Abstract
Evidence has been accumulating for a role for inflammation in the development of Alzheimer’s disease (AD), a progressive neurodegenerative disorder causing a common form of dementia in the elderly. C1q, part of the initiation component of the classical complement pathway (CCP), is associated with β-sheet, fibrillar amyloid plaques in AD brain. In vitro, β-amyloid peptide in fibrillar β-sheet conformation (fAβ) can activate CCP via interaction of specific negatively charged amino acids of the β-amyloid fibril with human C1q. Previous results using peptide inhibitors led to the hypothesis that a highly positively charged domain consisting of three arginine residues, such as that present in the N terminal collagen-like region of the human C1q A chain, may be critical for the activation event. However, mouse C1q A chain lacks two of three arginines in the corresponding C1q A chain collagen-like region. To test the hypothesis that this divergent activation domain results in a weaker C’ activation and thus may contribute to the lower neuronal loss observed in transgenic mouse models of AD, a partially humanized C1q A chain knock-in mouse was generated. The mouse C1q A chain gene was modified by homologous recombination to replace 4 residues in the 13–20 amino acid region to mimic the corresponding sequence from human A chain. No significant differences in the expression of C1q were found in sera from mice homozygous for the humanized C1q A chain compared to littermate wild type mice. Two distinct C1 activation assays demonstrated that activation by fAβ was not significantly different in the homozygous humanized C1q A chain mice. Activation of C1 by DNA, previously hypothesized to interact with this C1q A chain arginine-rich sequence was also not significantly different in the knock in mouse. Molecular modeling based on the published crystal structure of human C1q B chain globular head and a β-sheet model for fibrillar amyloid suggests an alternative arginine ladder in the globular head domain may provide the functional C1 activating interaction domains. The humanized C1q mouse generated here should provide a better animal model for assessing the mechanisms of C1 activation and the contribution of C1q to human health and disease.
Introduction
The complement system, which consists of more than 30 soluble and membrane bound proteins, represents a critical component of innate immunity. Complement can be activated via three different pathways: classical, lectin, and alternative, which differ by the nature of the recognition interaction. C1q is a subcomponent of C1, the first component of the classical complement pathway (CCP). Each C1q molecule is comprised of six copies of three polypeptide chains: the A-chain (223 residues), the B-chain (226 residues) and the C-chain (217 residues) (Reid, 1979). Each of these chains has a short three to nine residue amino-terminal region containing a cysteine residue, followed by a collagen-like region (CLR) of 81 residues and a carboxy-terminal globular head module of 135 residues (Sellar et al., 1991). Traditionally, the CCP is known to be activated via the binding of C1q to IgG or IgM-containing immune complexes (IC) which, in turn, leads to the activation of two serine protease proenzymes C1r and C1s. This activated C1 complex (C1q + 2C1r + 2C1s) enzymatically cleaves C4 and C2, thereby triggering the downstream cascade of the CCP. This activation leads to the release of signaling peptides (C3a and C5a) and the final formation of the terminal membrane attack complex (MAC) (Arlaud et al., 2002) providing major protective mechanisms against pathogens. Several molecules including beta-amyloid (Aβ), DNA, C-reactive protein, serum amyloid P, fibromodulin and specific strains of E. coli have been reported to bind to C1q and activate CCP through an antibody independent pathway (Jiang et al., 1992a;Sjoberg et al., 2005;Volanakis, 1982;Bristow and Boackle, 1986;Rogers et al., 1992;Tenner et al., 1984). Many of these antibody-independent activators are multimeric and some contain a high density of negative charges. The polyanionic nature of C1q interacting molecules suggests that C1q may recognize a specific pattern and /or spacing of charged residues or groups (Kishore et al., 2004). Studies using a peptide containing residues 14–26 of human C1q A chain showed that this highly positively charged peptide specifically inhibited C1q binding to Aβ, DNA, C-reactive protein, and serum amyloid P (Jiang et al., 1992a;Jiang et al., 1992b;Jiang et al., 1994;Ying et al., 1993).
β-amyloid protein has been implicated in the pathogenesis of Alzheimer’s disease (AD), a progressive neurodegenerative disorder that results in the loss of cognitive ability in the elderly. Genetic mutations causing over expression of the amyloid precursor protein (APP) lead to early or familial AD. Aβ1–42 peptide has been shown to be neurotoxic in vitro and it has the ability to trigger a local inflammatory response via activation of complement. In vitro, Aβ in β-sheet conformation can activate the CCP via binding of specifically spaced negative charges of the β-amyloid fibril to human C1q (Velazquez et al., 1997). Previous results using peptide inhibitors suggested that the domain 14–26 consisting of three arginine residues in the collagen-like region of the C1q A chain may be critical for the activation event (Jiang et al., 1994;Chen et al., 1996). However, studies assessing fAβ binding to isolated proteolytic fragments of C1q (Tacnet-Delorme et al., 2001;Jiang et al., 1994) and more recently to expressed individual domains of the C1q molecule (Kishore et al., 2003), also demonstrate fAβ binding to the C1q head regions.
Many murine transgenic models have been developed to study AD. However, none of these models fully recapitulates the entire neuropathological spectrum of the human disease (Wong et al., 2002). For example, in AD mouse models over expressing human amyloid precursor protein, despite abundant Aβ deposits comparable to that seen in AD, these mice do not demonstrate neurofibrillary tangles. In addition, neuronal loss is usually seen only after extensive deposition of Aβ in many of the mouse models (Calhoun et al., 1998;Hsiao et al., 1996), generally exceeding the levels observed in severe AD cases, although clearly behavioral deficits can be detected at earlier times. One possible explanation for the less extensive neuronal loss in these mice is that complement activation by fibrillar amyloid may be less robust, thus generating less inflammation. Murine C1q lacks two of the three arginine residues (R16 and R20) previously proposed as an interaction site with the negative charges (Asp7 and Glu11) of fibrillar Aβ (Jiang et al., 1994;Chen et al., 1996;Webster et al., 1999). Here, we describe the development and characterization of a partially humanized C1q A chain knock-in mouse which contains all three arginine residues as present in the human C1q A chain (R16, R19, and R20). Data show that the “humanized” C1q is equivalently expressed in sera and is equally efficient as native murine C1q in its ability to form functional C1 and to mediate activation of C1 by immune complexes. However, the ability of C1 containing humanized C1q to be activated by fibrillar Aβ does not differ from the C1 containing the wild type murine C1q. Molecular modeling of the crystal structure of the C1q globular domain (Gaboriaud et al., 2003) and fibrillar Aβ suggests that an alternative arginine ladder in the C1q B chain globular head domain, with similar geometric spacing as in the A chain region targeted here, may provide functional C1 activating interaction domains for fibrillar Aβ and other negatively charged antibody independent activators of the classical complement pathway.
Materials and Methods
1. Reagents
Purified human complement proteins C1, C4, C2 and C1-Inhibitor were obtained from Advanced Research Technologies (now Complement Technologies, Tyler, TX). Human C1q, Clr and Cls were purified according to published methods (Ziccardi and Cooper, 1976;Tenner et al., 1981;Valet and Cooper, 1974). Aβ1–42, obtained from Dr. C. Glabe (UC, Irvine), was synthesized as previously described (Burdick et al., 1992). For individual experiments, lyophilized aliquots of Aβ1–42 were solubilized at 4mg/ml in double distilled deionized water and then diluted with 2x buffer such that the final solution was 2mg/ml peptide in 0.016 M Tris-buffered saline (TBS, pH 7.4). Solutions were incubated at 4°C for 24hr, yielding β-pleated sheet peptide, as indicated by circular dichroism (as previously described by (Cribbs et al., 1997)). This fibrillar Aβ exhibits dose-dependent consumption of C4 (Velazquez et al., 1997). Mouse IgG was purified from normal mouse serum by octanoic acid/ammonium sulfate precipitation according to published methods (McKinney and Parkinson, 1987). IgG preparations were diluted to 10 mg/ml and heated at 63°C for 15 min to induce aggregation. Salmon sperm DNA was obtained from Invitrogen (Carlsbad, CA).
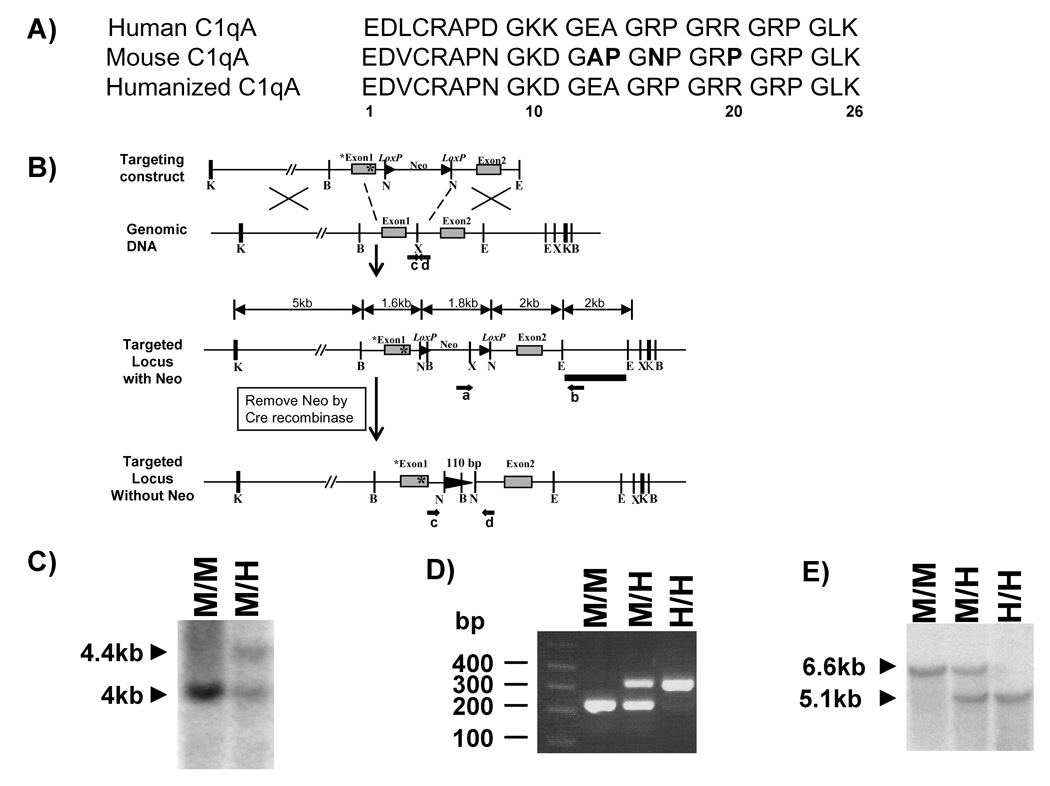
2. Generation of the humanized C1q A chain mice
The mouse C1qA chain gene was isolated from a 129/Sv genomic library. The original genomic sequence flanking mouse C1qA chain gene (11309bp Kpn I fragment) in pBluescript II KS (+) was a kind gift from Dr. Marina Botto. Mutations were introduced using Promega GeneEditor Mutagenesis system to replace four amino acids of mouse sequence (A13, P14, N16, P20) with human sequence (E13, A14, R16, R20) (Figure 1A). The primer used in mutagenesis was 5’-GCACCCAACGGGAAGGATGGAGAAGCAGGAAGACCTGGCCGCCGAGGGAGGCCGGGTCTCAAAGG-3’. Sequences carried the introduced mutations were selected by the loss of the “Sma I” restriction site and later were confirmed by sequencing (Automated DNA Sequencing Facility at UCI). The targeting vector was constructed by inserting a Neo cassette gene flanked by two lox P sites into a Xba I site located in the intron region between exon 1 and 2 of the C1q A chain gene via a Not I polylinker (5’-CTAGTGCGGCCGCA-3’) (Figure 1B). The vector was linearized with Asc I and introduced into the embryonic cell line ES14 by electroporation. ES cells that survived G418 selection were screened for correct targeting by the detection of a PCR fragment about 2.5kb in length using primer a (within Neo cassette): 5’-GGATGCGGTGGGCTCTATGGCTTCTG-3’ and primer b (outside the targeting construct): 5’-GTGTGATTGTGTTTGAGCCTGGGATTGTGG-3’. This correct targeting was later confirmed by Southern blot analysis of Xba I digested mouse tail genomic DNA (Figure 1C). The probe used for the Southern blot hybridization is an 1840 base pair EcoRI-EcoRI genomic sequence that is down stream of the C1q A chain exon2 gene and external to the targeting construct (Figure 1B). The detected band from Southern blot is ~4 kb for the wild type DNA and ~4.4kb for the mutant DNA. Of a total 183 ES14 colonies screened, 5 of them were positive for correct recombination of targeting vector. Subsequently, cells from 2 of the targeted ES clones were directly injected into C57BL/6J blastocysts to create chimeric mice, which carry the Neo cassette in the C1q A chain gene. These same two clones were separately transiently transfected with a construct encoding Cre recombinase to delete the Neo cassette via lox P sites before injecting into the blastocysts to create chimeras. Chimeric mice were crossed to C57BL/6J females (Jackson laboratory, Bar Harbor, ME) and tested for germline transmission of the partially humanized C1q A chain by PCR. The heterozygous mice with Neo cassette were further bred with mice of the EIIA-Cre stain (Lakso et al., 1996) obtained from the Jackson laboratory to remove the Neo cassette. With both approaches, the removal of Neo cassette was detected by PCR for a band with an extra 110bp in length compared to the wild type band using primer c: 5’-CCTGCTTTGACATTCCTCTTTCCCCTTTCT-3’ and d: 5’-CTCATAACTCCTCCCCTCTCGTCCTCCTCA-3’ (Figure 1B,D). The presence of Neo cassette led to no detection of the mutant band because of the maximum length limitation of the designed PCR setting. This PCR set up was also used for mouse genotyping as this extra length is always linked to the knocked-in mutations specifically detected by forward primer e (5’-CAACGGGAAGGATGGAGAAGCAGGAAGA-3’) and backward primer f (5’-GGAAAGAGGAATGTCAAAGCAGGAAAGAGA-3’) (Figure 1D and data not shown). The deletion of Neo cassette was also confirmed by Southern blot analysis of Bam HI digested mouse tail genomic DNA using the same 1840 base pair EcoRI-EcoRI genomic sequence as probe (Figure 1E). The wild type band is about 6.6 kb and the mutant band is about 5.1kb. The humanized C1q A chain mice used for the experiments were on the (129/Sv × C57BL/6J) hybrid genetic background backcrossed to C57BL/6J 2–7 generations. Sera from mice generated with either of the 2 targeted ES clones (D9 or D11) and with the CRE deletion performed in vitro or via breeding with the CRE deleter gave equivalent results in all assays.
Figure 1. Generation of knock-in mice containing partially humanized C1q A chain.
A) Comparison of the N-terminal first 26 amino acids between human and mouse C1q A chain sequences. Four amino acids (in bold) within the mouse sequence were converted to the corresponding human sequence and subsequently introduced into the targeting construct as indicated by * in Figure 1B.
B) Structure of the targeting vector containing the mutated C1q A chain gene, the wild-type mouse C1q A chain allele, the mutated allele after homologous recombination (targeted locus with Neo), and the mutated allele after deletion of the neomycin resistance cassette (Neo) by Cre recombinase (targeted locus without Neo). Exons are depicted by gray boxes. Restriction sites are as follows: Bam HI (B), Eco RI (E), Kpn I (K), Not I (N), and Xba I (X). The position of a probe (black bar) used for Southern blot analysis and the primers a and b (black arrows) used for PCR screening the targeted ES clones are shown.
C) Southern blot analysis of genomic DNA extracted from the wild type ES cells and the targeted ES cells containing Neo cassette. The DNA was digested with Xba I and subjected to hybridization with the probe depicted in B.
D) PCR genotyping of genomic DNA extracted from the tail of mice without Neo cassette using primer c and d as indicated in B.
E) Southern blot analysis of genomic DNA extracted from the tail of mice of the indicated genotypes after removal of Neo cassette. The DNA was digested with Bam HI and subjected to hybridization with the probe depicted in B.
3. Mouse sera
Normal mouse sera were obtained from 8–12 week old mice by cardiac puncture. Briefly, mice were anesthetized with Nembutal (Western Medical Supply, Arcadia, CA); blood was slowly withdrawn from the left ventricle with a 23 gauge needle and transferred to a glass tube pre-chilled on ice. The blood was clotted on ice for 60 minutes before spinning at 2000g for 10 minutes at 4°C. Serum was removed from the top layer of the sample. All sera and complement proteins were immediately distributed into small aliquots on ice and stored at −70°C until use.
4. Mouse C1q ELISA
Immulon 2-HB flat bottom removal well strips (DYNEX Technologies Inc., Chantilly, VA) were pre-coated with a polyclonal rabbit anti-mouse C1q (1151) antibody (Huang et al., 1999) at 5µg/ml in PBS buffer overnight at 4°C. The wells were then washed in PBS-0.05% Tween 20 and blocked with PBS-3% milk at 37°C for 1 hour. Serum samples were diluted in PBS-1% milk, added to the wells and incubated at room temperature (RT) for 2 hours. The mouse anti-mouse C1q detection antibody Jl-1 (Trouw et al., 2004), a kind gift from Dr. M. Daha (Leiden, The Netherlands), was added at 0.5µg/ml to the wells and incubated at RT for 1 hour. [While the 1151 polyclonal and J1-1 monoclonal antibody to mouse C1q recognized the humanized C1q molecule, the anti mouse C1q monoclonal 7H8 (Cell Sciences, Canton, MA) was reactive with wild type but not the humanized C1q.] Mouse IgG was detected using HRP-donkey anti-mouse IgG (Jackson laboratory) at a 1:1000 dilution in PBS-1% milk followed by OPD substrate (Sigma Co., St Louis, MO). The change in absorbance at 405nm between 0 and 45 min was read with a Thermomax microplate reader (Molecular Devices, Sunnyvale, CA).
5. Hemolytic assays
Sheep erythrocytes (Colorado Serum Co., Denver, CO) were sensitized using rabbit anti-sheep erythrocyte IgG (Colorado Serum Co.) for the C1q hemolytic assay and C4-sensitized sheep erythrocytes (EAC4b) prepared for C1 hemolytic assay as previously described (Whaley and North, 1997). Two microliters of mouse sera diluted in GVB (4.8mM sodium barbital, 0.1% gelatin, 142mM NaCl, 1mM MgCl2 and 0.15mM CaCl2 at pH 7.3) was combined with human C1q deficient serum reagent (300µl) as previously described (Tenner et al., 1981). Eighty microliters of EA at 5 × 108EA/ml was added and incubated at 37°C for 30min. Lysis was halted by the addition of 1.6 ml ice-cold GVB. Solutions were then centrifuged at 800g at 4°C for 3min and the optical density at 412 nm of the supernatants were obtained as an index of released hemoglobin. The activity of mouse C1q, expressed as the Z-value (the dilution at which 63% of the target cells are lysed) was determined by linear regression analysis of hemolytic data derived from the means of duplicate serial dilutions of each sample as previously described (Tenner et al., 1981).
A C1 hemolytic assay previously described (Webster et al., 1999) was employed to compare the ability of the altered C1q molecule to complex with mouse native C1r and C1s to form a functional C1 capable of activating the classical complement pathway. Briefly, complement activators were diluted in DGVB (2 mM sodium barbital, pH 7.2, 57mM NaCl, 166mM dextrose, 1 mM MgCl2, 0.15 mM CaCl2, and 0.1% gelatin) and combined with an equal volume of mouse serum diluted 2.5-fold with DGVB. Reaction mixtures were incubated at 30°C for 30 min. C1 consumption was assayed by addition of 100µl of samples serially diluted in DGVB to 50µl EAC4b targets (5 × 107/mL final concentration) and incubated at 30°C for 10 min. After centrifugation at 800g for 3 min (RT), buffer was removed and targets were resuspended in an equal volume of DGVB at RT. Purified C2 was added to a final concentration of 200–1000 U/ml (dependent on assayed specific hemolytic activity) and incubated at 30°C for 4 min. Normal guinea pig serum (Cedarlane, Ontario, Canada) diluted 1:50 in GVBE (GVB containing 20mM EDTA) was added, and solutions were incubated at 30°C for 30 min. After centrifugation at 800g at 4°C for 3 min, the optical density at 412 nm of the supernatants were obtained as an index of released hemoglobin. The consumption of C1 (a reflection of C1 activation) was determined by linear regression analysis of the means of duplicate serial dilutions of each sample as previously described (Whaley and North, 1997).
6. Mouse C1q purification
Mouse C1q was purified from mouse serum by the method for human C1q purification published previously (Tenner et al., 1981) with some modifications. Pooled mouse serum from humanized A-chain knock-in homologous mice (H/H) or the wild type (M/M) was adjusted to contain 25mM EDTA, and directly applied to a column of BioRex 70 (Bio-Rad, Hercules, CA) (1ml resin/2.5 ml serum) equilibrated with 50mM sodium phosphate buffer pH7.3, 82mM NaCl, 20mM EDTA. C1q was eluted by application of a salt gradient to 300mM NaCl. Fractions positive for C1q by hemolytic titer were pooled. In order to further purify the material, pooled fractions were dialyzed into TBS (50mM Tris pH 7.3, 150mM NaCl) and applied to a Heparin-agarose column (Sigma) (1mL resin/10mL start material) equilibrated with TBS. C1q was eluted by application of a salt gradient to 400mM NaCl. Fractions positive for C1q by hemolytic titer were pooled, concentrated by ultrafiltration and adjusted to 150mM NaCl using 50mM Tris pH 7.3.
7. C1 activation assay using purified C1 components
C1 activation was measured using purified C1 components (C1q, C1r, and C1s) and C1 inhibitor as described previously (Ziccardi and Cooper, 1976) with minor modifications. Briefly, C1 was reconstituted by incubation of human or mouse Clq, and human Clr and Cls in a molar ratio of approximately 1:2:2 in the presence of 1.5mM calcium and 120–140 µg/ml C1 Inhibitor on ice for 5 minutes. Buffer control or various complement activators were then added and incubated at 30°C for 30min. The reaction was terminated by adding an equal volume of 2x reducing SDS loading buffer and boiling. The samples were then subjected to 8% SDS-polyacrylamide gel electrophoresis and transferred to PVDF. Activation of C1 is determined by Western blot analysis using a polyclonal antibody against human C1s (Quidel, San Diego, CA). The extent of activation was quantified as the percent of C1s cleaved (59,000 Mr) under each condition. Bands were analyzed using densitometry software (Scion Image).
8. Circular dichroism
Circular dichroism was recorded at ambient temperature using a Jasco J720 spectropolarimeter. Data were collected at 0.5nm intervals, with 4 scans averaged, but not smoothed. A cell of 0.5mm path length was used. Spectra were obtained with amyloid peptides in Tris buffered saline, pH 7.4 at 50µM.
9. Molecular modeling
Coordinates of the human C1q B chain (PDB ID 1PK6) globular domain were accessed as published by Gaboriaud and colleagues (Gaboriaud et al., 2003). The previously generated model for the β amyloid peptide N-terminus (Webster et al., 1999) was used in conjunction with the molecular docking program, Chimera (http://www.cgl.ucsf.edu/chimera/), to construct a potential interaction between these two proteins.
Results
1. Generation of humanized C1q A chain knock in mice
A peptide consisting of residues 14–26 of the human C1q A chain has been shown to inhibit fibrillar Aβ induced CCP activation (Jiang et al., 1994;Chen et al., 1996). Molecular modeling of this corresponding region of the C1q A chain demonstrated appropriate spacing for interaction of R16 and R19 or R19 and R20 with aspartate and glutamate of Aβ when constrained to β-sheet conformation (Velazquez et al., 1997). The corresponding region of the mouse C1q A chain lacks two of the three arginines (R16 and R20) (Figure 1A). Since intact functional C1q had not been recombinantly expressed, an in vivo model was generated to test if insertion of these arginines into the mouse C1q molecule would enhance fibrillar Aβ induced CCP activation, providing a more human-like mouse model for AD. The mouse C1q A chain gene was modified to replace residues in the 13–20 amino acid region to mimic the corresponding sequence from human A chain by homologous recombination (Figure 1B). Chimeric mice were generated by blastocyst injection of targeted ES cells containing the Neo cassette or targeted EC cells with the Neo cassette removed by transient transfection and expression of Cre recombinase. Knocked in mutations from two separately derived ES clones (with or without Neo cassette) were transmitted to the germ line successfully. All groups of mice were viable, with normal growth and reproduction rates, and exhibited no overt gross phenotypes, although the inclusion of Neo cassette within the intron between exon 1 and 2 resulted in a complete deficiency of C1q in serum (data not shown). The mice containing the Neo cassette flanked by lox P sites were further bred with C57BL/6 transgenic mice expressing Cre recombinase protein in early embryos under the control of the adenovirus EIIA early promoter to remove the Neo cassette except for a 110 base pair insertion in the intron region between exon 1 and 2 of mouse C1q A gene. Homozygous humanized C1q A chain mice (H/H) were generated subsequently by interbreeding of the heterozygous (M/H) mice. Correct homologous recombination in the knock in mice was confirmed by Southern blot analysis of tail genomic DNA digested with Bam HI (Figure 1E).
2. Protein and function levels of the humanized C1q in serum are equal to wild type mouse C1q
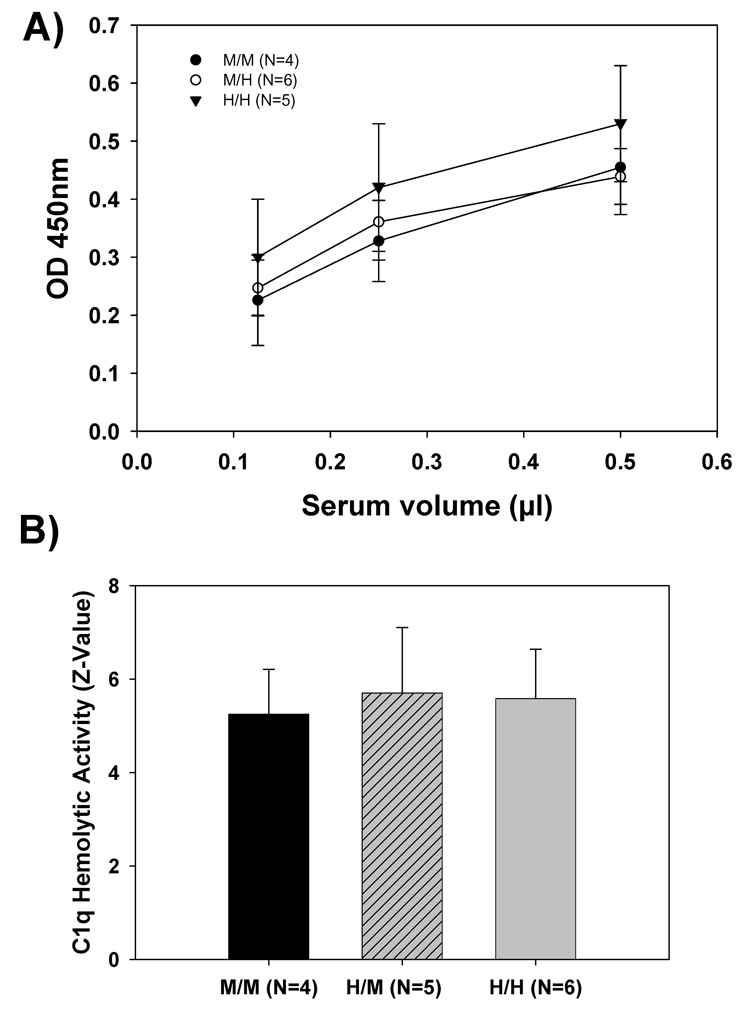
Protein concentration of wild type and humanized mouse C1q protein in serum was first assessed using a semiquantitative Western blot analysis using a rabbit polyclonal antibody against mouse C1q (#1151). A comparable level of mouse C1q was detected in humanized mice (M/H or H/H) as in wild type (M/M) mice (data not shown). For more quantitative analysis, an ELISA assay for mouse C1q that also detected the partially humanized mouse C1q molecule was developed. Figure 2A shows that no significant difference in the serum concentration of C1q was seen in heterozygous (M/H), homozygous (H/H) C1q A mice, and wild type C1q A animals (M/M).
Figure 2. Mouse serum C1q protein levels and hemolytic activities in wild type mice and humanized C1q A chain knock in mice.
A) Serum from three genotypes of mice, wild type (M/M), heterozygous (M/H), and humanized (H/H) was diluted 200, 400, and 800 fold with PBS-1% milk and the C1q protein level measured using a mouse C1q sandwich ELISA as described in Materials and Methods. The data presented are the mean ± SD of average values for 4–6 animals per genotype (sample assayed in duplicate).
(B) Human C1q-deficient serum was reconstituted with dilutions of sera from M/M, M/H, and H/H mice (all bled at 8–10 weeks of age) as a source of C1q. Hemolytic activity was measured as described in Materials and Methods and expressed as Z-values. Data presented are the mean ± SD of activity from 4, 5 and 6 mice respectively, from three independent experiments.
The ability of the altered C1q to support C1 activation was first measured using a C1q hemolytic assay. In this assay, serially diluted mouse sera, as a source for C1q, is added to a C1q-depleted human serum. This reconstituted serum is then tested for its ability to activate the CCP resulting in lysis of antibody coated erythrocytes. The concentration of functional C1q in sera from both homozygous (H/H) and heterozygous (M/H) mice were compared with that from the wild type (M/M) mice by determining the Z-value (serum dilution at which 63% of target cells is lysed) of each mouse serum sample tested. The mean Z-value of the three mouse genotypes demonstrated no significant difference among the groups (Figure 2B).
3. Consumption of C1 hemolytic activity of the humanized C1q A chain mice
The C1r2C1s2 proenzyme complex of C1 associates with the collagen tail region of C1q (Reid et al., 1977). To assure equivalent formation of the mouse C1 complex with C1q containing the humanized A chain, C1 baseline activity was measured and compared between the wild type (M/M) and the homozygous humanized C1q A chain (H/H) mice. C1 baseline activities varied among individual animals. However, no significant difference was found between these two mouse genotypes, suggesting that the presence of the knocked-in human sequence in the mouse C1q A chain did not interfere with the association and activation properties of the C1qC1r2C1s2 proenzyme complex when EAIgG was the activator (Figure 3A). Upon addition of mouse aggregated IgG, a soluble activator also known to bind to the globular head region of the C1q molecule and to activate murine C1 (Webster et al., 1999), the percent of C1 consumed did not significantly differ between the wild type mice (M/M) [55%±3% (N=4)] and the humanized C1q A chain mice (H/H) [59%±6% (N=5)], again indicating that the knocked in mutations did not grossly alter the ability of the C1q globular head to bind to its activator and induce activation of the proenzymes C1r and C1s (Figure 3B).
Figure 3. C1 activity and mouse aggregated IgG induced C1 consumption in wild type mice and humanized C1q A chain knock in mice.
A) C1 activity was measured by its ability to cause the lysis of antibody coated rabbit erythrocytes as described in Materials and Methods. Each mouse serum was assayed in duplicate and with multiple dilutions to obtain a z-value for each animal per experiment. Values for each genotype from 4 independent experiments were averaged. Data are the mean ± SD of the averaged z-values from a total of 4 wild type (M/M) and 5 homozygous knock in (H/H) mice. No significant difference has been found between the groups.
B) Activation of mouse C1 by aggregated mouse IgG (15µg/ml) was determined as described in Materials and Methods and expressed as percentage of C1 consumed. Data are the mean ± SD from a total of 4 (M/M) and 5 (H/H) mice tested in one experiment.
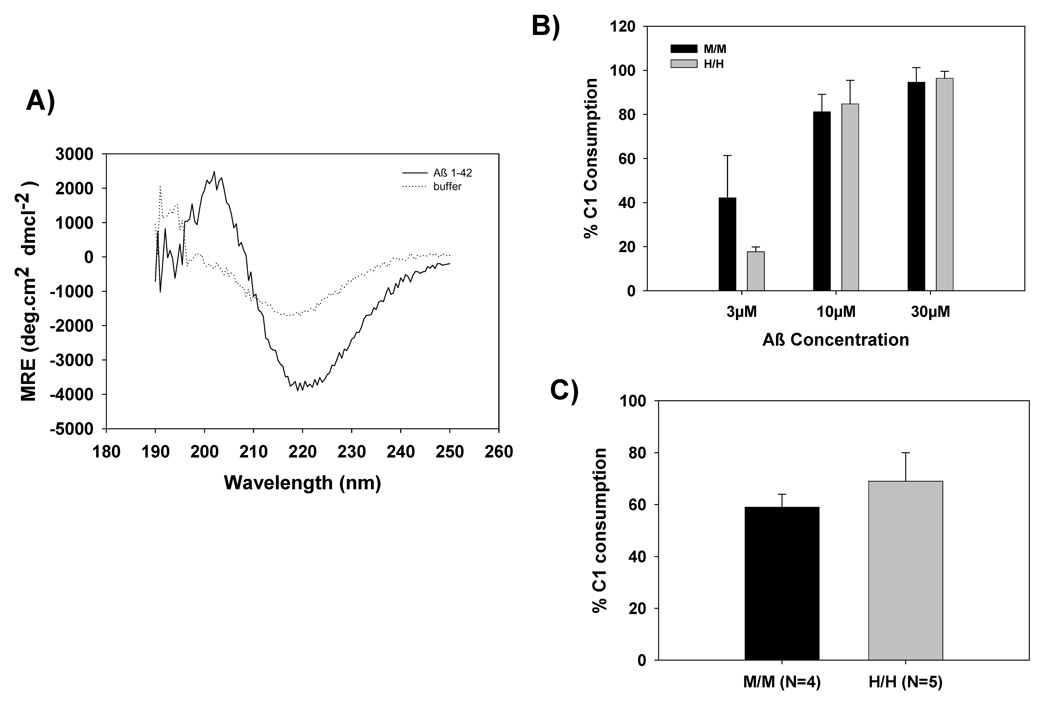
Previous studies using a peptide containing residues 14–26 within the collagen tail of the human C1q A chain as an inhibitor suggested that this region was involved in binding to fibrillar Aβ, DNA, C-reactive protein, and subsequently activating the CCP via a non-traditional antibody-independent fashion (Jiang et al., 1992a;Jiang et al., 1994;Chen et al., 1996;Jiang et al., 1992b). Therefore, C1 consumption by fibrillar Aβ was compared using sera from littermate wild type and homozygous humanized C1q A mice. Preliminary experiments using various doses of fibrillar Aβ demonstrated a concentration dependent activation of C1 in murine sera from both genotypes (data not shown) with no activation resulting from incubation with monomeric Aβ (consistent with previous reports). The β-sheet structure of the fibrillar Aβ was confirmed by circular dichroism, with a mean residue ellipticity of >3000 at 218nm (Figure 4A). Fibrillar Aβ at concentrations of 3, 10 and 30µM were selected to compare M/M and H/H mice. As depicted in Figure 4B, the humanized C1q A mice (H/H) did not show an enhanced C1 consumption compared to that of the wild type mice (M/M). DNA has been suggested to be another activator of the CCP which activates C1 via binding the collagen like domain of C1q. Based on preliminary dose response experiments, 0.6 µg/ml DNA resulted in submaximal activation of C1 and thus was chosen as an optimal concentration of DNA to compare C1 activation. No significant difference in DNA-dependent C1 consumption was detected between the wild type sera and sera from mice expressing humanized C1q [59%±5% (N=4) with the wild type mice (M/M) vs. 69%±11% (N=5) with the humanized C1q A mice (H/H)] (Figure 4C).
Figure 4. Fibrillar Aβ and DNA induce C1 consumption in wild type mice and humanized C1qA chain knock in mice.
A) Degree of β-sheet conformation was determined using Circular Dichroism (CD). Mean residue ellipticity (MRE) of fibrillar Aβ at 50µM vs. buffer was determined between wavelengths of 190nm–250nm.
B) Activation of mouse C1 by fibrillar Aβ 1–42 (3, 10, and 30µM) as determined by percentage of C1 consumption. Data are averages from three experiments using sibling M/M and H/H sera.
C) Salmon sperm DNA (0.6µg/ml) was used as an activator of mouse C1 and activation was determined by percentage of C1 consumption. Data are the mean ± SD of 4 M/M or 5 H/H mice tested in one experiment.
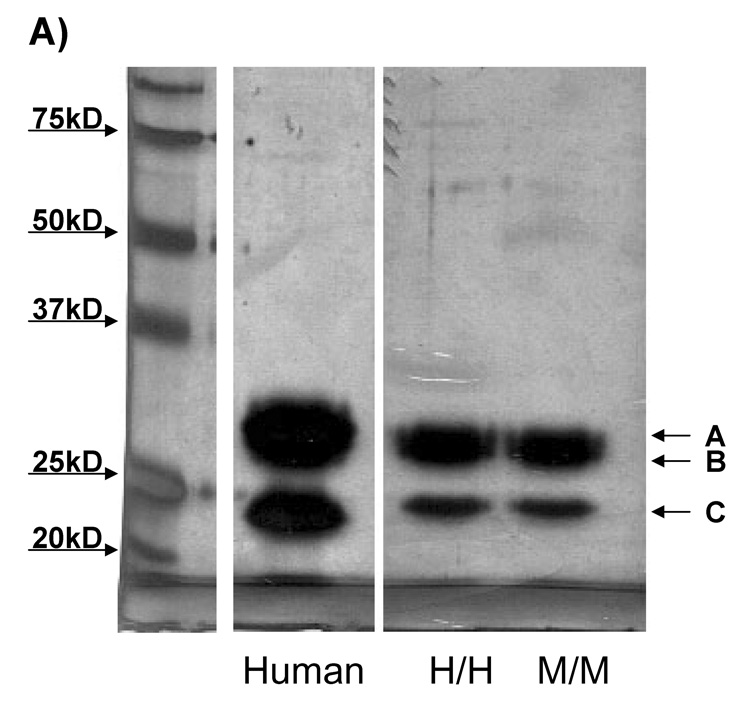
4. Activation of in vitro reconstituted C1 using C1q purified from wild type and humanized chain mice
C1 assembled from the purified components C1q, C1r and C1s has been shown to have activation properties similar to serum C1. Use of purified components eliminates confounding contributions by serum components. Thus, C1q purified from murine sera from wild type or the C1q A chain knock in (Figure 5A) mouse was added to purified C1r and C1s and the ability of the resultant C1 to be activated by fibrillar Aβ was assessed by detection of the 59,000 Mr cleavage product of C1s indicative of activated C1 (Cooper and Ziccardi, 1977). No significant difference was detected in the activation of C1 containing murine C1q or humanized C1q by fibrillar amyloid or aggregated IgG (Figures 5B and 5C). Importantly, C1q purified from mouse serum gives comparable C1 activation by Aβ and/or aggregated IgG as does C1q purified from human serum when compared in the same experiment and with different concentrations of activator (data not shown).
Figure 5. Purified C1q from wild type and C1q A chain knock in mice are equally effective in activation of C1 by fibrillar Aβ.
A) Mouse wild type or humanized C1q was purified from murine serum as described in Materials and Methods. Purity was analyzed by 10% SDS PAGE gel under reducing conditions followed by silver staining. 2.5 µg of protein was loaded per lane, and compared to purified human C1q. Values of molecular weight standards (far left lane) are indicated by the arrows to the left of the gel. A, B, and C indicate the C1q A, B, and C chain, respectively.
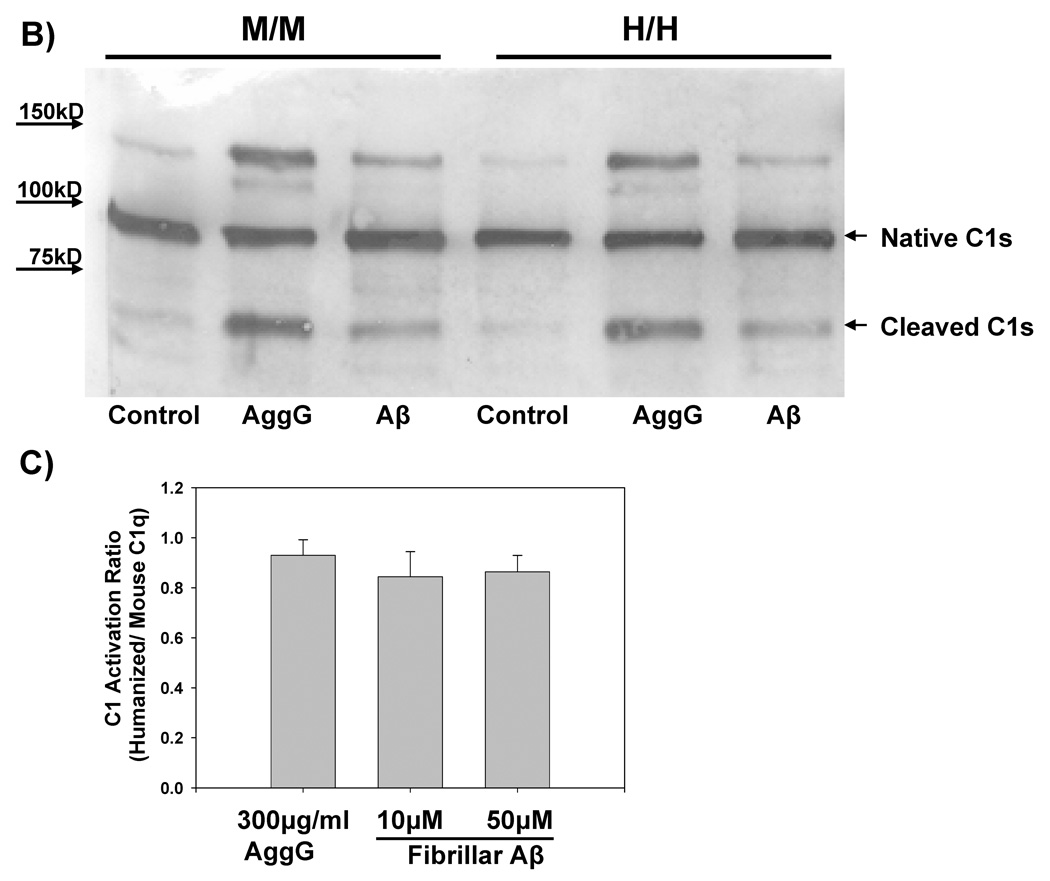
B) C1 activation was measured using cleaved C1s as an activation indicator. Purified mouse C1q or humanized C1q at 50µg/ml was combined with purified human C1r and C1s in 1:2:2 molar ratios to form a functional C1 complex. After addition of C1 Inhibitor, C1 was combined with fAβ (5µM) or aggregated IgG (AgG, 75µg/ml) or buffer as a control (C) and incubated for 30 min at 30°C. Reactions were subjected to SDS-PAGE under reducing conditions, followed by Western blot and detection of unactivated Cls (~90k Mr) and activated Cls (59k Mr) with anti C1s as described in Materials and Methods.
C) C1 reconstituted as described in B containing mouse or humanized C1q was combined with 10µM or 50µM fAβ or aggregated IgG (AgG, 300µg/ml) along with C1 inhibitor and incubated as in B. Percent activation of C1s was determined by comparing percent cleaved C1s to total native C1s by densitometry using Scion Image Pro. C1 activation ratio is the value obtained by dividing the percent cleaved C1s obtained in complex with humanized C1q by the percent cleaved C1s obtained in complex with wild type mouse C1q. Results are the mean ± SD from 4 experiments.
5. Molecular modeling of C1q head-fibrillar Aβ interaction
A previous molecular model of residues 14–24 of human C1q mapped onto the collagen triple helix backbone demonstrated that the spacing of arginines 16 and 19 as well as 19 and 20 was compatible with electrostatic interactions with negative charges Asp7 and Glu11 in the β-amyloid peptide when confined to an antiparallel β-sheet conformation (Velazquez et al., 1997). Using the coordinates for the human C1q globular domains originally published by Gaboriaud, and colleagues, the 3-dimensional spacing of Arg 101, 114 and 129 in the B chain is nearly identical to that postulated for Arg16, 19 and 20 in the human C1q A chain, even though the residues on the B chain are distant in primary linear sequence. Using the Chimera program, molecular docking of this region with the N-terminal region of Aβ in the β-sheet conformation, supports the postulated electrostatic interaction proposed as an alternative site (Tenner, 2001), and supported by the published data of both the Grenoble group and the group in Oxford (Tacnet-Delorme et al., 2001;Kishore et al., 2003;Kojouharova et al., 2003). Theses arginine residues are strictly conserved in the murine C1q B chain also.
Discussion
A novel murine model has been constructed by introducing a coding region into the C1q A chain gene that changes 4 amino acids in the collagen-like region of the molecule such that it is identical to the human sequence previously proposed to be a site for antibody independent activation of C1 (Jiang et al., 1992a;Chen et al., 1996;Jiang et al., 1994). The level of expression of this humanized C1q molecule was not altered nor was the ability of the molecule to complex with C1r and C1s to form a functional C1 macromolecule compromised either in serum or after isolation and reconstitution with purified C1r and C1s. Both in serum and in purified forms the “humanized” murine C1 displayed comparable ability to be activated by immune complexes. Contrary to the initial hypothesis, there was no significant difference in the ability of fAβ to activate C1 containing humanized mouse C1q compared to wild type mouse C1q.
Significant correlative data suggest that inflammation may contribute to the pathogenesis and neuronal dysfunction in Alzheimer's disease (Patel et al., 2005;Rozemuller et al., 2005;Akiyama et al., 2000). Fibrillar Aβ interaction with C1q has been shown to activate the classical complement pathway, which leads to the generation of activation fragments capable of recruiting and activating glial cells. C1q deficient mouse models transgenic for the human APP protein have the age specific characteristic amyloid pathology of the corresponding C1q sufficient animals, but show significantly lower microglial and astrocyte recruitment, and attenuated loss of markers of neuronal integrity (Fonseca et al., 2004). However, these animal models, and those used by many other investigators, are not a perfect mimic of the human disease. A difference in the arginine rich region of the C1q collagen-like region was hypothesized as contributing to a decrease complement activation and subsequent lower level of inflammation and neuronal loss seen in the mouse models relative to humans with AD. While it remains to be seen if the presence of this humanized C1q in an animal model of AD will more closely mimic the human disease, the in vitro results presented here suggest that the arginine ladder in this region of the C1q molecule does not play a prominent role in fAβ- dependent CCP activation.
Interestingly, the crystal structure of the C1q head regions solved by Gaboriaud and colleagues (Gaboriaud et al., 2003) demonstrate a region within the B chain of the C1q globular domain that has similar spatial arrangement of arginines matching that of the arginine ladder in the collagen-like region of the A chain. Molecular modeling of this region (Figure 6) was compatible with an interaction with the Asp7 and Glu11 of the amyloid peptide, residues that have been repeatedly shown to be critical for the activation of C1 by fibrillar amyloid peptide (Velazquez et al., 1997;Tacnet-Delorme et al., 2001). Clearly, the peptide inhibitors (based on the A chain sequence) used in earlier studies would inhibit any interaction site sharing the appropriate spacing of positive charges as the result of protein folding, and thus, such inhibitor studies have limitations in deducing definitive conclusions about interaction sites. Thus, we propose a model in which fAβ can activate C1 through interactions with the arginine rich B chain globular region (Arg101, Arg114 and Arg129). This model is consistent with the in vitro data of others implicating the globular domain as the site of the amyloid C1 activation interaction (Tacnet-Delorme et al., 2001;Kishore et al., 2003). Nevertheless, future studies will be needed to validate whether this “B chain” site plays the major role in fAβ mediated C1 activation in vivo.
Figure 6. Proposed model of arginine residues in the C1q B chain globular domain interacting with fibrillar Aβ.
The C1q B chain model was generated directly from the coordinates of the crystal structure of C1q globular head (Gaboriaud et al., 2003). Only the arginine side chains (R101, R114, R129) of the C1q B-chain and the aspartic and glutamic acids of the amyloid peptide are shown as space filling models. One possible interaction with Asp7 and Glu11 of the Aβ fibril constrained in an antiparallel β-sheet conformation is illustrated.
Several other studies have suggested that some nonimmunoglobulin activators of C1 activate C1 via interactions within the C1q collagen-like stalk region. A more recent report by Tissot et al. clearly indicates that DNA can bind to the collagen domain of C1q (Tissot et al., 2003). Here the DNA-dependent activation of normal mouse C1q lacking two of the three positive charges in this region was not significantly different than the humanized C1q. This result may reflect the contribution of amino acids 76–92 in the A chain collagen like domain (also positively charged) which have been previously shown to bind SAP and CRP (Ying et al., 1993;Jiang et al., 1992b;Jiang et al., 1992a), however, an alternative mechanism is the ability of the B chain “arginine ladder” present in each of the 6 globular heads of C1q to constrain the C1q molecule inducing the activation of C1r and subsequently C1s, the proteases that propagate the subsequent complement cascade. One other non-immunoglobulin activator of C1q, the extracellular matrix protein fibromodulin, was shown to bind C1q via the head regions by both electron microscopy and binding to the isolated globular head domain of C1q (Sjoberg et al., 2005). Further definition of the amino acid residues involved in that interaction remain to be determined. Since the relative contribution of interactions to the “activation of C1” can not be definitively assessed using proteolytic fragments of C1q, this genetically altered C1q is and will be useful in testing if the arginine-rich sequence in the collagen-like region of the C1q A chain contributes significantly to the activation of C1 by any of the antibody independent activators (Sim et al., 2007).
Animal models are an essential component of assessing the relevance of molecular and biochemical pathways in disease processes. The development of more human-like murine models will facilitate translation of mechanisms defined in animal models to human processes as well as facilitate testing of potential therapies that may promote normal developmental processes or block disease inducing events (Sarvari et al., 2003;Craft et al., 2005;Stevens et al., 2007).
Acknowledgements
This work is supported by NIH NS 35144, AG 00538-25 and NIH Training Grants AG00096-21 (M.L.) and NS-007444 (R.R.A.). We thank Dr. Michele Musacchio and the University of California, Irvine, Transgenic Mouse Facility for ESC targeting and generation of the C1q A chain knock in mouse. The authors also thank Drs. Saskia Milton and Charles Glabe (UC, Irvine, CA) for providing the synthetic human Aβ peptide, Dr. Marina Botto (Imperial College, London) for the plasmid containing the murine C1q A chain sequence, Dr. Mo Daha (University Hospital Leiden, Nl) for the gift of JL-1 anti C1q antibody, Dr. Thomas Poulos (UC, Irvine, CA) for the molecular modeling in Figure 6.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Akiyama H, Barger S, Barnum S, Bradt B, Bauer J, Cole GM, Cooper NR, Eikelenboom P, Emmerling M, Fiebich BL, Finch CE, Frautschy S, Griffin WS, Hampel H, Hull M, Landreth G, Lue L, Mrak R, Mackenzie IR, McGeer PL, O'Banion MK, Pachter J, Pasinetti G, Plata-Salaman C, Rogers J, Rydel R, Shen Y, Streit W, Strohmeyer R, Tooyoma I, Van Muiswinkel FL, Veerhuis R, Walker D, Webster S, Wegrzyniak B, Wenk G, Wyss-Coray T. Inflammation and Alzheimer's disease. Neurobiol. Aging. 2000;21:383–421. doi: 10.1016/s0197-4580(00)00124-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arlaud GJ, Gaboriaud C, Thielens NM, Rossi V. Structural biology of C1. Biochem. Soc. Trans. 2002;30:1001–1006. doi: 10.1042/bst0301001. [DOI] [PubMed] [Google Scholar]
- 3.Bristow CL, Boackle R. Evidence for the binding of human serum amyloid P component to C1q and Fab. Mol. Immunol. 1986;23:1045. doi: 10.1016/0161-5890(86)90003-9. [DOI] [PubMed] [Google Scholar]
- 4.Burdick D, Soreghan B, Kwon M, Kosmoski J, Knauer M, Henschen A, Yates J, Cotman C, Glabe C. Assembly and aggregation properties of synthetic Alzheimer's A4/beta amyloid peptide analogs. J. Biol. Chem. 1992;267:546–554. [PubMed] [Google Scholar]
- 5.Calhoun ME, Wiederhold KH, Abramowski D, Phinney AL, Probst A, Sturchler-Pierrat C, Staufenbiel M, Sommer B, Jucker M. Neuron loss in APP transgenic mice [letter] Nature. 1998;395:755–756. doi: 10.1038/27351. [DOI] [PubMed] [Google Scholar]
- 6.Chen S, Frederickson RC, Brunden KR. Neuroglial-mediated immunoinflammatory responses in Alzheimer's disease: complement activation and therapeutic approaches. Neurobiol. Aging. 1996;17:781–787. doi: 10.1016/0197-4580(96)00103-0. [DOI] [PubMed] [Google Scholar]
- 7.Cooper NR, Ziccardi RJ. Reconstitution of C1 in Native, Proenzyme Form and Its Use in a Quantitative C1 Activation Test. J. Immunol. 1977;119:1664–1667. [PubMed] [Google Scholar]
- 8.Craft JM, Watterson DM, Van Eldik LJ. Neuroinflammation: a potential therapeutic target. Expert. Opin. Ther. Targets. 2005;9:887–900. doi: 10.1517/14728222.9.5.887. [DOI] [PubMed] [Google Scholar]
- 9.Cribbs DH, Velazquez P, Soreghan B, Glabe CG, Tenner AJ. Complement activation by cross-linked truncated and chimeric full-length β-amyloid. NeuroReport. 1997;8:3457–3462. doi: 10.1097/00001756-199711100-00009. [DOI] [PubMed] [Google Scholar]
- 10.Fonseca MI, Zhou J, Botto M, Tenner AJ. Absence of C1q leads to less neuropathology in transgenic mouse models of Alzheimer's disease. J Neurosci. 2004;24:6457–6465. doi: 10.1523/JNEUROSCI.0901-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gaboriaud C, Juanhuix J, Gruez A, Lacroix M, Darnault C, Pignol D, Verger D, Fontecilla-Camps JC, Arlaud GJ. The crystal structure of the globular head of complement protein C1q provides a basis for its versatile recognition properties. J Biol. Chem. 2003;278:46974–46982. doi: 10.1074/jbc.M307764200. [DOI] [PubMed] [Google Scholar]
- 12.Hsiao KK, Chapman P, Nilsen S, Eckman C, Harigaya Y, Younkin S, Yang F, Cole G. Correlative memory deficits, Aβ elevations, and amyloid plaques in transgenic mice. Science. 1996;274:99–102. doi: 10.1126/science.274.5284.99. [DOI] [PubMed] [Google Scholar]
- 13.Huang J, Kim LJ, Mealey R, Marsh HC, Jr, Zhang Y, Tenner AJ, Connolly ES, Jr, Pinsky DJ. Neuronal protection in stroke by an sLex-glycosylated complement inhibitory protein. Science. 1999;285:595–599. doi: 10.1126/science.285.5427.595. [DOI] [PubMed] [Google Scholar]
- 14.Jiang H, Burdick D, Glabe CG, Cotman CW, Tenner AJ. β-amyloid activates complement by binding to a specific region of the collagen-like domain of the C1q A chain. J. Immunol. 1994;152:5050–5059. [PubMed] [Google Scholar]
- 15.Jiang H, Cooper B, Robey FA, Gewurz H. DNA binds and activates complement via a specific sequence on the human C1q A chain. J. Biol. Chem. 1992a;267:25597–25601. [PubMed] [Google Scholar]
- 16.Jiang H, Robey FA, Gewurz H. Localization of sites through which CRP binds and activates complement to residues 14–26 and 76–92 of the human C1q A chain. J. Exp. Med. 1992b;175:1373–1379. doi: 10.1084/jem.175.5.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kishore U, Gaboriaud C, Waters P, Shrive AK, Greenhough TJ, Reid KB, Sim RB, Arlaud GJ. C1q and tumor necrosis factor superfamily: modularity and versatility. Trends Immunol. 2004;25:551–561. doi: 10.1016/j.it.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 18.Kishore U, Gupta SK, Perdikoulis MV, Kojouharova MS, Urban BC, Reid KB. Modular organization of the carboxyl-terminal, globular head region of human C1q A, B, and C chains. J Immunol. 2003;171:812–820. doi: 10.4049/jimmunol.171.2.812. [DOI] [PubMed] [Google Scholar]
- 19.Kojouharova MS, Tsacheva IG, Tchorbadjieva MI, Reid KB, Kishore U. Localization of ligand-binding sites on human C1q globular head region using recombinant globular head fragments and single-chain antibodies. Biochim. Biophys. Acta. 2003;1652:64–74. doi: 10.1016/j.bbapap.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 20.Lakso M, Pichel JG, Gorman JR, Sauer B, Okamoto Y, Lee E, Alt FW, Westphal H. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc. Natl. Acad. Sci. U. S. A. 1996;93:5860–5865. doi: 10.1073/pnas.93.12.5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKinney MM, Parkinson A. A simple, non-chromatographic procedure to purify immunoglobulins from serum and ascites fluid. J. Immunol. Methods. 1987;96:271–278. doi: 10.1016/0022-1759(87)90324-3. [DOI] [PubMed] [Google Scholar]
- 22.Patel NS, Paris D, Mathura V, Quadros AN, Crawford FC, Mullan MJ. Inflammatory cytokine levels correlate with amyloid load in transgenic mouse models of Alzheimer's disease. J. Neuroinflammation. 2005;2:9. doi: 10.1186/1742-2094-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reid KBM. Complete amino acid sequences of the three collagen-like regions present in subcomponent C1q of the first component of human complement. Biochem. J. 1979;179:367–371. doi: 10.1042/bj1790367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reid KBM, Sim RB, Faiers AP. Inhibition of the Reconstitution of the Haemolytic Activity of the First Component of Human Complement by a Pepsin-Derived Fragment of Subcomponent C1q. Biochem. J. 1977;161:239–245. doi: 10.1042/bj1610239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rogers J, Schultz J, Brachova L, Lue L-F, Webster S, Bradt B, Cooper NR, Moss DE. Complement activation and β-amyloid-mediated neurotoxicity in Alzheimer's disease. Res. Immunol. 1992;143:624–630. doi: 10.1016/0923-2494(92)80046-n. [DOI] [PubMed] [Google Scholar]
- 26.Rozemuller AJ, van Gool WA, Eikelenboom P. The neuroinflammatory response in plaques and amyloid angiopathy in Alzheimer's disease: therapeutic implications. Curr. Drug Targets. CNS. Neurol. Disord. 2005;4:223–233. doi: 10.2174/1568007054038229. [DOI] [PubMed] [Google Scholar]
- 27.Sarvari M, Vago I, Weber CS, Nagy J, Gal P, Mak M, Kosa JP, Zavodszky P, Pazmany T. Inhibition of C1q-beta-amyloid binding protects hippocampal cells against complement mediated toxicity. J Neuroimmunol. 2003;137:12–18. doi: 10.1016/s0165-5728(03)00040-7. [DOI] [PubMed] [Google Scholar]
- 28.Sellar GC, Blake DJ, Reid KBM. Characterization and organization of the genes encoding the A-, B-, and C-chains of human complement subcomponent C1q. Biochem. J. 1991;274:481–490. doi: 10.1042/bj2740481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sim RB, Kishore U, Villiers CL, Marche PN, Mitchell DA. C1q binding and complement activation by prions and amyloids. Immunobiology. 2007;212:355–362. doi: 10.1016/j.imbio.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 30.Sjoberg A, Onnerfjord P, Morgelin M, Heinegard D, Blom AM. The extracellular matrix and inflammation. Fibromodulin activates the classical pathway of complement by directly binding C1q. J. Biol. Chem. 2005 doi: 10.1074/jbc.M504828200. [DOI] [PubMed] [Google Scholar]
- 31.Stevens B, Allen NJ, Vazquez LE, Howell GR, Christopherson KS, Nouri N, Micheva KD, Mehalow AK, Huberman AD, Stafford B, Sher A, Litke AM, Lambris JD, Smith SJ, John SW, Barres BA. The classical complement cascade mediates CNS synapse elimination. Cell. 2007;131:1164–1178. doi: 10.1016/j.cell.2007.10.036. [DOI] [PubMed] [Google Scholar]
- 32.Tacnet-Delorme P, Chevallier S, Arlaud GJ. Beta-amyloid fibrils activate the C1 complex of complement under physiological conditions: evidence for a binding site for A beta on the C1q globular regions. J. Immunol. 2001;167:6374–6381. doi: 10.4049/jimmunol.167.11.6374. [DOI] [PubMed] [Google Scholar]
- 33.Tenner AJ. Complement in Alzheimer's disease: opportunities for modulating protective and pathogenic events. Neurobiol. Aging. 2001;22:849–861. doi: 10.1016/s0197-4580(01)00301-3. [DOI] [PubMed] [Google Scholar]
- 34.Tenner AJ, Lesavre PH, Cooper NR. Purification and radiolabeling of human C1q. J. Immunol. 1981;127:648–653. [PubMed] [Google Scholar]
- 35.Tenner AJ, Ziccardi RJ, Cooper NR. Antibody-independent C1 activation by E. coli. J. Immunol. 1984;133:886–891. [PubMed] [Google Scholar]
- 36.Tissot B, Daniel R, Place C. Interaction of the C1 complex of complement with sulfated polysaccharide and DNA probed by single molecule fluorescence microscopy. Eur. J Biochem. 2003;270:4714–4720. doi: 10.1046/j.1432-1033.2003.03870.x. [DOI] [PubMed] [Google Scholar]
- 37.Trouw LA, Groeneveld TW, Seelen MA, Duijs JM, Bajema IM, Prins FA, Kishore U, Salant DJ, Verbeek JS, van Kooten C, Daha MR. Anti-C1q autoantibodies deposit in glomeruli but are only pathogenic in combination with glomerular C1q-containing immune complexes. J Clin. Invest. 2004;114:679–688. doi: 10.1172/JCI21075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Valet G, Cooper NR. Isolation and characterization of the proenzyme form of the C1r subunit of the first complement component. J. Immunol. 1974;112:1667–1673. [PubMed] [Google Scholar]
- 39.Velazquez P, Cribbs DH, Poulos TL, Tenner AJ. Aspartate residue 7 in amyloid β-protein is critical for classical complement pathway activation: Implications for Alzheimer's disease pathogenesis. Nature Med. 1997;3:77–79. doi: 10.1038/nm0197-77. [DOI] [PubMed] [Google Scholar]
- 40.Volanakis JE. Complement activation by C-reactive protein complexes. Ann. N. Y. Acad. Sci. 1982;389:235–250. doi: 10.1111/j.1749-6632.1982.tb22140.x. [DOI] [PubMed] [Google Scholar]
- 41.Webster SD, Tenner AJ, Poulos TL, Cribbs DH. Mouse C1q A-chain sequence alters beta-amyloid-induced complement activation. Neurobiol. Aging. 1999;20:297–304. doi: 10.1016/s0197-4580(99)00020-2. [DOI] [PubMed] [Google Scholar]
- 42.Whaley K, North J. Haemolytic assays for whole complement activity and individual components. In: Dodds AW, Sim RB, editors. Complement: A practical approach. Oxford, UK: Oxford University Press; 1997. pp. 19–47. [Google Scholar]
- 43.Wong PC, Cai H, Borchelt DR, Price DL. Genetically engineered mouse models of neurodegenerative diseases. Nat. Neurosci. 2002;5:633–639. doi: 10.1038/nn0702-633. [DOI] [PubMed] [Google Scholar]
- 44.Ying S-C, Gewurz AT, Jiang H, Gewurz H. Human serum amyloid P component oligomers bind and activate the classical complement pathway via residues 14–26 and 76–92 of the A chain collagen-like region of C1q. J. Immunol. 1993;150:169–176. [PubMed] [Google Scholar]
- 45.Ziccardi RJ, Cooper NR. Physicochemical and Functional Characterization of the C1r Subunit of the First Complement Component. J. Immunol. 1976;116:496–503. [PubMed] [Google Scholar]