Abstract
Mitochondrial genomes generally encode a minimal set of tRNAs necessary for protein synthesis. However, a number of eukaryotes import tRNAs from the cytoplasm into their mitochondria. For instance, Saccharomyces cerevisiae imports cytoplasmic tRNAGln into the mitochondrion without any added protein factors. Here, we examine the existence of a similar active tRNA import system in mammalian mitochondria. We have used subcellular RNA fractions from rat liver and human cells to perform RT-PCR with oligonucleotide primers specific for nucleus-encoded tRNACUGGln and tRNAUUGGln species, and we show that these tRNAs are present in rat and human mitochondria in vivo. Import of in vitro transcribed tRNAs, but not of heterologous RNAs, into isolated mitochondria also demonstrates that this process is tRNA-specific and does not require the addition of cytosolic factors. Although this in vitro system requires ATP, it is resistant to inhibitors of the mitochondrial electrochemical gradient, a key component of protein import. tRNAGln import into mammalian mitochondria proceeds by a mechanism distinct from protein import. We also show that both tRNAGln species and a bacterial pre-tRNAAsp can be imported in vitro into mitochondria isolated from myoclonic epilepsy with ragged-red fiber cells if provided with sufficient ATP (2 mM). This work suggests that tRNA import is more widespread than previously thought and may be a universal trait of mitochondria. Mutations in mitochondrial tRNA genes have been associated with human disease; the tRNA import system described here could possibly be exploited for the manipulation of defective mitochondria.
Keywords: human, MERFF
Different protein-synthesizing systems exist in the cytoplasm and organelles (mitochondria and chloroplasts) of eukaryotic cells. Reflecting the evolutionary origin of the organelle, the mitochondrial system is bacteria-like (1, 2). For instance, the tRNAs and aminoacyl-tRNA synthetases present in the mitochondria are closely related to their bacterial counterparts. The mitochondrial genome of higher eukaryotes encodes all of the tRNA species necessary for protein synthesis (3), whereas the aminoacyl-tRNA synthetases are nucleus-encoded and imported into the organelle. The mitochondrial protein-synthesizing system is important for the synthesis of a number of proteins for multienzyme complexes involved in oxidative phosphorylation (1). Since the original suggestion 40 years ago (4), tRNA import into mitochondria has been demonstrated in Tetrahymena, trypanosomatids, yeast, plants, and marsupials (reviewed in refs. 5 and 6). The detailed nature of the different tRNA import mechanisms is not yet known; however, ATP hydrolysis is always required. In the yeast system, the importance of some mitochondrial membrane proteins as well as of specific cytosolic proteins (enolase and lysyl-tRNA synthetase) has been established (reviewed in refs. 5 and 6). After the early observation of the presence of a cytoplasmic tRNACUULys in Saccharomyces cerevisiae mitochondria (7), import of cytoplasmic tRNACUGGln was also demonstrated (8). Further studies showed that these tRNAs are imported by different mechanisms (8–10). The question of import into human mitochondria has never been addressed for its own sake. It was believed that under normal physiological conditions, tRNA import into human mitochondria does not take place (5). Motivated by the desire to complement mitochondrial tRNA gene mutations linked to diseases, it was established that yeast tRNALys (11) and human tRNALys (12) could be imported into human mitochondria under special conditions or with the help of additional factors.
Is tRNA import a normal occurrence in human cells? In all known cases, the eukaryotic mitochondrial genomes encode one glutamine tRNA species with the anticodon UUG, tRNAUUGGln, for translating the corresponding CAA and CAG codons according to the wobble rules (13). However, should the first anticodon base of this tRNA be modified to 2-thiouracil, a frequent occurrence in bacteria, then CAG codon recognition may suffer (14). In this case, full decoding capacity could be maintained if cytoplasmic tRNACUGGln were imported into the mitochondrion. This is exactly the situation that we encountered in our analysis of S. cerevisiae mitochondria (8). Here, we extend this experimental pursuit to human and rat mitochondria.
Results
tRNAGln Is Imported into Rat and Human Mitochondria.
To explore the possibility of tRNA import in mammals, mitochondria were isolated from rat liver and HeLa cells by isotonic lysis and Percoll gradient centrifugation (15). The purified mitochondria were extensively treated with micrococcal nuclease to destroy nucleic acids that might nonspecifically bind to the mitochondrial surface during isolation (16). We then performed reverse transcription (RT)-PCR analyses of total mitochondrial RNA with oligonucleotide primers specific for the nucleus-encoded tRNAGln (ntRNAGln) isoacceptors (Fig. 1).
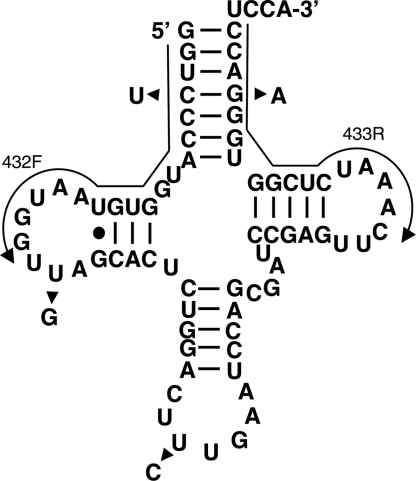
Fig. 1.
Secondary structure of the nucleus-encoded tRNAGln from rat. Arrowheads indicate the single-nucleotide differences between the six different isoacceptors found in the genome. Arrows indicate the position of the two oligonucleotide primers used for RT-PCR amplification (432F and 433R). These two primers can only discriminate four of the possible six isoacceptors. The same primers were used for the analysis of nucleus-encoded tRNAGln from humans.
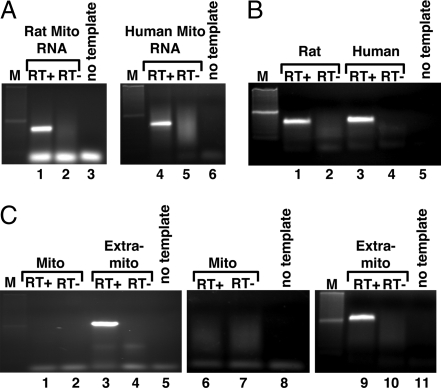
These reactions yielded PCR products consistent with the import of these tRNAs from the cytoplasm (Fig. 2A, lanes 1 and 4). We also isolated total RNA from the extramitochondrial cellular fraction, which contains a mixture of cytoplasmic and nuclear RNAs (nuclei burst during our mitochondrial purification procedure). This fraction was also analyzed by RT-PCR as a positive control for ntRNAGln amplification and yielded products whose size was identical to those obtained from the total mitochondrial RNA (Fig. 2B, lanes 1 and 3). Additionally, control RT-PCRs with oligonucleotides specific for an extramitochondrial RNA, U6 RNA, showed no detectable levels of U6 in our mitochondrial fractions (Fig. 2C, lanes 1 and 6). However, when similar reactions were performed with the total nonmitochondrial fractions, a clear amplification product was observed (Fig. 2C, lanes 3 and 9). No PCR products were observed throughout these experiments when “mock” controls were performed in the absence of reverse transcriptase, ruling out DNA contamination in our RNA fractions as the source of the observed PCR products. The identity of the U6 RT-PCR product was confirmed by sequencing (data not shown).
Fig. 2.
Nucleus-encoded tRNAGln is imported into rat and human mitochondria. (A) RT-PCR with primers specific for the nucleus-encoded tRNAGln (ntRNAGln) isoacceptors and total RNA isolated from rat (Left) or human (Right) mitochondria. These primers are unable to amplify the endogenous mitochondria-encoded tRNAUUGGln (mttRNAUUGGln) and can only discriminate between four of the possible six nucleus-encoded isoacceptors. (B) Similar RT-PCR as in A but with extramitochondrial RNA, which includes a mixture of cytoplasmic as well as nuclear RNA generated from the same preparation as that used to isolate the mitochondria in A. (C) RT-PCR with U6-specific oligonucleotides (extramitochondrial marker) with the same RNA fractions as in A and B. RT+ and RT− refer to cDNA synthesis reactions performed in the presence or absence of reverse transcriptase, respectively. No template is a mock reaction in which no cDNA was added, also serving as a negative control. Mito, RNA isolated from purified mitochondria. M, a 100-bp DNA ladder used as size markers during electrophoresis.
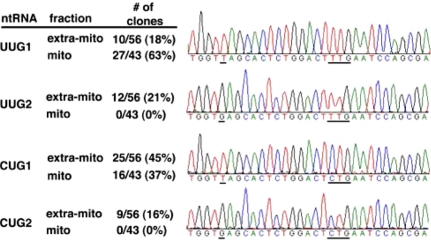
We then analyzed the identity of the mitochondrial and cytoplasmic tRNAGln products. After cloning and sequencing a number of independent clones from the RT-PCR products (Fig. 3), we found that tRNAUUG1Gln and tRNACUG1Gln, two of the possible four nucleus-encoded isoacceptors that can be distinguished by our primers, were found in mitochondria (Fig. 3). The presence of a mitochondrial-encoded tRNAUUGGln was also established by RT-PCR and sequencing, when primers specific for the mitochondria-encoded tRNA were used (data not shown). These experiments demonstrate that our mitochondrial fractions are devoid of any significant extramitochondrial contamination and are consistent with the idea that: (i) the nucleus-encoded tRNAGln isoacceptors are imported from the cytoplasm and (ii) both the mitochondrial and nucleus-encoded tRNAUUGGln isoacceptors coexist in mammalian mitochondria.
Fig. 3.
Distribution of imported tRNAGln species in rat mitochondrial RNA fractions. (Left) Table summarizing the total distribution of each isoacceptor in the various fractions. Similar numbers were obtained for human mitochondria (data not shown). (Right) DNA sequencing traces representative of the different products obtained from the RT-PCR experiments in (Fig. 2). The single-nucleotide differences that can be discerned with the primers used are underlined. Also underlined are the anticodon sequences.
In Vitro Import into Mammalian Mitochondria Does Not Require Soluble Cytosolic Factors.
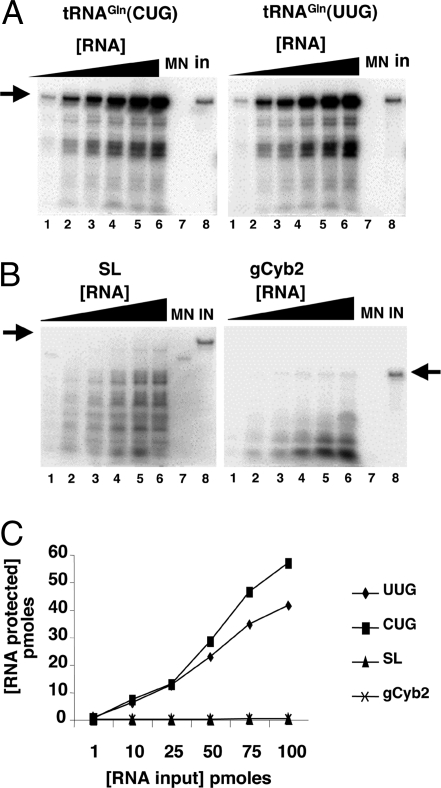
In vitro import of tRNAs into isolated mitochondria has been demonstrated in a number of systems including plants, yeast, and trypanosomatids (17–21). We examined the in vitro import of various RNAs into isolated rat mitochondria by a nuclease protection assay (see Materials and Methods) (Fig. 4) and showed that radioactively labeled transcripts corresponding to the two imported tRNA isoacceptors are efficiently imported into mitochondria in vitro (Fig. 4 A and C). We also purified native tRNAGln from rat liver as described in ref. 22. These tRNAs showed import behavior identical to that observed with the in vitro transcripts (data not shown), ruling out the possibility that posttranscriptional modifications play a significant role in this specific import system. A time course experiment showed that the system reached an import plateau after 20 min of incubation under import conditions, where 10 min of incubation was well within the linear range of the assay (data not shown). Therefore the latter incubation time has thus been used throughout our experiments. We then performed similar import assays with two unrelated in vitro transcribed RNAs, the spliced leader RNA (SL RNA) and the gCyb2 guide RNA from trypanosomes (110 and 56 nucleotides in length, respectively). These RNAs were chosen based on both their length (close to the length of a tRNA) and the fact that they are highly structured. Neither substrate supported in vitro import (Fig. 4B). These observations demonstrate that our in vitro system can discriminate tRNA substrates from heterologous highly structured RNAs.
Fig. 4.
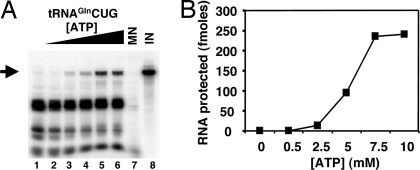
Nucleus-encoded tRNAGln is efficiently and specifically imported into isolated rat liver mitochondria in vitro. (A) Increasing concentrations of radioactively labeled ntRNACUGGln (Left) or ntRNAUUGGln transcripts (Right) were incubated with constant concentrations of Percoll-purified mitochondria as described in Materials and Methods. (B) Reactions similar to those in A but with two heterologous RNAs as specificity controls for in vitro import, where SL and gCyb2 refer to spliced leader and gRNA transcripts from trypanosomatids. These RNAs do not exist in mammalian cells. (C) A plot of the results from A and B where pmoles of input RNA protected are plotted vs. the input. MN (lanes 7), control reactions treated with micrococcal nuclease in the absence of mitochondria. These reactions serve as a control for MN digestion. IN, input lane of the original tRNA used in the import experiments and serving as a quantitation standard. The black triangle denotes increasing concentrations of a given reagent used in the assay (at 1, 10, 25, 50, 80, and 100 pmol of input, lanes 1–6 in A and B). The arrow marks the position of the full-length RNA after gel electrophoresis as described in Materials and Methods.
Import into Mammalian Mitochondria Is ATP-Dependent.
In all other cases known, tRNA import into mitochondria is an ATP-requiring process (8, 21, 23, 24). We have tested the ATP requirements for tRNAGln import into rat mitochondria and showed that import also requires ATP (Fig. 5 A and B). Before import, the mitochondria were incubated under conditions that deplete endogenous ATP because without depletion a small fraction of the tRNA is still imported, presumably due to the large amount of ATP generated in these mitochondria (data not shown). A similar basal level of tRNA import in the absence of added ATP has been described for the in vitro import of tRNAs into isolated plant mitochondria (17). However, the same is not true with isolated human mitochondria, perhaps reflecting relative differences in the steady-state ATP levels in our isolated organelles.
Fig. 5.
Import of tRNA requires ATP. (A) Constant concentrations of radioactively labeled tRNACUGGln or tRNAUUGGln (data not shown) were incubated in the absence (lane 1) or in the presence of increasing ATP (at 0.5, 1, 2, 5, 10, and 15 mM final concentration, lanes 2–8) yielding identical results. (B) Plot of the protected tRNA fraction following import vs. ATP concentration. MN and IN (lanes 7 and 8) are as described in the legend to Fig. 4.
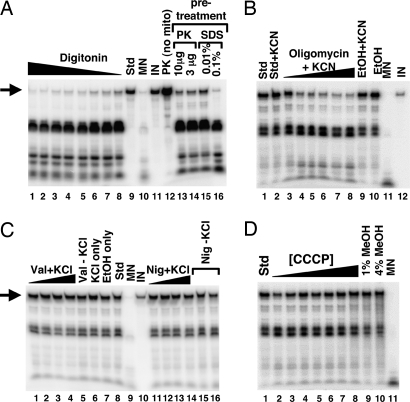
Pretreatment of the mitochondria with digitonin, a detergent that at low concentrations (i.e., <2% final volume) can selectively remove the mitochondrial outer membrane, led to inhibition of import (Fig. 6A, lanes 1–8). These digitonin concentrations are not sufficient to completely disrupt mitochondrial integrity but selectively solubilize the outer membrane (25). Pretreatment of mitochondria with proteinase K also led to a marked decrease in import efficiency (Fig. 6A, lanes 13 and 14). Similar decreases were observed when the mitochondria were treated with the ionic detergent SDS (Fig. 6A, lanes 15 and 16). Taken together, the data show that tRNA import is a protein-mediated process that requires intact mitochondria.
Fig. 6.
tRNA import occurs by a mechanism distinct from protein import. (A) The import of tRNACUGGln was examined in purified mitochondria pretreated with digitonin (at 0.01, 0.1, 0.5, 1, and 5% final concentration; lanes 1–8, respectively) and compared with a standard import reaction (Std, lane 9) without pretreatment. In addition, mitochondria were pretreated with proteinase K (PK, lanes 13 and 14) or mildly treated with SDS (lanes 15 and 16). PK (no mito), incubation of the labeled transcript in the presence of an amount of proteinase K equivalent to that used in the assay in lane 13 but in the absence of mitochondria, demonstrating that the lack of protection in lanes 13 and 14 cannot be ascribed to degradation of the tRNA by proteinase K. (B) Mitochondria were also treated with increasing concentrations of oligomycin (1.3, 3.2, 31.7, 63.3, 95, and 126 nM; lanes 3–8, respectively) to inhibit ATPase activity in the presence of KCN. Import of tRNA under these conditions was compared with a standard import reaction (Std, lane 1) and a standard reaction with KCN alone (lane 2). In addition, tRNAs were also imported into mitochondria treated with ethanol (EtOH) (lane 10) or KCN + ethanol (EtOH+KCN, lane 9) alone serving as a control for the various cofactors and solvents. (C) Mitochondria were also treated with increasing concentrations of the potassium ionophore valinomycin (1, 5, 10, and 20 μM; lanes 1–4, respectively) in the presence of KCl (70 mM final concentration). The highest level of valinomycin used represents 100× the concentration required to collapse the membrane potential (data not shown). Val+KCl (lane 5), KCl only (lane 6), and EtOH only (lane 7) refer to import reactions in the presence of these various components used as cofactors or solvents in the valinomycin experiment. Incubation of tRNA alone with any of these components does not inhibit MN cleavage (data not shown). Similar reactions were performed in reactions where valinomycin was replaced by nigericin (Nig) (1, 5, 10, and 20 μM; lanes 11–14), a potent inhibitor of the pH gradient. nig (−) KCL (lanes 15 and 16) refers to an import reaction in the presence of nigericin but in the absence of KCl. Control reactions without mitochondria and in the presence of nigericin showed that nigericin does not inhibit MN cleavage (data not shown). (D) tRNA import assays with mitochondria treated with the uncoupler CCCP at concentrations well above what is required to destroy the electrochemical gradient (0.1, 0.25, 0.5, 0.75, 1.0, 1.5, and 2 mM; lanes 2–8, respectively). Because CCCP is dissolved in methanol, control import reactions in the presence of methanol are also shown (lanes 9 and 10). Throughout the figure, arrows denote the full-length tRNA substrate, and the black triangle represents increasing or decreasing concentrations of a given chemical. MN and IN, control reactions for nuclease cleavage and as a quantitation standard, respectively.
Import of tRNA into Mammalian Mitochondria Is Distinct from Protein Import.
The observed ATP requirement and the need for an intact outer membrane raise the question of whether ATP generation by the mitochondria is a key component of tRNA import, as has been described in Leishmania. We then treated mitochondria with the ATPase inhibitor oligomycin (26). We found that, even at low concentrations, this antibiotic inhibited tRNA import (Fig. 6B, lanes 3–8). Therefore, in the presence of added ATP, the function of the mitochondrial ATPase is still required for import. This effect of course could be indirect, where inhibition of the ATPase affects the membrane potential or the electrochemical gradient across the inner membrane and in turn affects import.
A membrane potential is needed for the import of a number of tRNAs into isolated plant mitochondria (17) as well as import of tRNALys into yeast mitochondria (20), but it is not required in Leishmania or in the import of tRNAGln into yeast mitochondria (8, 21). We then performed import assays in the presence of the potassium ionophore valinomycin. This antibiotic is known to collapse the membrane potential ΔΨ in the presence of potassium ions. Even when valinomycin is added in vast excess (100 times the concentration required to collapse ΔΨ in the presence of potassium chloride) (21), it has no effect on the tRNA import efficiency in rat mitochondria (Fig. 6C, compare lanes 1–4 with lane 8).
Alternatively, it is possible that the transport of a negatively charged molecule across the mitochondrial membrane could be driven by the inherent pH gradient across the membrane. We also performed import assays in the presence of nigericin, a known inhibitor of the ΔpH (27). We found that again this antibiotic had little effect on import efficiency (Fig. 6C, lanes 11–14). Because the ΔΨ and the ΔpH values are tightly linked, it is possible that in the presence of either inhibitor the electrochemical gradient is partially maintained by a concomitant increase in ΔΨ or ΔpH, depending on which component is being inhibited. However, incubation of the mitochondria with the potent uncoupler carbonylcyanide m-chlorophenylhydrazone (CCCP) (28) had no effect on tRNA import, demonstrating that neither the electrochemical gradient nor its components ΔΨ and ΔpH play a significant role in tRNAGln import in our system (Fig. 6D, lanes 2–8). Interestingly, a membrane potential is absolutely required for protein import into the mitochondrial matrix (29). These data indicate that tRNAGln import into mammalian mitochondria is facilitated by a pathway distinct from that used predominantly for protein import. However, this conclusion does not rule out the possibility that protein import factors may participate in tRNAGln import.
Rescue of tRNA Import into MERRF Mitochondria by Addition of Exogenous ATP.
A number of mutations in the mitochondrial tRNA genes of humans are associated with myoclonic epilepsy with ragged red fibers (MERRF) (30). Generally, these mutations lead to a drastic reduction in the synthesis of mitochondrial proteins with concomitant reduction in mitochondrial respiration and thus reduced ability to efficiently generate ATP. It was recently shown that a cytosolic tRNA could be delivered into mitochondria of cytoplasmic hybrids (cybrids) from patients suffering from MERFF, if these cells were incubated with the putative RNA import complex (RIC) from Leishmania (12).
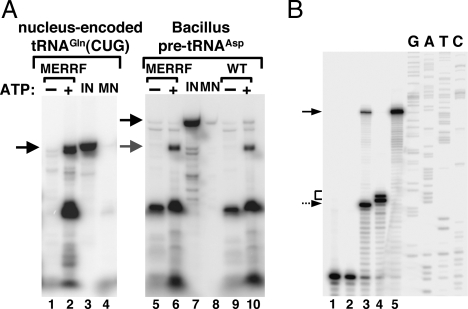
In light of the findings described here, we decided to examine the possibility that the observed complementation could be partly the result of the ability of the Leishmania RIC to complement the ATP deficiency of these cells. Indeed, subunits of the respiratory ATPase are part of the RIC complex (12). In such a scenario, the added RIC would help generate enough ATP so that the endogenous tRNA import machinery could then naturally drive the import of tRNAs. To test this hypothesis, mitochondria were isolated from wild-type and MERRF cybrid cells and used in our in vitro import assay. As can be seen in Fig. 7A (lane 2) the presence of 2 mM ATP rescued the ability of MERRF mitochondria to import tRNA and restored levels comparable with those observed for wild-type organelles (compare Fig. 7A, lanes 5 and 6 with lanes 9 and 10).
Fig. 7.
Addition of ATP rescues the ability of diseased mitochondria to import tRNAGln. (A) Radioactively labeled tRNACUGGln was incubated with mitochondria isolated from wild-type (WT) human cells, and their import behavior was compared with that isolated from cybrid cells originally generated from patients with a mitochondrial defect that causes MERRF. Mitochondria were Percoll-purified as described in Materials and Methods. Import experiments were performed in the absence (−) or presence (+) of ATP. MN, control reactions treated with micrococcal nuclease in the absence of mitochondria. These reactions serve as a control for MN digestion. IN, input lane of the original tRNA used in the import experiments and serving as a quantitation standard. Black arrows mark the position of the full-length RNA, and the gray arrow marks the position of the mature tRNA processed in the organelle following import. All import reactions were analyzed by gel electrophoresis as described in Materials and Methods. Lanes 1–4 show import reactions using a transcript corresponding to the nucleus-encoded human tRNACUGGln. Lanes 5–10 show reactions similar to those in lanes 1–4, except that pre-tRNAAsp from Bacillus was used in the import reactions to demonstrate transport of the tRNA into the matrix. (B) A reaction similar to that in lane 6 of A was analyzed by a primer extension assay to map the 5′ end of the products generated after import as described in ref. 21. Lanes 1 and 2 are negative control reactions performed in the absence of an RNA template or the absence of reverse transcriptase. Lane 3, in vitro cleavage reaction with Escherichia coli RNase P (140 pmol) incubated with the precursor tRNAAsp (40 pmol) as described in ref. 21, which yields a mature tRNA and serves as a size marker. Lane 4, result of an RNA import reaction with MERRF mitochondria and unlabeled tRNAAsp. Lane 5, full-length pre-tRNA serving as a control and size marker. G, A, T, and C denote an unrelated sequencing ladder used as size markers for the reaction. The dashed arrow corresponds to the 5′ end of pre-tRNAAsp after treatment with RNase P. The solid arrow marks the position of the pre-tRNAAsp, and the bracket marks the position of the cleavage product generated by the mitochondrial matrix localized nuclease activity found in mammalian mitochondria and described in ref. 31. This activity has been shown to digest bacterial pre-tRNAs at 2–4 nucleotides from the authentic 5′ end of the mature product (31).
We have shown previously that Bacillus subtilis pre-tRNAAsp could be imported into isolated Leishmania mitochondria, where the presequence was efficiently processed by an RNase P-like activity present in the mitochondrial matrix. This experiment provided evidence for the translocation of this tRNA into the matrix. We performed a similar test with both wild-type and MERRF mitochondria. Pre-tRNAAsp was efficiently imported into both in an ATP-dependent manner. Furthermore, upon import, pre-tRNA was processed to generate a product of a size consistent with that of a tRNA lacking the presequence (Fig. 7A, compare lanes 6 and 10).
Previous reports have described an RNase P-like activity in mammalian mitochondria that can efficiently remove the 5′ leader from a pre-tRNA (31). When bacterial pre-tRNAs were tested, this activity did not cleave precisely, leaving 2–3 extra nucleotides at the 5′ end of the substrate (31). We took advantage of this observation to substantiate further our in vitro import into MERRF mitochondria. We performed the in vitro import assay with unlabeled pre-tRNAAsp. After import, we performed a primer extension assay with a radioactive primer specific for this tRNA. The observed import product corresponded to the 5′ matured tRNA with 2–3 additional nucleotides at the 5′ end (Fig. 7B, lane 4) compared with a control reaction where pre-tRNAAsp was incubated with bacterial RNase P in vitro (Fig. 7B, lane 3). These results are consistent not only with the behavior of the RNase P-like activity of mammalian mitochondria (31) but also with what has been observed for the import of bacterial pre-tRNAs into the mitochondria of Leishmania (21).
Discussion
Our studies have shown that tRNACUGGln is imported naturally in vivo into human and rat mitochondria and that in vitro this import requires both addition of ATP and a functional ATPase (Fig. 6B). Thus, no “extraneous' components are needed for tRNAGln import. This finding prompts the questions: (i) What is the use of the imported tRNAs, and are they essential for mitochondrial function? (ii) Why are there different mechanisms of tRNA import?
Mitochondria contain a smaller number of tRNA species than does the cytoplasm (13, 32). This might be explained by the fact that an unmodified U in the first position of the tRNA anticodon allows such a tRNA species to decode four codons, which are degenerate in the third position (32). However, should the first anticodon nucleotide of the mitochondria-encoded tRNAUUGGln be thiolated, the ability of the tRNA to interact efficiently with the glutamine codon GAG may be compromised (14). Therefore, import from the cytoplasm of a second different glutamine isoacceptor (tRNACUGGln) would be beneficial for optimal protein synthesis. Import of cytoplasmic glutaminyl-tRNA synthase is not necessary; the mitochondrion contains a glutamyl-tRNAGln amidotransferase for acylation of the tRNAGln isoacceptors (see ref. 33). The idea of tRNA modification and a changed coding response is also supported by the adaptation of the mitochondrial translational apparatus via the import of a mutant yeast tRNALys (34). Our findings also give rise to the possibility that mammalian tRNA import may include additional tRNA species whose identity might be uncovered if the degree of anticodon modifications in mitochondrial tRNAs were known.
Despite differences in the number and types of factors associated with tRNA import in various organisms, tRNAs can mechanistically be imported in two different ways: either via the protein import pathway [e.g., coimport with cytoplasmic aminoacyl-tRNA synthetase (9) or via a pathway independent from protein import that does not require cytosolic factors]. Future studies may reveal other pathways. However, the disparate nature of the import mechanisms suggests that import evolved independently in different systems. Alternatively, differences among various systems may suggest import as a dynamic process that has adapted to intracellular environmental changes provided by a particular organism. The findings presented here that MERRF mitochondria are able to import tRNAs in vitro when provided with sufficient ATP highlight the fact that multiple import pathways could operate in a single organism. This observation is thus reminiscent of the yeast tRNA import system where one mechanism requires cytosolic factors (tRNALys import) whereas the other appears to occur independently of them (tRNAGln import). The import system described here is also analogous to that described with Leishmania tarentolae mitochondria (21), where neither a membrane potential nor the presence for cytosolic factors is necessary for in vitro import.
A number of mitochondrial diseases have been tightly linked to mutations in mitochondrial tRNAs. Our findings that mammalian mitochondria actively import tRNAs make the prospect of mitochondrial therapy feasible. However, the observed lack of tRNA import into mitochondria from MERFF patients (12) combined with our ability to rescue tRNA import in vitro by adding ATP suggest that unless alternative ATP sources are provided in vivo, both native and surrogate import systems may fail as potential therapies. Import systems may themselves be compromised by mutations that affect certain mitochondrial functions such as the ability to synthesize enough ATP.
Materials and Methods
In Vitro Import Assays.
For import assays in all of the studies above, mitochondria from rat liver and/or HeLa cells were isolated as described in ref. 15. For the import assays, native tRNAGln was treated with calf intestinal alkaline phosphatase (Invitrogen) followed by phenol extraction and ethanol precipitation. The tRNA was then 5′-end-labeled with [γ-32P]ATP and T4 polynucleotide kinase (Invitrogen) followed by gel purification on a 7 M urea, 8% polyacrylamide gel. In vitro RNA import assays were performed in a 20-μl reaction volume containing 100,000 cpm of 5′-end-labeled tRNA, 1 μg of mitochondria, 0.6 M sorbitol, 20 mM Tris·HCl (pH 8.0), 5 mM ATP, 2 mM DTT, 20 mM MgCl2, and 2 mM EDTA. After incubation at 27°C for 10 min, 100 units of micrococcal nuclease (MN) (Roche) and 5 mM CaCl2 were added, and the reaction was incubated an additional 30 min to digest the tRNAs that were not imported into the mitochondria. MN was then inhibited by the addition of 10 mM EGTA (pH 8.0). To isolate protected tRNAs, the mitochondria were washed with 0.6 M sorbitol, 20 mM Tris-HCl (pH 8.0), pelleted, resuspended in 90 μl of 10 mM Tris-HCl (pH 8.0), 1 mM EDTA, and 0.1% SDS, and extracted with 0.1 ml of water-saturated phenol pH 4.5 followed by ethanol precipitation. The radioactively labeled tRNAs were separated by electrophoresis through a 7 M urea, 6% acrylamide gel. After electrophoresis, the gels were dried onto Whatman 3MM chromatography paper and visualized with the Storm PhosphorImager imaging system.
RT-PCR.
RNA was isolated from total, MN-treated mitochondrial fractions and/or cytosolic fractions by the guanidinium thiocyanate/phenol/chloroform extraction method (35). After purification, RNA was treated with RQ1 RNase-free DNase (Promega). Primers AGGTCCTACCCGGATTCGAACCGGG reverse transcription and PCR forward primer, and GGTCCTATAGTGTAGTGGTTATCAC PCR reverse primer were used for RT-PCRs. The RT primer was annealed to 5 μg of total mitochondrial RNA at 50°C followed by the addition of reverse transcriptase. The RT reaction was carried out as described in the SuperScriptTM II reverse transcriptase first-strand synthesis protocol (Invitrogen). Following RT, 1 μl of the 20-μl RT reaction was used as a template in a PCR with the forward and reverse primers (above). PCRs were performed by using Taq polymerase following manufacturer's instructions and included a 30-s 95°C denaturation step, 55°C annealing for 30 s and 30 s of 72°C elongation for a total of 30 cycles. Controls included a mock reaction where the RT was left out of the reaction and used as a negative control to test for DNA contamination in the RNA samples. RT-PCR products were cloned by using the TOPO method according to the manufacturers' instructions (Invitrogen).
Acknowledgments.
We thank A. K. Hopper, M. Ibba, and all members of the Reichert, Söll, and Alfonzo groups for useful comments. This work was supported by Deutsche Forschungsgemeinschaft Grant SFB 594 (to A.S.R.) and by grants from the National Institute of General Medical Sciences (to D.S.) and the National Science Foundation (to J.D.A.).
Footnotes
The authors declare no conflict of interest.
References
- 1.Rorbach J, Soleimanpour-Lichaei R, Lightowlers RN, Chrzanowska-Lightowlers ZM. How do mammalian mitochondria synthesize proteins? Biochem Soc Trans. 2007;35:1290–1291. doi: 10.1042/BST0351290. [DOI] [PubMed] [Google Scholar]
- 2.Schneider A. Does the evolutionary history of aminoacyl-tRNA synthetases explain the loss of mitochondrial tRNA genes? Trends Genet. 2001;17:557–559. doi: 10.1016/s0168-9525(01)02439-8. [DOI] [PubMed] [Google Scholar]
- 3.Schneider A, Marechal-Drouard L. Mitochondrial tRNA import: Are there distinct mechanisms? Trends Cell Biol. 2000;10:509–513. doi: 10.1016/s0962-8924(00)01854-7. [DOI] [PubMed] [Google Scholar]
- 4.Suyama Y. The origins of mitochondrial ribonucleic acids in Tetrahymena pyriformis. Biochemistry. 1967;6:2829–2839. doi: 10.1021/bi00861a025. [DOI] [PubMed] [Google Scholar]
- 5.Mirande M. The ins and outs of tRNA transport. EMBO Rep. 2007;8:547–549. doi: 10.1038/sj.embor.7400989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tarassov I, et al. Import of nuclear DNA-encoded RNAs into mitochondria and mitochondrial translation. Cell Cycle. 2007;6:2473–2477. doi: 10.4161/cc.6.20.4783. [DOI] [PubMed] [Google Scholar]
- 7.Martin RP, Schneller JM, Stahl AJ, Dirheimer G. Import of nuclear deoxyribonucleic acid coded lysine-accepting transfer ribonucleic acid (anticodon CUU) into yeast mitochondria. Biochemistry. 1979;18:4600–4605. doi: 10.1021/bi00588a021. [DOI] [PubMed] [Google Scholar]
- 8.Rinehart J, Krett B, Rubio MA, Alfonzo JD, Söll D. Saccharomyces cerevisiae imports the cytosolic pathway for Gln-tRNA synthesis into the mitochondrion. Genes Dev. 2005;19:583–592. doi: 10.1101/gad.1269305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tarassov I, Entelis N, Martin RP. Mitochondrial import of a cytoplasmic lysine-tRNA in yeast is mediated by cooperation of cytoplasmic and mitochondrial lysyl-tRNA synthetases. EMBO J. 1995;14:3461–3471. doi: 10.1002/j.1460-2075.1995.tb07352.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Entelis N, et al. A glycolytic enzyme, enolase, is recruited as a cofactor of tRNA targeting toward mitochondria in Saccharomyces cerevisiae. Genes Dev. 2006;20:1609–1620. doi: 10.1101/gad.385706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kolesnikova OA, et al. Nuclear DNA-encoded tRNAs targeted into mitochondria can rescue a mitochondrial DNA mutation associated with the MERRF syndrome in cultured human cells. Hum Mol Genet. 2004;13:2519–2534. doi: 10.1093/hmg/ddh267. [DOI] [PubMed] [Google Scholar]
- 12.Mahata B, Mukherjee S, Mishra S, Bandyopadhyay A, Adhya S. Functional delivery of a cytosolic tRNA into mutant mitochondria of human cells. Science. 2006;314:471–474. doi: 10.1126/science.1129754. [DOI] [PubMed] [Google Scholar]
- 13.Sprinzl M, Vassilenko KS. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 2005;33:D139–D140. doi: 10.1093/nar/gki012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Agris PF, Vendeix FA, Graham WD. tRNA's wobble decoding of the genome: 40 years of modification. J Mol Biol. 2007;366:1–13. doi: 10.1016/j.jmb.2006.11.046. [DOI] [PubMed] [Google Scholar]
- 15.Pallotti F, Lenaz G. Isolation and subfractionation of mitochondria from animal cells and tissue culture lines. Methods Cell Biol. 2001;65:1–35. doi: 10.1016/s0091-679x(01)65002-7. [DOI] [PubMed] [Google Scholar]
- 16.Kapushoc ST, Alfonzo JD, Rubio MA, Simpson L. End processing precedes mitochondrial importation and editing of tRNAs in Leishmania tarentolae. J Biol Chem. 2000;275:37907–37914. doi: 10.1074/jbc.M007838200. [DOI] [PubMed] [Google Scholar]
- 17.Delage L, Dietrich A, Cosset A, Marechal-Drouard L. In vitro import of a nuclearly encoded tRNA into mitochondria of Solanum tuberosum. Mol Cell Biol. 2003;23:4000–4012. doi: 10.1128/MCB.23.11.4000-4012.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mahapatra S, Ghosh T, Adhya S. Import of small RNAs into Leishmania mitochondria in vitro. Nucleic Acids Res. 1994;22:3381–3386. doi: 10.1093/nar/22.16.3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yermovsky-Kammerer AE, Hajduk SL. In vitro import of a nuclearly encoded tRNA into the mitochondrion of Trypanosoma brucei. Mol Cell Biol. 1999;19:6253–6259. doi: 10.1128/mcb.19.9.6253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kolesnikova OA, et al. Suppression of mutations in mitochondrial DNA by tRNAs imported from the cytoplasm. Science. 2000;289:1931–1933. doi: 10.1126/science.289.5486.1931. [DOI] [PubMed] [Google Scholar]
- 21.Rubio MA, Liu X, Yuzawa H, Alfonzo JD, Simpson L. Selective importation of RNA into isolated mitochondria from Leishmania tarentolae. RNA. 2000;6:988–1003. doi: 10.1017/s1355838200991519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alfonzo JD, Blanc V, Estevez AM, Rubio MA, Simpson L. C to U editing of the anticodon of imported mitochondrial tRNATrp allows decoding of the UGA stop codon in Leishmania tarentolae. EMBO J. 1999;18:7056–7062. doi: 10.1093/emboj/18.24.7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tarassov IA, Martin RP. Mechanisms of tRNA import into yeast mitochondria: an overview. Biochimie. 1996;78:502–510. doi: 10.1016/0300-9084(96)84756-0. [DOI] [PubMed] [Google Scholar]
- 24.Brubacher-Kauffmann S, Marechal-Drouard L, Cosset A, Dietrich A, Duchene AM. Differential import of nuclear-encoded tRNAGly isoacceptors into Solanum tuberosum mitochondria. Nucleic Acids Res. 1999;27:2037–2042. doi: 10.1093/nar/27.9.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suzuki H, et al. Characterization of rat TOM40, a central component of the preprotein translocase of the mitochondrial outer membrane. J Biol Chem. 2000;275:37930–37936. doi: 10.1074/jbc.M006558200. [DOI] [PubMed] [Google Scholar]
- 26.Linnett PE, Beechey RB. Inhibitors of the ATP synthethase system. Methods Enzymol. 1979;55:472–518. doi: 10.1016/0076-6879(79)55061-7. [DOI] [PubMed] [Google Scholar]
- 27.Kaback HR. Molecular biology and energetics of membrane transport. J Cell Physiol. 1976;89:575–593. doi: 10.1002/jcp.1040890414. [DOI] [PubMed] [Google Scholar]
- 28.Cunarro J, Weiner MW. Mechanism of action of agents which uncouple oxidative phosphorylation: Direct correlation between proton-carrying and respiratory-releasing properties using rat liver mitochondria. Biochim Biophys Acta. 1975;387:234–240. doi: 10.1016/0005-2728(75)90106-1. [DOI] [PubMed] [Google Scholar]
- 29.Neupert W, Herrmann JM. Translocation of proteins into mitochondria. Annu Rev Biochem. 2007;76:723–749. doi: 10.1146/annurev.biochem.76.052705.163409. [DOI] [PubMed] [Google Scholar]
- 30.Florentz C, Sohm B, Tryoen-Tóth P, Pütz J, Sissler M. Human mitochondrial tRNAs in health and disease. Cell Mol Life Sci. 2003;60:1356–1375. doi: 10.1007/s00018-003-2343-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rossmanith W, Tullo A, Potuschak T, Karwan R, Sbisa E. Human mitochondrial tRNA processing. J Biol Chem. 1995;270:12885–12891. doi: 10.1074/jbc.270.21.12885. [DOI] [PubMed] [Google Scholar]
- 32.Heckman JE, Sarnoff J, Alzner-DeWeerd B, Yin S, RajBhandary UL. Novel features in the genetic code and codon reading patterns in Neurospora crassa mitochondria based on sequences of six mitochondrial tRNAs. Proc Natl Acad Sci USA. 1980;77:3159–3163. doi: 10.1073/pnas.77.6.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sheppard K, et al. From one amino acid to another: tRNA-dependent amino acid biosynthesis. Nucleic Acids Res. 2008;36:1813–1825. doi: 10.1093/nar/gkn015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kamenski P, et al. Evidence for an adaptation mechanism of mitochondrial translation via tRNA import from the cytosol. Mol Cell. 2007;26:625–637. doi: 10.1016/j.molcel.2007.04.019. [DOI] [PubMed] [Google Scholar]
- 35.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]