Abstract
Carboxysomes are organelle-like polyhedral bodies found in cyanobacteria and many chemoautotrophic bacteria that are thought to facilitate carbon fixation. Carboxysomes are bounded by a proteinaceous outer shell and filled with ribulose 1,5-bisphosphate carboxylase/oxygenase (RuBisCO), the first enzyme in the CO2 fixation pathway, but exactly how they enhance carbon fixation is unclear. Here we report the three-dimensional structure of purified carboxysomes from Synechococcus species strain WH8102 as revealed by electron cryotomography. We found that while the sizes of individual carboxysomes in this organism varied from 114 to 137 nm, surprisingly, all were approximately icosahedral. There were on average ∼250 RuBisCOs per carboxysome, organized into 3-4 concentric layers. Some models of carboxysome function depend on specific contacts between individual RuBisCOs and the shell, but no evidence of such contacts was found: no systematic patterns of connecting densities or RuBisCO positions against the shell's presumed hexagonal lattice could be discerned, and simulations showed that packing forces alone could account for the layered organization of RuBisCOs.
Keywords: carboxysomes, electron cryotomography, bacterial ultrastructure, Calvin cycle
Introduction
The primary point of entry of carbon into the biosphere is the Calvin-Benson-Bassham cycle of autotrophic organisms such as cyanobacteria, which are among the most ancient and abundant organisms on earth and account for a large fraction of global carbon fixation. In the first step of this pathway, the fixation of molecular CO2 is catalyzed by the enzyme ribulose 1,5-bisphosphate carboxylase/oxygenase (RuBisCO). RuBisCO is a notoriously slow and inefficient enzyme. It has a low affinity for CO2 and also catalyzes the unproductive and energy-requiring fixation of O2 (photorespiration)1. Thus cyanobacteria use a sophisticated carbon concentrating mechanism2 whose main elements seem to be: i) a mechanism for the active transport of HCO3− and CO2 into the cytoplasm3; ii) localized carbonic anhydrase activity to convert HCO3− into CO2; and iii) the carboxysome, a polyhedral cellular compartment bounded by a proteinaceous shell that sequesters RuBisCO into a distinct microenvironment4.
Carboxysomes are present in all known cyanobacteria and are divided into two distinct classes, α- or cso-type and β- or ccm-type, on the basis of the form of RuBisCO (1A or 1B) that they encapsulate5. Some chemoautotrophic proteobacteria also contain carboxysomes5 and similar polyhedral bodies (dubbed “enterosomes”) and homologs of carboxysome genes have now been found in enteric bacteria as well, suggesting that these structures may be of more ancient origin and that such compartments may be used more generally in bacteria to regulate a variety of metabolic pathways6.
Our knowledge of the structure of carboxysomes has come mainly from conventional electron microscopy (EM) and X-ray crystallography. EM studies revealed that carboxysomes are polyhedral bodies, proposed to be either icosahedra7 or pentagonal dodecahedra8 of variable size containing a paracrystalline arrangement of RuBisCO. The latter study suggested that carboxysomes are hollow compartments with a single layer of RuBisCOs attached to the inner layer of the protein shell. The crystal structures of two β-carboxysome shell proteins, CcmK2 and CcmK4, that share significant sequence homology with the α-carboxysomal CsoS1 protein, were recently solved9. These proteins crystallized into hexamers surrounding a central, charged pore, suggesting the possibility that they create a specialized barrier that may regulate the flow of metabolites into and out of the carboxysome. The crystal structure of one α-carboxysome shell protein, CsoS3, is also available, and this protein was shown to have carbonic anhydrase activity10, 11.
While it is clear that carboxysomes offer some metabolic advantage, the mechanism is still unknown. One model is that they simply increase the local concentration of both RuBisCO and CO2, while perhaps also decreasing the concentration of competing O2. It has been hypothesized that some further special property of the carboxysome, like the structure of the protein shell or the arrangement of the internal RuBisCOs, functions as a selectively permeable barrier, either limiting CO2 outward diffusion, O2 inward diffusion, or both12. It has also been suggested that perhaps the carboxysome somehow maintains its internal RuBisCOs in an active conformation, or in a conformation that favors the carboxylation reaction over the competing oxygenation reaction12. Observed sequence homology between the β-carboxysomal shell protein CcmM and the RuBisCO small subunit led to the suggestion that specific contacts might exist between the shell and RuBisCO, and that this could arrange carboxysomal RuBisCOs in a preferred orientation or possibly assist in carboxysome biogenesis13,14.
In an effort to shed further light on the structure and function of carboxysomes, we produced 3-D reconstructions of purified carboxysomes from the α-cyanobacterium, Synechococcus species strain WH8102. We chose this organism because of its importance in the global carbon cycle and to complement other ongoing efforts that use it as a model system15-17. Because carboxysomes vary in size, techniques that rely on averaging signals from identical objects (X-ray and electron crystallography, nuclear magnetic resonance spectroscopy and single particle electron microscopy) are unsuitable. Instead we used electron cryotomography (ECT), the highest resolution technique available today for the study of such heterogeneous samples18. In ECT, specimens are preserved in a near-native, “frozen-hydrated” state and their 3-D structure is reconstructed to a few nm resolution from a series of EM images recorded while the specimen is tilted incrementally around one or two orthogonal axes19. Measurements and observations of the size, shape, and internal organization of S. species strain WH8102 carboxysomes are reported, along with analyses that look for evidence of potential interactions between the shell and individual RuBisCOs. While this paper was in the final stages of preparation, a complementary electron cryotomographic study of carboxysomes from a different species, Halothiobacillus neapolitanus, appeared20. The important similarities and differences are considered in the Discussion.
Results
Sample preparation and data collection
Carboxysomes were purified from S. species strain WH8102 as described previously17 and plunge-frozen in thin films across EM grids (see Materials and Methods). Three single- and eight dual-tilt image series were collected. Each series had in the field of view one to five intact carboxysomes surrounded by what appeared to be dissassembled fragments. As a result, the embedding ice was often thicker than the diameter of carboxysomes. Fig. 1 shows one of the images from a single-tilt series and a slice from the corresponding reconstruction. Supp. Movie 1 shows an entire dual-tilt image series, the corresponding reconstruction and the major results in 3-D.
Fig. 1. Example image and reconstruction slice.

(a) The nominally untilted image from a single-axis tilt series. (b) 6.7 nm-thick slice through the corresponding (undenoised) 3-D reconstruction. Scale bar 100 nm.
Icosahedral shape
From the reconstructed tomograms, 26 carboxysomes were extracted, denoised, and segmented, revealing in every case a polyhedron with 20 triangular faces (an icosahedron). Initially, in some cases it was unclear whether the observed icosahedra were regular because the top and bottom surfaces were poorly resolved due to the missing “wedge” or “pyramid” of data inherent in ECT (the tilt ranges were at best +/− 66°). In this respect, when all other experimental parameters such as total dose and defocus were held equal, reconstructed carboxysomes from dual-tilt series were better resolved as expected, and appeared more exactly icosahedral (see also 19). To more carefully assess the architecture of the carboxysomes, in each case the regions of the shell that were clearly resolved were fit and compared to complete, regular icosahedra (Fig. 2). They matched very well in all cases, though the vertices and edges of the protein shells were more rounded than the geometrically perfect fitted icosahedra. The thickness of the protein shells was ∼4 nm, in good agreement with reported values for α-carboxysomes21 and the determined crystal structures of protein shell homologs9, but thinner than that reported for β-carboxysomes from S. species strain PCC 7942 (5-6 nm, as visualized by EM in frozen-hydrated intact cells, but by the Hilbert Differential Contrast method22).
Fig. 2. Carboxysomes are icosahedral.
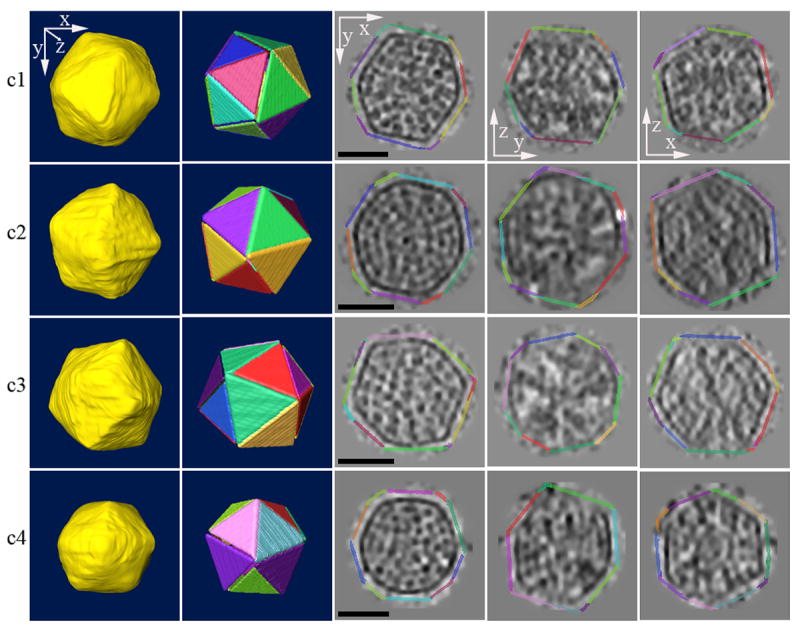
Four individual reconstructed carboxysomes are shown (one per row). From left to right appear the manually segmented surface, the best-fitting regular icosahedron, and three orthogonal central slices (xy, yz, and xz) through the denoised carboxysomes. The central slices are surrounded by the fit icosahedra, enlarged slightly and with the same color-coded faces to allow comparison of the actual shells with the corresponding cross sections of regular icosahedron. The anisotropic resolution is due to the limited tilt range accessible in ECT. Scale bar 50 nm.
Size distribution
The diameters of the 26 carboxysomes (defined here as the longest dimension of each geometrically perfect fitted icosahedron, or 1.9 times the edge length) varied from 114 nm to 137 nm, with an average of 123 ± 5 nm. Interestingly, the Synechocystis shell proteins CcmK2 and CcmK4 crystallized as hexamers, which are the basic building blocks of icosahedra9. Icosahedra of different sizes can be built from hexagonal and pentagonal units in a way described by the so-called T number23. Prompted by the sequence similarities between these proteins and the S. species strain WH8201 major shell protein CsoS1 (32% and 49% identity with CcmK4 and CcmK2, respectively), and the fact that the carboxysomes we observed were indeed icosahedral, we calculated the range of T-numbers that would correspond to our observed shells, assuming they were in fact assemblies of hexamers with the same dimensions as those seen in the crystals (4 nm edge length). The carboxysomes we imaged spanned a range of hypothetical T numbers from 73 to 103 (see Fig. 3 – where the sizes corresponding to particular T numbers are indicated by vertical arrows), and 855 hexamers would be required to form the complete protein shell of the average-sized carboxysome (123 nm diameter).
Fig. 3. Histogram of carboxysome diameters.

Each bin is 1 nm wide. The diameter of icosahedra with all possible T-numbers in the same size range built from hexagons of edge length 4 nm (corresponding to crystal structure PDB 2AIB of CcmK2) are also shown.
Organization of RuBisCOs
The interiors of 14 of the 26 reconstructed carboxysomes were densely packed with distinct protein-like densities. Because (i) the vast majority of these had the same approximate size as RuBisCO, (ii) proteomic analysis of purified carboxysomes such as these had demonstrated that 62% of the total protein is RuBisCO24, and (iii) RuBisCO is known to be packed inside the carboxysome25, 26, we assumed that these densities were in fact RuBisCOs. It was clear from simple visual inspection that the RuBisCOs were arranged in three and sometimes four concentric layers. The other 12 carboxysomes had fewer RuBisCOs and less internal order. These were probably just damaged by the purification and grid freezing procedures17 (which clearly disrupted many shells, as evidenced by the plentiful proteinaceous debris on the grids), but we cannot be sure because the missing wedge of data always makes certain faces of the shell appear to be missing. An alternative is that these were assembly intermediates or even mature carboxysomes with a different organization. Future studies of intact cells should clarify this point.
Radial density profiles were calculated for the 14 carboxysomes whose interiors were ordered. The origin of each carboxysome was taken as the center of the fit icosahedron. Radial densities were calculated within both spherical shells and nested icosahedral shells, but the results were essentially indistinguishable. Each radial profile had 4 to 5 peaks. The outermost peak corresponded to the shell and was the highest in every case. Three and sometimes four smaller internal peaks appeared, corresponding to ordered layers of RuBisCO. As carboxysome sizes were variable and the spacing between the layers were presumed to scale accordingly, in order to compare profiles, the relative density as a function of percent radius was normalized, plotted and averaged (Fig. 4). The internal peaks appeared at 81, 60, and 38% of the radius. In an average-sized carboxysome with radius 62 nm, this would correspond to peaks at 50, 37, and 24 nm from the center.
Fig. 4. Radial density profiles.
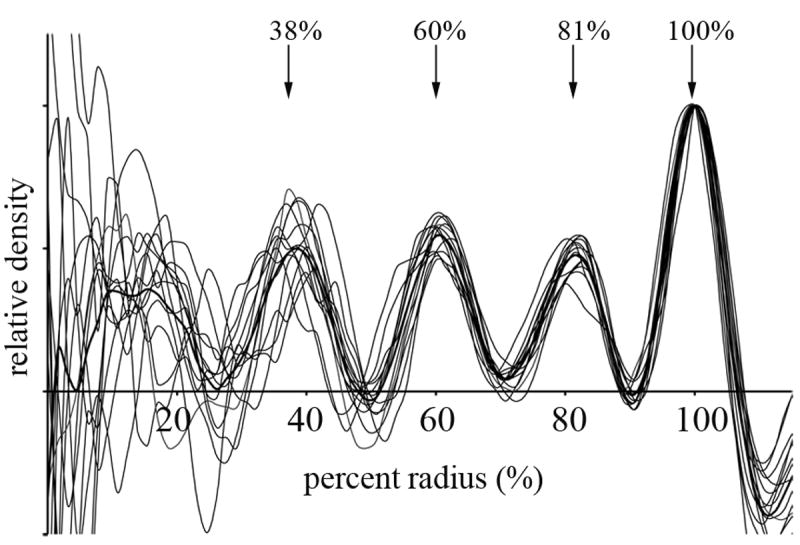
Radial density profiles of individual carboxysomes (thin lines) and their average (thick line), scaled to normalize the densities of the outermost peaks (the shell). The positions of the well-defined peaks (the three internal RuBisCO layers and the carboxysome shell) are marked with arrows. For absolute scale, the average radius was 62 nm.
Number of RuBisCOs per carboxysome
The number of RuBisCOs contained within each carboxysome was estimated by “template matching”27; i.e. the (undenoised) volumes of the carboxysomes were searched for densities that correlated well with the known, low-pass filtered crystal structure of RuBisCO28. Because there were no abrupt discontinuities in the spectrum of cross-correlation coefficients reported, no clear threshold value that would distinguish true RuBisCOs from surrounding material was apparent. Further, among the set of higher coefficients, many were too close together to be separate RuBisCOs and many were false positives clustered on the shell. We therefore discarded peaks on the shell and systematically selected peaks one by one from the highest coefficient to the lowest, each time discarding all other peaks within 11 nm (the approximate diameter of RuBisCO) until there were no more spaces large enough for a RuBisCO left. As a control, peak-to-peak minimum distances of 10 or 12 nm were also tried, but by visual inspection these values clearly produced too many false positives or negatives, respectively. As seen in Fig. 5, this strategy located most of the densities that were consistent with the known size of RuBisCO.
Fig. 5. Identification of individual RuBisCOs through template-matching.

Densities identified as RuBisCO by template matching followed by a customized peak search are circled in red on 6.7 nm slices through the (a) undenoised and (b) denoised carboxysome. (c) 3-D representation of the same carboxysome in which the densities have been replaced by the RuBisCO template.
The average number of RuBisCOs found per carboxysome was 232 ± 18, with values ranging from 207 to 269. Overall, the number of RuBisCOs per carboxysome increased with the size of the carboxysome (Fig. 6). Treating each RuBisCO as a sphere of diameter 11 nm and dividing by the volume of the average fitted regular icosahedron, about a quarter (27%) of the carboxysome volume was occupied by RuBisCOs.
Fig. 6. Number of RuBisCOs per carboxysome as a function of carboxysome volume.

The number of RuBisCOs was assessed by template matching followed by a customized peak-search algorithm, and the volume calculated as that of the best fitting regular icosahedron.
Relationship of RuBisCO layers to carboxysome shell
Several considerations suggested that specific interactions between the shell and individual RuBisCOs might exist. First, the RuBisCOs were found to be organized in concentric layers just inside the shell. Second, some models for the function of carboxysomes posit that RuBisCO is held in an activated conformation or in a specific arrangement within the carboxysome shell29-31. Third, one carboxysomal protein (CcmM) was found to have sequence homology to the small subunit of RuBisCO, and could therefore conceivably domain swap with or otherwise bind the other subunits of RuBisCO13, 14.
We therefore looked for evidence of specific contacts between the shell and individual RuBisCOs. Many RuBisCOs were in fact either touching or very near the shell and many weak densities were visible between RuBisCOs and the shell, but neither phenomenon was systematic, strong, or clearly resolved enough to be conclusive. Two additional analyses were performed to check for evidence of specific contacts: RuBisCOs were mapped onto the presumed hexagonal lattice of the underlying shell, and simulations were run to test whether simple packing forces alone could have caused the RuBisCO layering.
i. Mapping RuBisCOs onto the presumed shell lattice
If shell proteins contacted RuBisCOs specifically, the positions of the RuBisCOs might reflect the underlying shell lattice. An in-house program was therefore developed to project the density within the outermost RuBisCO layer onto the shell (Fig. 7). Individual RuBisCOs were clearly recognizable as dense patches (about 7 per face), but when the presumed hexagonal lattice of the shell was superimposed (about 40 hexagons per face for the T=81 icosahedron shown), no relationship between the two patterns was discerned. Nevertheless, this picture did highlight how many more pores probably exist in the shell than RuBisCOs. Based on the total number of RuBisCOs per carboxysome (232) and presumed hexagons in the shell 855, there would be on average 3 to 4 shell pores per RuBisCO.
Fig. 7. Relative arrangement of RuBisCOs and the presumed hexagonal lattice of the shell.
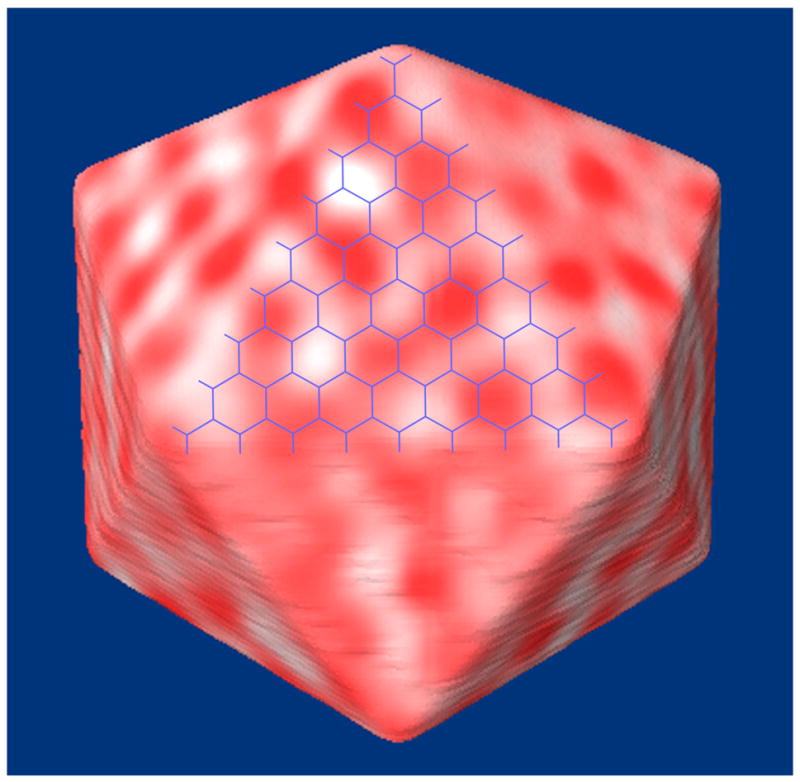
All the densities (red) in the outermost layer of RuBisCO were projected on the faces of the icosahedron fit to the carboxysome shell. The presumed hexagonal lattice (with hexagonal spacing of 7 nm) for the corresponding T=81 icosahedron is shown in blue. No correspondence was discerned.
This projection procedure also provided a second way to roughly estimate the number of RuBisCOs per carboxysome and per layer. Because of the missing pyramid (or wedge) of data, the boundaries of the RuBisCOs were not isotropically resolved, and thus their projections onto certain faces of the carboxysome were clearer than others. By estimating the average number of RuBisCOs per face where they were most clearly resolved, and assuming that all regions would be equally packed, we calculated the total number of RuBisCOs in the three layers to be 149 ± 13, 85 ± 8, and 37 ± 4, respectively, for a total of 271 ± 25 per carboxysome (standard errors arose from considering all 14 well-ordered carboxysomes). Thus the number of RuBisCOs in each layer diminished by approximately a factor of 2. These estimates were uniformly ∼20% higher than those obtained through template matching, but it is unclear which method was more nearly correct.
ii. Emergence of layers through random packing
The second way we checked for evidence of specific contacts between the shell and RuBisCOs was to test whether packing forces alone could have given rise to the observed layers. We ran a series of simulations in which RuBisCOs were modeled as semi-hard spheres with diameters spanning the range of dimensions present in RuBisCO (from 11 to 13.3 nm), moving freely within an icosahedral container 123 nm in diameter. For each sphere diameter, the total number of spheres was varied from 124 to 344 in steps of 20 to more than span the range of experimentally determined RuBisCOs per carboxysome. In order to compare the results with the experimentally reconstructed carboxysomes, the simulation was repeated 14 times for each set of parameters and the average radial density was plotted (Fig. 8).
Fig. 8. Spontaneous generation of nested layers in icosahedral packing simulations.

Icosahedral containers of average size (123 nm) were filled with randomly diffusing hard spheres of various diameters spanning the range of molecular dimensions present in RuBisCO. The resulting radial average density profiles from 14 independent packing simulations are shown in each case. (a) Profiles for 304 spheres of different diameters. The spheres spontaneously pack into layers in all cases. (b) Profiles for different numbers of 12.3 nm diameter spheres. As the number of spheres gets larger, the layers move slightly closer to the container boundary. (c) The average profile from the experimentally reconstructed carboxysomes (including an additional outer peak corresponding to the shell) (black) and the profile of the closest matching simulation, a 123 nm container filled with three hundred and four 12.3 nm spheres (red).
For each diameter tested, peaks in the average radial density plot developed in the same general pattern. When the number of diffusing spheres was low (124-144), a single peak was observed next to the icosahedral boundary. As the number of spheres was increased (164-184), the density within this outer layer grew and another peak began to emerge as well. Higher numbers of spheres caused 1-2 additional peaks to appear at even smaller radii (Fig. 8b and Supp. Movie 2). As the number of spheres and layers increased, their spacing also decreased slightly, and the outermost layer shifted closer to the boundary. The best match of the peaks to the experimental curves was obtained using 304 spheres with diameters of 12.3 nm (Fig. 8c).
Discussion
This study established the 3-D structure of a cyanobacterial carboxysome and the number and organization of RuBisCOs within it. This was accomplished with ECT, a technique that allowed the carboxysomes to be visualized in a nearly-native, “frozen-hydrated” state, thus avoiding the artifacts associated with conventional EM. In confirmation of previous analyses of Nitrobacter winogradskyi carboxysomes7, we found that S. species carboxysomes were icosahedral, and their observed dimensions (114 to 137 nm) were similar to those reported in H. neapolitanus8 and N. winogradskyi7. It remains to be established whether all carboxysomes and the related enterosomes (such as those associated with ethanolamine breakdown in Salmonella typhimurium32 and Escherichia coli33 and propanediol utilization in Salmonella typhimurium6, 34) are similarly icosahedral. Such consistency would not be surprising, since the two Synechocystis shell proteins that have been crystallized multimerize into hexagons, the structural building blocks of icosahedra, and sequence alignments of all the major shell proteins show strict conservation of the residues at the protein-protein interfaces9. Nevertheless, the smooth and broad distribution of sizes observed here was unusual for biological icosahedra. While it is known that a single protein can assemble into icosahedra with different T-numbers35, these shells must also be quite flexible.
The internal RuBisCOs were clearly organized into concentric layers. Their total number was estimated in two ways: template matching and projection onto icosahedral shells, yielding 232 ± 18 and 271 ± 25, respectively. It is unclear which method is more nearly correct: template matching followed by our peak selection algorithm clearly introduced some false positives and neglected some false negatives; but the projection method depended on visual estimates of the number of RuBisCOs per icosahedral face, the assumption that all the faces would be similar, and that all the major densities were in fact RuBisCOs. We conclude that there were on average very roughly 250 RuBisCOs per carboxysome.
Several considerations, including (i) the layered organization of RuBisCOs and their spacing, (ii) proposals in the literature that a preferential organization of RuBisCOs is mediated by interactions with the protein shell13, 14, 29, and (iii) models of carboxysomes that suggest such interactions, prompted us to look for evidence of specific contacts between the shell and RuBisCOs. None was found: no systematic connecting densities appeared in the reconstructions, the pattern of RuBisCO positions did not appear to be related to the presumed hexagonal lattice of the shell, and simulations showed that concentric layers like those observed arise spontaneously when volume-excluding objects are packed into icosahedral containers. The spontaneous emergence of layers agreed with what would have been predicted by related packing studies36.
As the work presented here was in the final stages of preparation for publication, a complementary study by Schmid et al. appeared reporting ECT analysis of the related carboxysomes from H. neapolitanus20. H. neapolitanus carboxysomes were also found to be icosahedral, occupy a range of sizes, and contain concentric layers of RuBisCOs. Concerned with potential T-numbers and underlying hexagonal shell lattices, we measured diameters as the largest distance across fit regular icosahedra, from which edge lengths can be derived. Schmid et al. measured diameters as the position of the outermost peak in 1-D (spherically averaged) radial density profiles. When for the purposes of comparison, we followed the Schmid et al. procedure, the S. species carboxysomes we imaged here were just slightly larger (101 ± 3 nm) than those in H. neapolitanus (average ∼96 nm), but the degree of heterogeneity (spanning ∼20 nm) was quite similar. These differences are, however, within the few percent magnification uncertainty present in electron microscopy without embedded calibration standards.
In addition to technical differences between the two studies (voltage, energy filtering, dual-axis tilting, use of gold fiducials for alignment, etc.), the most important difference in the approaches taken was that Schmid et al. grouped carboxysomes into classes based on size and then averaged them, producing maps with subtly different patterns of density and holes in the shell faces. This led them to interpret their different size classes as carboxysomes with different subunit arrangements and to speculate that the holes they saw in their averages were authentically variable pore patterns. We think it highly unlikely that the shell proteins arrange themselves in a fundamentally different pattern in the variably sized carboxysomes and suggest that the pore patterns in their averages were artifactual (noise). While we agree that the size variation is probably due in part to different T numbers, we would further speculate that the continuous size distribution also indicates flexibility in the shell contacts. If so, class averages would be hard to interpret. Schmid et al. also assert that the five-fold symmetric apexes of the carboxysomes are thicker than the faces and suggest that the shell-associated carbonic anhydrases may bind there; however we did not observe any such thickening.
These results contribute information relevant to several models. First, it is significant that over half of the RuBisCOs are found packed immediately adjacent to the shell, where a carbonic anhydrase is now known to exist10. Thus the local concentrations of RuBisCO and CO2 would indeed be expected to be very high just inside the shell. Second, we found no evidence for specific contacts between the shell and RuBisCOs that might activate or order them12, 29. Third, concerning transport of substrates, it has been proposed that the charged pores of the shell proteins regulate the passage of metabolites9. Recent studies suggest that enterosomes may play a similar role, preventing the leakage of gaseous metabolites such as acetaldehyde37. Here we report that on average each RuBisCO is served by about 3 to 4 shell pores, but that the protein-protein contacts in the shell are likely quite flexible, and may themselves also allow passage of metabolites. Fourth, we observed that the inner layers of RuBisCOs were broader and less organized than the layers closer to the protein shell. This does not support the model of carboxysome biogenesis in which RuBisCOs assemble into a polyhedral structure before the protein shell is added38. Finally, our observation that RuBisCOs filled the entire volume of the carboxysome, coupled with the recent revelation that CsoS3 is a shell-associated carbonic anhydrase10, contradicts the model that carboxysomes contain a centrally located carbonic anhydrase that provides CO2 to peripheral RuBisCOs organized to function as a barrier to CO2 diffusion30. More information is still clearly needed to elucidate the function and mechanism of this important molecular machine.
Materials and methods
Carboxysome purification
Carboxysomes were purified as described previously17. Briefly, S. species cells were grown for 15 days and collected by centrifugation. The cell pellet was resuspended in 25 mM N-Tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid (TES)-NaOH, pH 7.0, containing 1 mM EDTA (ethylenediaminetetraacetate) and 1mM PMSF (phenylmethylsulfonyl fluoride) and passed through a French press system (Emulsiflex-C5TM; Avestin Inc., Ottawa, Ontario). The lysates were cleared of large cellular debris by centrifugation at 12 000 g and then the supernatant was centrifuged further at 40 000 g to pellet the carboxysomes. The carboxysome pellet was resuspended in the above TES buffer also containing 20 mM MgSO4 and 33% (v/v) YPERS™, centrifuged again at 40 000 g, and resuspended once more in TES buffer.
Electron microscopy
The solution of purified carboxysomes was combined in a ratio of 4:1 with 10 nm colloidal gold (Ted Pella, 20x conc.) and plunge-frozen onto holey carbon grids (R1.2/1.3, Quantifoil; or lacey carbon, Ted Pella) with a Vitrobot (FEI, Netherlands). The grids were imaged with a 300 kV FEG “G2 Polara” TEM (FEI, Netherlands) using a 2048 × 2048 GIF CCD. Energy-filtered (20 eV slit width) single- and dual-tilt image series were acquired with the UCSF tomography package39 at either 34 or 27.5kx magnification (CCD pixel sizes corresponding to 6.7 and 8.2 Å, respectively). The data used for analysis came from 8 dual-tilt and 3 single-tilt tomograms, collected with total doses ranging from 120 to 180 electrons/Å2; defocus values of −10, −12, −14, −15 or −16 μm (corresponding to the first CTF zero at 44.4, 48.6, 52.5, 54.3, and 56.1 Å, respectively); and an angular range of +/− 66° in fixed steps of 2 or 3°.
Data processing and analysis
Tomographic reconstructions were generated with IMOD40. Individual carboxysomes were selected, denoised41 and aligned to a standard icosahedron using Bsoft42. The shapes of the outer shells were segmented and visualized using Amira (Mercury Computer Systems) after the carboxysome reconstructions were subjected to 200 cycles of denoising. The segmentation was used to create a solid object that was subsequently oriented with respect to a standard icosahedron (having x, y, and z as the 2-fold axes). Icosahedral and spherical radial density plots of the oriented, non-denoised carboxysomes were calculated with custom-written Amira (Mercury Computer Systems) modules.
The number of RuBisCOs and their locations within carboxysomes were determined using a template-matching program27 with the crystal structure of RuBisCO as a reference (PDB 1RCX)28. The carboxysome outer shell segmentation was used as a mask to eliminate the cross-correlation peaks that were present on the shell. Cross-correlation peaks were further selected by imposing an 11 nm inter-peak minimum distance, corresponding to the diameter of RuBisCO. The projection of densities in icosahedral shells onto surfaces was performed with custom Amira modules developed in-house. The numbers of RuBisCOs per face were estimated by measuring their average separation as well as simple counting.
Modeling and simulation
The carboxysome was modeled as an icosahedron of diameter 123 nm, the average experimentally measured carboxysome diameter. RuBisCOs were modeled as spherical particles with fixed diameter. Initially, particles were placed sequentially at random but non-overlapping locations (Random Sequential Addition). This algorithm is unable to create random configurations that are denser than a saturation limit, which is about 38% 36. With RuBisCO spheres of diameter 12.3 nm, this corresponds to about 214 spheres per carboxysome. In order to achieve higher RuBisCO packing densities, particles were placed at random (and potentially overlapping) initial locations within the icosahedron. An energy function meant only to impose volume occlusion was constructed with a single step-function force repelling pairs of overlapping spheres. Thus at each time step, a force was applied to spheres whose centers were closer than a RuBisCO diameter, pushing them directly away from each other. The overlapping spheres in the initial configuration seeded the model with a set of starting velocities and the predicted movements of the spheres were then simulated. The magnitude of the force and time step were set empirically (in abstract units of mass, distance, and time) so that spheres “bounced” off of each other and typically traveled several time steps in one direction before hitting the next particle. The spontaneous emergence of layers was insensitive to perturbations in the force or time step used. The simulation was performed at constant temperature, i.e. the average particle velocity was normalized every 100 steps to reduce the cumulative impact of integration errors. The locations of the particles after 5000 time steps were recorded and used for subsequent analysis. The procedure was repeated 14 times each for packing densities of 124-344 spheres in steps of 20 and RuBisCO diameters of 11, 11.5, 12.3 and 13.3 nm (i.e. 168 sets of coordinates for each RuBisCO diameter).
Supplementary Material
First, a dual-axis image tilt-series is shown of one field of carboxysomes, surrounded by what are believed to be broken carboxysomes. The small dark dots are gold fiducials and the large arc-like boundary is the edge of a hole in the carbon support. Next, the corresponding 3-D reconstruction is shown slice-by-slice. Then a single carboxysome is enlarged and shown in detail, so that individual RuBisCOs, and even their central channels, can be clearly seen. A manual segmentation of the carboxysome shell appears and is then compared to an ideal icosahedron. Finally, the results of the template matching search for RuBisCOs is shown, highlighting their organization into concentric layers.
The icosahedral container is represented by a simple line drawing and the diffusing spheres appear as colored balls whose size changes as they move closer or farther from the viewer. The front of the container and its contents are clipped out of the view for clarity. Thus some balls appear or disappear as they move into or out of that region. Additional balls are added during the simulation, resulting in the gradual development of concentric layers, as seen in the radial density profile at bottom (plotted from the container's center to its edge from left to right).
Acknowledgments
This work was supported by NIH grants P01 GM66521 and R01 AI067548 to GJJ, DOE grant DE-FG02-04ER63785 to GJJ, a Searle Scholar Award to GJJ, and gifts to Caltech from the Ralph M. Parsons Foundation, the Agouron Institute, and the Gordon and Betty Moore Foundation. We thank Dr. Wolfgang Baumeister's group at the Max Planck Institute for Biochemistry for the most current version of the MolMatch template matching software and Dr. Hong Zhuo for discussion of heterogeneity in the sizes of icosahedral viruses, and Dr. Todd Yeates for pointing out an error in our original calculation of T numbers.
Abbreviations
- EM
electron microscopy
- 3-D
three-dimensional
- ECT
electron cryotomography
- RuBisCO
ribulose 1,5-bisphosphate carboxylase/oxygenase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tcherkez GGB, Farquhar GD, Andrews TJ. Despite slow catalysis and confused substrate specificity, all ribulose bisphosphate carboxylases may be nearly perfectly optimized. Proc Nat Acad Sci USA. 2006;103:7246–7251. doi: 10.1073/pnas.0600605103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Badger MR, Price GD, Long BM, Woodger FJ. The environmental plasticity and ecological genomics of the cyanobacterial CO2 concentrating mechanism. J Exp Bot. 2006;57:249–265. doi: 10.1093/jxb/eri286. [DOI] [PubMed] [Google Scholar]
- 3.Price GD, Sültemeyer D, Klughammer B, Ludwig M, Badger MR. The function of the CO2 concentrating mechanism in several cyanobacterial strains: a review of general physiological characteristics, genes, proteins, and recent advances. Can J Bot. 1998;76:973–1002. [Google Scholar]
- 4.Heinhorst S, Cannon G, Shively JM. Carboxysomes and carboxysome-like inclusions. In: Shively JM, editor. Microbiology Monographs, Complex intracellular structures in prokaryotes. Vol. 2. Springer; Berlin, Heidelberg, New York: 2006. pp. 141–166. [Google Scholar]
- 5.Badger MR, Hanson D, Price GD. Evolution and diversity of CO2 concentrating mechanisms in cyanobacteria. Funct Plant Biol. 2002;29:161–173. doi: 10.1071/PP01213. [DOI] [PubMed] [Google Scholar]
- 6.Bobik TA. Polyhedral organelles compartmenting bacterial metabolic processes. Appl Microbiol Biotechnol. 2006;70:517–525. doi: 10.1007/s00253-005-0295-0. [DOI] [PubMed] [Google Scholar]
- 7.Peters KR. Charakterisierung eines phagenähnlichen Partikels aus Zellen von Nitrobacter. Arch Microbiol. 1974;97:129–140. doi: 10.1007/BF00403052. [DOI] [PubMed] [Google Scholar]
- 8.Holthuijzen YA, van Breeman JFL, Konings WN, van Bruggen EFJ. Electron microscopic studies of carboxysomes of Thiobacillus neapolitanus. Arch Microbiol. 1986;144:258–262. [Google Scholar]
- 9.Kerfeld CA, Sawaya MR, Tanaka S, Nguyen CV, Phillips M, Beeby M, Yeates TO. Protein Structures Forming the Shell of Primitive Bacterial Organelles. Science. 2005;309:936–938. doi: 10.1126/science.1113397. [DOI] [PubMed] [Google Scholar]
- 10.So AKC, Espie GS, Williams EB, Shively JM, Heinhorst S, Cannon GC. A Novel Evolutionary Lineage of Carbonic Anhydrase (ε Class) Is a Component of the Carboxysome Shell. J Bacteriol. 2004;186:623–630. doi: 10.1128/JB.186.3.623-630.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sawaya MR, Cannon GC, Heinhorst S, Tanaka S, Williams EB, Yeates TO, Kerfeld CA. The structure of carbonic anhydrase from the carboxysomal shell reveals a distinct subclass with one active site for the price of two. J Biol Chem. 2006;281:7546–7555. doi: 10.1074/jbc.M510464200. [DOI] [PubMed] [Google Scholar]
- 12.Cannon GC, Bradburne CE, Aldrich HC, Baker SH, Heinhorst S, Shively JM. Microcompartments in Prokaryotes: Carboxysomes and Related Polyhedra. Appl Environ Microbiol. 2001;67:5351–5361. doi: 10.1128/AEM.67.12.5351-5361.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Price GD, Howitt SM, Harrison K, Badger MR. Analysis of a genomic DNA region from the cyanobacterium Synechococcus sp. strain PCC7942 involved in carboxysome assembly and function. J Bacteriol. 1993;175:2871–2879. doi: 10.1128/jb.175.10.2871-2879.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ludwig M, Sültemeyer D, Price GD. Isolation of ccmKLMN genes from the marine cyanobacterium Synechococcus sp. PCC7002 and evidence that CcmM is essential for carboxysome assembly. J Phycol. 2000;36:1109–1118. [Google Scholar]
- 15.Heffelfinger GS, Martino A, Gorin A, Xu Y, Rintoul MD, 3rd, Geist A, Al-Hashimi HM, Davidson GS, Faulon JL, Frink LJ, Haaland DM, Hart WE, Jakobsson E, Lane T, Li M, Locascio P, Olken F, Olman V, Palenik B, Plimpton SJ, Roe DC, Samatova NF, Shah M, Shoshoni A, Strauss CE, Thomas EV, Timlin JA, Xu D. Carbon Sequestration in Synechococcus Sp.: From Molecular Machines to Hierarchical Modeling. OMICS. 2002;6:305–330. doi: 10.1089/153623102321112746. [DOI] [PubMed] [Google Scholar]
- 16.Palenik B, Brahamsha B, Larimer FW, Land M, Hauser L, Chain P, Lamerdin J, Regala W, Allen EE, McCarren J, Paulsen I, Dufresne A, Partensky F, Webb EA, Waterbury J. The genome of a motile marine Synechococcus. Nature. 2003;424:1037–1042. doi: 10.1038/nature01943. [DOI] [PubMed] [Google Scholar]
- 17.Gonzales AD, Light YK, Zhang Z, Iqbal T, Lane TW, Martino A. Proteomic analysis of the CO2-concentrating mechanism in the open-ocean cyanobacterium Synechococcus WH8102. Can J Bot. 2005;83:735–745. [Google Scholar]
- 18.Lucic V, Förster F, Baumeister W. Structural studies by electron tomography: From cells to molecules. Annu Rev Biochem. 2005;74:833–865. doi: 10.1146/annurev.biochem.73.011303.074112. [DOI] [PubMed] [Google Scholar]
- 19.Iancu CV, Wright ER, Benjamin J, Tivol WF, Dias DP, Murphy GE, Morrison RC, Heymann JB, Jensen GJ. A “flip-flop” rotation stage for routine dual-axis electron cryotomography. J Struct Biol. 2005;151:288–297. doi: 10.1016/j.jsb.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 20.Schmid MF, Paredes AM, Khant HA, Soyer F, Aldrich HC, Chiu W, Shively JM. Structure of Halothiobacillus neapolitanus Carboxysomes by Cryo-electron Tomography. J Mol Biol. 2006;364:526–535. doi: 10.1016/j.jmb.2006.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shively JM, Ball FL, Kline BW. Electron Microscopy of the Carboxysomes (Polyhedral Bodies) of Thiobacillus neapolitanus. J Bacteriol. 1973;116:1405–1411. doi: 10.1128/jb.116.3.1405-1411.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaneko Y, Danev R, Nagayama K, Nakamoto H. Intact carboxysomes in a cyanobacterial cell visualized by Hilbert differential contrast transmission electron microscopy. J Bacteriol. 2006;188:805–808. doi: 10.1128/JB.188.2.805-808.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caspar DLD, Klug A. Physical principles in the construction of regular viruses. Cold Spring Harbor Symp Quant Biol. 1962;27:1–24. doi: 10.1101/sqb.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- 24.Cannon GC, Shively JM. Characterization of a homogeneous preparation of carboxysomes from Thiobacillus neapolitanus. Arch Microbiol. 1983;134:52–59. [Google Scholar]
- 25.Shively JM, Ball F, Brown DH, Saunders RE. Functional organelles in prokaryotes: polyhedral inclusions (carboxysomes) of Thiobacillus neapolitanus. Science. 1973;182:584–586. doi: 10.1126/science.182.4112.584. [DOI] [PubMed] [Google Scholar]
- 26.Badger MR, Price GD. Carbonic anhydrase activity associated with the cyanobacterium Synechococcus PCC7942. Plant Physiol. 1989;89:51–60. doi: 10.1104/pp.89.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Böhm B, Frangakis AS, Hegerl R, Nickell S, Typke D, Baumeister W. Toward detecting and identifying macromolecules in a cellular context: Template matching applied to electron tomograms. Proc Nat Acad Sci USA. 2000;97:14245–14250. doi: 10.1073/pnas.230282097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taylor TC, Andersson I. The structure of the complex between RuBisCO and its natural substrate ribulose 1,5-bisphosphate. J Mol Biol. 1997;265:432–444. doi: 10.1006/jmbi.1996.0738. [DOI] [PubMed] [Google Scholar]
- 29.Holthuijzen YA, van Breemen JFL, Kuenen JG, Konings WN. Protein composition of the carboxysomes of Thiobacillus neapolitanus. Arch Microbiol. 1986;144:398–404. [Google Scholar]
- 30.Reinhold L, Kosloff R, Kaplan A. A model for inorganic carbon fluxes and photosynthesis in cyanobacterial carboxysomes. Can J Bot. 1991;69:984–988. [Google Scholar]
- 31.Schwarz R, Reihold L, Kaplan A. Low activation state of ribulose-1,5-bisphosphate carboxylase/oxygenase in carboxysome-defective Synechococcus mutants. Plant Physiol. 1995;108:183–190. doi: 10.1104/pp.108.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kofoid E, Rappleye C, Stojiljkovic I, Roth J. The 17-gene ethanolamine (eut) operon of Salmonella typhimurium encodes five homologues of carboxysome shell proteins. J Bacteriol. 1999;181:5317–5329. doi: 10.1128/jb.181.17.5317-5329.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shively JM, Bradburne CE, Aldrich HC, Bobik TA, Mehlman JL, Jin S, Baker SH. Sequence homologs of the carboxysomal polypeptide CsoS1 of the thiobacilli are present in cyanobacteria and enteric bacteria that form carboxysomes-polyhedral bodies. Can J Bot. 1998;76:906–916. [Google Scholar]
- 34.Bobik TA, Havemann GD, Busch RJ, Williams DS, Aldrich HC. The propanediol utilization (pdu) operon of Salmonella enterica serovar Typhimurium LT2 includes genes necessary for formation of polyhedral organelles involved in coenzyme B(12)-dependent 1,2-propanediol degradation. J Bacteriol. 1999;181:5967–5975. doi: 10.1128/jb.181.19.5967-5975.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Böttcher B, Wynne SA, Crowther RA. Determination of the fold of the core protein of hepatitis B virus by electron cryomicroscopy. Nature. 1997;386:88–91. doi: 10.1038/386088a0. [DOI] [PubMed] [Google Scholar]
- 36.Torquato S. Random Heterogeneous Materials: Microstructure and Macroscopic Properties. Springer-Verlag; New York, Berlin, Heidelberg: 2002. [Google Scholar]
- 37.Penrod JT, Roth JR. Conserving a Volatile Metabolite: A Role for Carboxysome-Like Organelles in Salmonella enterica. J Bacteriol. 2006;188:2865–2874. doi: 10.1128/JB.188.8.2865-2874.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Orús MI, Rodríguez ML, Martínez F, Marco E. Biogenesis and Ultrastructure of Carboxysomes from Wild Type and Mutants of Synechococcus sp. Strain PCC 7942. Plant Physiol. 1995;107:1159–1166. doi: 10.1104/pp.107.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng QS, Braunfeld MB, Sedat JW, Agard DA. An improved strategy for automated electron microscopic tomography. J Struct Biol. 2004;147:91–101. doi: 10.1016/j.jsb.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 40.Mastronarde DN. Dual-axis tomography: an approach with alignment methods that preserve resolution. J Struct Biol. 1997;120:343–352. doi: 10.1006/jsbi.1997.3919. [DOI] [PubMed] [Google Scholar]
- 41.Frangakis AS, Hegerl R. Noise reduction in electron tomographic reconstructions using nonlinear anisotropic diffusion. J Struct Biol. 2001;135:239–250. doi: 10.1006/jsbi.2001.4406. [DOI] [PubMed] [Google Scholar]
- 42.Heymann JB, Belnap DM. Bsoft: Image and molecular modeling in electron microscopy. J Struct Biol. 2007;157:3–18. doi: 10.1016/j.jsb.2006.06.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
First, a dual-axis image tilt-series is shown of one field of carboxysomes, surrounded by what are believed to be broken carboxysomes. The small dark dots are gold fiducials and the large arc-like boundary is the edge of a hole in the carbon support. Next, the corresponding 3-D reconstruction is shown slice-by-slice. Then a single carboxysome is enlarged and shown in detail, so that individual RuBisCOs, and even their central channels, can be clearly seen. A manual segmentation of the carboxysome shell appears and is then compared to an ideal icosahedron. Finally, the results of the template matching search for RuBisCOs is shown, highlighting their organization into concentric layers.
The icosahedral container is represented by a simple line drawing and the diffusing spheres appear as colored balls whose size changes as they move closer or farther from the viewer. The front of the container and its contents are clipped out of the view for clarity. Thus some balls appear or disappear as they move into or out of that region. Additional balls are added during the simulation, resulting in the gradual development of concentric layers, as seen in the radial density profile at bottom (plotted from the container's center to its edge from left to right).


