). The inability to detect SdhA in the early phases of infection likely is due to the low level expression of this protein (9).
Deletion of sdhA in Internalized Bacteria Led to Cell Death.
SdhA functions to protect infected macrophages from apoptosis (9), but whether deletion of this gene in actively replicating bacteria also leads to cell death is unknown. We therefore examined the apoptotic status of infected macrophages after IPTG-induced deletion of sdhA. One hour after uptake, we added IPTG to a subset of infected cells, and samples withdrawn at different time points were analyzed for apoptosis. During the entire experimental duration, <3% of the uninfected cells appeared apoptotic (data not shown). In infections using the wild-type strain or in uninduced infections using the sdhA knock-in strain, the fraction of apoptotic cells increased slightly over the 16-h incubation but never exceeded 10% (Fig. 5A). Similar to an earlier study (9), in infections using the sdhA mutant, up to 50% of the infected cells appeared apoptotic at 8 h after infection (Fig. 5A). However, further incubation to 16 h resulted in a drop of the cell death rates to ≈40% (Fig. 5A). Importantly, deletion of sdhA in intracellular bacteria led to cell death, and the rates peaked at 8 h after induction. A lower (27%) cell death rate was detected when the induction was extended to 16 h (Fig. 5A). The decrease in apoptosis rates in prolonged infections most likely was due to the loss of infected cells (ref. 9 and Y.L., and Z.-Q.L., unpublished data).
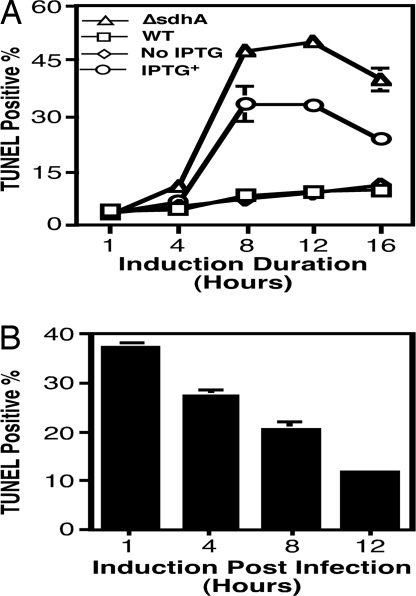
Fig. 5.
Deletion of sdhA during infection led to cell death. (A) Extent of cell death caused by the sdhA mutation and by in vivo gene deletion. Macrophages infected at an MOI of 1 were withdrawn at various time points after phagocytosis, and cell death was assessed by TUNEL staining. (B) Deletion of sdhA in replicating bacteria caused less cell death. IPTG was added to infections at the indicated times after phagocytosis; samples were processed for TUNEL staining after an additional 8 h of incubation.
We also determined whether deletion of sdhA in actively replicating bacteria led to cell death by initiating gene deletion at several points after bacterial uptake. Approximately 38% of the infected cells were apoptotic when IPTG was added at 1 h after uptake followed by an additional 8 h of incubation (Fig. 5B). The percentage of apoptotic infected cells decreased when the infection was allowed to proceed for longer time before the initiation of gene deletion (Fig. 5B). Cell death occurred in cells containing either small (one to two bacteria) or medium (approximately five bacteria) vacuoles (Fig. S5). Thus, deletion of sdhA in replicating L. pneumophila also led to cell death, although to a lesser extent. Taken together, these data indicated that in mouse macrophages L. pneumophila needs the SdhA protein, and thus the Dot/Icm transporter, to maintain a replicative vacuole even when the bacteria are actively replicating.
Discussion
The experimental versatility of the Cre/loxP system has been exploited for both temporal and spatial control of gene deletions in various murine systems, leading to the elucidation of the functions of many proteins in diseases and development (17). Similarly, the combination of site-specific recombinase and the in vivo expression technology (known as IVET) (18) has led to the identification of genes important for different phases of infection (2). In some cases, the requirement of a certain gene at a specific infection stage has been elucidated by using the in vivo inducible gene expression method (19). Such analyses are of particular importance in the clarification of possible cross-talk between virulence determinants that appear to control different stages of infection. However, few studies have used site-specific recombination systems for inducible gene deletion in prokaryotic cells to analyze the temporal requirement of specific genes or gene sets. In this report, we have developed such a system to demonstrate that during bacterial intracellular growth, the Dot/Icm type IV transporter is required by L. pneumophila for a longer time than previously thought.
Consistent with its constitutive expression pattern (8), we showed that the SdhA protein was required for at least 8 h for L. pneumophila to complete a productive infection. Considering the fact that it took at least 2 h of IPTG induction for gene deletion to occur in most of the bacterial cells, the relatively lower cell death rates, and thus substantial multiplication, very likely were a result of the activity of SdhA synthesized before gene deletion (Fig. 5A). Similarly, SdhA contributed by multiple bacterial cells could account for the low cell death rates observed in infections induced after bacterial replication had begun (Fig. 5B). Clearly, macrophages infected by the sdhA mutant or the knock-in strain (induced) did not immediately become apoptotic. The several hours of delay may be necessary for the putative cell-death-inducing factors to accumulate to a critical level or such toxic proteins may be induced only when the bacteria have entered active growth phase.
Several lines of evidence suggested that the stability of the IcmQ protein is accounted for by the observed short time need for the Dot/Icm transporter after gene disruption. Gene deletion and Cm treatment clearly led to a lower IcmQ protein level, but this reduction did not translate into the abolishment of intracellular growth, because the transporter was still capable of transferring effectors into host cells (Fig. 3). It is most likely that the decrease in IcmQ protein levels is accounted for by dilution associated with cell division (Figs. 2 B and C and 3E). These results are consistent with an earlier study showing that a single Dot/Icm-competent L. pneumophila cell was able to initiate and maintain a vacuole to support full replication of a coresiding Dot/Icm-deficient bacterium (4). Similarly, lower levels of DotA did not cause significant loss of intracellular growth (7). These data suggested that effectors of the Dot/Icm system are very efficient in the establishment and maintenance of the replicative vacuoles. Although we did not exhaustively examine every single Dot/Icm protein, an earlier study on DotA (3) and our analysis of IcmQ suggest that this transporter is extremely stable, probably due to the lack of a mechanism for disassembling the apparatus.
Some type IV secretion systems have been shown to remain active after conditions for expression of their component genes are no longer favorable. In Agrobacterium tumefaciens, the transfer of the tumor-inducing plasmid continues for many hours after removing the inducing signals (20). After being assembled, a stable transporter would be beneficial to the microorganism when null mutations occurred in its component genes. A functional transporter would allow the transfer of the genetic elements to complete or the production of a large number of progenies to occur. Such outcomes could increase the probability of repairing the mutated gene by recombination or other mechanisms. Whether other type IV secretion systems involved in pathogenicity exhibit similar stability remains to be determined.
In summary, we have developed a genetic system that allows inducible deletion of specific bacterial genes even when the bacteria are at a specialized development stage or are associated with their host. As long as the biological membranes of the model organism are permeable to IPTG and the organism does not efficiently metabolize this compound, this method should be useful for analyzing the temporal requirement of microbial factors in other systems.
Materials and Methods
Bacterial Strains and Plasmids.
All L. pneumophila strains used in this study were derivative of the Philadelphia 1 strain Lp02 (21) and are listed in Table S1. In-frame deletion mutants of L. pneumophila were described previously (13, 22). Plasmids designed to introduce floxed L. pneumophila genes into the corresponding mutant were constructed by a multistep cloning procedure detailed in the SI Materials and Methods. Similarly, a series of plasmids for inducible expression of the Cre recombinase were constructed as described in the SI Materials and Methods. For each strain, the efficiency of IPTG-induced gene deletion in bacteria grown in broth or in macrophages was empirically determined by testing several of these plasmids in the knock-in strain. In each case, a cre-expressing plasmid that not only exerted tight control in the absence of IPTG (did not detectably affect the growth of bacteria on media containing kanamycin) but also allowed efficient IPTG-induced gene deletion was used.
Infection, Gene Deletion, and Intracellular Growth Analysis.
L. pneumophila strains grown to appropriate phases were used for induction or for infection at indicated multiplicities of infection (MOIs). Intracellular growth of L. pneumophila was analyzed in previously published procedures (23). In all experiments, IPTG was used at a final concentration of 1 mM. For in vitro induction, samples withdrawn at the time points indicated in Fig. 1D were subjected to extensive washes (five times) to remove the inducer, and the diluted bacteria were plated onto appropriate charcoal yeast extract plates. To determine the time required for efficient gene deletion in phagocytosed bacteria, we washed infected cells with PBS eight times to remove extracellular bacteria and then added IPTG. When required, Cm and kanamycin were used at 5 μl/ml and 20 μl/ml, respectively.
Immunoprecipitation, Trichloroacetic Acid (TCA) Precipitation, and Immunoblotting.
Details for protein sample preparation, antibody, immunoprecipitation, digitonin fractionation, TCA precipitation, and immunodetection are in the SI Materials and Methods.
Immunostaining, TUNEL Staining, and Fluorescence Microscopy.
The translocation of LidA was determined by immunostaining with a LidA-specific antibody (7). The association of calnexin with Legionella-containing vacuoles was assayed as previously described (8). The cell death status of infected cells was assessed by TUNEL staining as described previously (9, 24). Samples were inspected and scored under a fluorescence microscopy.
Supplementary Material
Acknowledgments.
We thank Dr. Ralph Isberg (Tufts University, Boston) for the bacterial mutants and antibodies specific for IcmQ, SdhA, and LidA. This work was supported by American Heart Association Grant 0535451Z and National Institutes of Health Grant R01AI069344 (to Z.-Q. L).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0801055105/DCSupplemental.
References
- 1.Ninio S, Roy CR. Effector proteins translocated by Legionella pneumophila: Strength in numbers. Trends Microbiol. 2007;15:372–380. doi: 10.1016/j.tim.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 2.Angelichio MJ, Camilli A. In vivo expression technology. Infect Immun. 2002;70:6518–6523. doi: 10.1128/IAI.70.12.6518-6523.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roy CR, Berger KH, Isberg RR. Legionella pneumophila DotA protein is required for early phagosome trafficking decisions that occur within minutes of bacterial uptake. Mol Microbiol. 1998;28:663–674. doi: 10.1046/j.1365-2958.1998.00841.x. [DOI] [PubMed] [Google Scholar]
- 4.Coers J, Monahan C, Roy CR. Modulation of phagosome biogenesis by Legionella pneumophila creates an organelle permissive for intracellular growth. Nat Cell Biol. 1999;1:451–453. doi: 10.1038/15687. [DOI] [PubMed] [Google Scholar]
- 5.Mintz CS, Chen JX, Shuman HA. Isolation and characterization of auxotrophic mutants of Legionella pneumophila that fail to multiply in human monocytes. Infect Immun. 1988;56:1449–1455. doi: 10.1128/iai.56.6.1449-1455.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gal-Mor O, Zusman T, Segal G. Analysis of DNA regulatory elements required for expression of the Legionella pneumophila icm and dot virulence genes. J Bacteriol. 2002;184:3823–3833. doi: 10.1128/JB.184.14.3823-3833.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conover GM, Derre I, Vogel JP, Isberg RR. The Legionella pneumophila LidA protein: A translocated substrate of the Dot/Icm system associated with maintenance of bacterial integrity. Mol Microbiol. 2003;48:305–321. doi: 10.1046/j.1365-2958.2003.03400.x. [DOI] [PubMed] [Google Scholar]
- 8.Liu Y, Luo ZQ. The Legionella pneumophila effector SidJ is required for efficient recruitment of endoplasmic reticulum proteins to the bacterial phagosome. Infect Immun. 2007;75:592–603. doi: 10.1128/IAI.01278-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laguna RK, et al. A Legionella pneumophila-translocated substrate that is required for growth within macrophages and protection from host cell death. Proc Natl Acad Sci USA. 2006;103:18745–18750. doi: 10.1073/pnas.0609012103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ingmundson A, Delprato A, Lambright DG, Roy CR. Legionella pneumophila proteins that regulate Rab1 membrane cycling. Nature. 2007;450:365–369. doi: 10.1038/nature06336. [DOI] [PubMed] [Google Scholar]
- 11.Chen J, et al. Legionella effectors that promote nonlytic release from protozoa. Science. 2004;303:1358–1361. doi: 10.1126/science.1094226. [DOI] [PubMed] [Google Scholar]
- 12.Molmeret M, et al. Activation of caspase-3 by the Dot/Icm virulence system is essential for arrested biogenesis of the Legionella-containing phagosome. Cell Microbiol. 2004;6:33–48. doi: 10.1046/j.1462-5822.2003.00335.x. [DOI] [PubMed] [Google Scholar]
- 13.Dumenil G, Isberg RR. The Legionella pneumophila IcmR protein exhibits chaperone activity for IcmQ by preventing its participation in high-molecular-weight complexes. Mol Microbiol. 2001;40:1113–1127. doi: 10.1046/j.1365-2958.2001.02454.x. [DOI] [PubMed] [Google Scholar]
- 14.Horwitz MA, Silverstein SC. Intracellular multiplication of Legionnaires' disease bacteria (Legionella pneumophila) in human monocytes is reversibly inhibited by erythromycin and rifampin. J Clin Invest. 1983;71:15–26. doi: 10.1172/JCI110744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dorer MS, Kirton D, Bader JS, Isberg RR. RNA interference analysis of Legionella in Drosophila cells: Exploitation of early secretory apparatus dynamics. PLoS Pathog. 2006;2:e34. doi: 10.1371/journal.ppat.0020034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Derre I, Isberg RR. LidA, a translocated substrate of the Legionella pneumophila type IV secretion system, interferes with the early secretory pathway. Infect Immun. 2005;73:4370–4380. doi: 10.1128/IAI.73.7.4370-4380.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Branda CS, Dymecki SM. Talking about a revolution: The impact of site-specific recombinases on genetic analyses in mice. Dev Cell. 2004;6:7–28. doi: 10.1016/s1534-5807(03)00399-x. [DOI] [PubMed] [Google Scholar]
- 18.Mahan MJ, Slauch JM, Mekalanos JJ. Selection of bacterial virulence genes that are specifically induced in host tissues. Science. 1993;259:686–688. doi: 10.1126/science.8430319. [DOI] [PubMed] [Google Scholar]
- 19.Dancz CE, Haraga A, Portnoy DA, Higgins DE. Inducible control of virulence gene expression in Listeria monocytogenes: Temporal requirement of listeriolysin O during intracellular infection. J Bacteriol. 2002;184:5935–5945. doi: 10.1128/JB.184.21.5935-5945.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Su S, Khan SR, Farrand SK. Induction and loss of Ti plasmid conjugative competence in response to the acyl-homoserine lactone quorum-sensing signal. J Bacteriol. 2008 doi: 10.1128/JB.01684-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berger KH, Isberg RR. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol Microbiol. 1993;7:7–19. doi: 10.1111/j.1365-2958.1993.tb01092.x. [DOI] [PubMed] [Google Scholar]
- 22.Vogel JP, Andrews HL, Wong SK, Isberg RR. Conjugative transfer by the virulence system of Legionella pneumophila. Science. 1998;279:873–876. doi: 10.1126/science.279.5352.873. [DOI] [PubMed] [Google Scholar]
- 23.Luo ZQ, Isberg RR. Multiple substrates of the Legionella pneumophila Dot/Icm system identified by interbacterial protein transfer. Proc Natl Acad Sci USA. 2004;101:841–846. doi: 10.1073/pnas.0304916101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Banga S, et al. Legionella pneumophila inhibits macrophage apoptosis by targeting pro-death members of the Bcl2 protein family. Proc Natl Acad Sci USA. 2007;104:5121–5126. doi: 10.1073/pnas.0611030104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.