Abstract
Systemic lupus erythematosus (SLE) is an autoimmune multisystem inflammatory disease characterized by the production of pathogenic autoantibodies. Previous genetic studies have suggested associations with HLA Class II alleles, complement gene deficiencies, and Fc receptor polymorphisms; however, it is likely that other genes contribute to SLE susceptibility and pathogenesis. Here, we report the results of a genome-wide microsatellite marker screen in 105 SLE sib-pair families. By using multipoint nonparametric methods, the strongest evidence for linkage was found near the HLA locus (6p11-p21) [D6S257, logarithm of odds (lod) = 3.90, P = 0.000011] and at three additional regions: 16q13 (D16S415, lod = 3.64, P = 0.000022), 14q21–23 (D14S276, lod = 2.81, P = 0.00016), and 20p12 (D20S186, lod = 2.62, P = 0.00025). Another nine regions (1p36, 1p13, 1q42, 2p15, 2q21–33, 3cent-q11, 4q28, 11p15, and 15q26) were identified with lod scores ≥1.00. These data support the hypothesis that multiple genes, including one in the HLA region, influence susceptibility to human SLE.
Systemic lupus erythematosus (SLE) is an idiopathic autoimmune disease in which self-reactive autoantibodies cause disease either by directly binding to self-antigens (e.g., antiphospholipid syndrome, immune cytopenias) or following the deposition of antibody–antigen immune complexes in blood vessels leading to vasculitis and tissue damage (1). The estimated prevalence of SLE in the U.S. is ≈45/100,000, with the peak incidence in women ages 20–40 (2). The familial clustering of SLE, together with higher rates of concordance in monozygotic compared with dizygotic twins, suggests an important role for genetic predisposition to systemic lupus (3–6).
Although the clinical manifestations of systemic lupus are heterogeneous, SLE can be distinguished from a number of related syndromes (drug-induced lupus, discoid lupus, subacute cutaneous lupus) and other diseases by careful clinical examination and laboratory testing. Diagnostic criteria for SLE have been developed by the American College of Rheumatology that are both highly specific and sensitive (7, 8). Because of the likelihood that multiple genes are responsible for susceptibility to SLE, and with the sibling relative risk ratio (λs) (9) for SLE ≈10–20 (10), we elected to use a sibling (sib)-pair family approach to map potential genetic loci.
MATERIALS AND METHODS
Families.
The recruitment of families for this study has been described (11). All patients met the 1997 American College of Rheumatology revised criteria for the diagnosis of SLE (7, 8).
Samples and Genotyping.
Genomic DNA was isolated from peripheral blood cells by using standard conditions. Genotyping was performed by using an Applied Biosystems fluorescently labeled human linkage mapping set (version 1.0, panels 1–18; version 2.0, panels 19–27). PCR (32 cycles) was performed on an ABI 877 Catalyst robotic workstation [5 μl reactions; 8 ng of genomic DNA, 2.5 mM MgCl2, 0.2 mM dNTPs (Pharmacia), 1.65 pmol of 5′ and 3′ primers, 0.2 units of Amplitaq Gold DNA polymerase (Perkin–Elmer) in 1× PCR Buffer II (Perkin–Elmer)].
Pooled amplification products were electrophoresed through 5% polyacrylamide gels (FMC) for 2 hr at 3,000 V by using an ABI 377 DNA Sequencer. Semiautomated fragment sizing was performed by using genescan 2.1 software (ABI) followed by allele calling with genotyper 2.0 software (ABI). Each genotype was reviewed manually by two members of the research team to confirm the accuracy of allele calling. Poor performing markers (a total of 17) were excluded from the analysis. The overall data drop-out rate for the ≈127,500 genotypes analyzed was <1.5%.
Data Analysis.
Nonparametric multipoint analysis was performed with genehunter plus (12), a modified version of genehunter (13), by using the “all” statistic. Allele frequencies for the parameter file were generated from the unaffected parental genotypes for the cohort. Marker map positions were obtained from the sex-averaged maps compiled by J. Weber (Marshfield Clinic, Marshfield, WI) (www.marshmed.org/genetics/).
RESULTS AND DISCUSSION
Families were recruited by advertising for “Sisters with Lupus” (11), and, as a result, our sample was highly enriched for female patients (female:male 219:1), compared with the population estimate for lupus of ≈90% female. Selected clinical and demographic data of the SLE patients in this study are shown in Table 1.
Table 1.
Clinical and demographic features of 220 SLE patients
| Age at diagnosis, ±SD | 31 ± 11 years |
| Duration of disease, ±SD | 12 ± 7 years |
| Sex, female:male | 219:1 |
| Ethnicity, % | |
| Caucasian | 80 |
| Hispanic | 8 |
| African-American | 5 |
| Asian | 3 |
| Mixed heritage | 4 |
| Laboratory clinical features*, % | |
| ANA-positive† | 98 |
| Anti-dsDNA-positive | 46 |
| Arthritis | 85 |
| Skin involvement | 91 |
| Pleuritis | 53 |
| Hematologic | 47 |
| Renal disease | 30 |
| CNS lupus | 22 |
| Pericarditis | 18 |
| More than one miscarriage | 13 |
| Medication history, % | |
| Corticosteroids | 77 |
| Antimalarials | 65 |
| Cytotoxic drugs | 28 |
| Intravenous steroids | 19 |
Data represents the percentage of SLE patients having the indicated laboratory/clinical features and medication history at any time during the course of their disease.
Four individuals negative for ANA tested positive for anti-dsDNA antibodies and otherwise fulfilled criteria. These data are comparable with those described in other large series of SLE patients (31). ANA, antinuclear antibodies; dsDNA, double-stranded DNA; CNS, central nervous system.
The sample studied in this report comprises the first 105 SLE sib-pair families collected (102 families with 2 SLE sibs, 2 families with 3 SLE sibs, and 1 family with 4 SLE sibs). All available parents (including three affected) were collected, and, in the absence of parents, an unaffected sibling was sampled to assist in reconstruction of parental genotypes. In three families an additional affected-first degree relative was included in the analysis. The final study cohort totaled 220 SLE patients and 155 unaffected parents or sibs. The racial composition of the 105 families studied was as follows: 84 Caucasian, 8 Hispanic, 6 African-American, 3 Asian, and 4 of mixed heritage.
Family members were genotyped with 341 highly polymorphic markers across the 22 autosomes, at an average intermarker distance of 9.7 centimorgans (cM). Mendelian inheritance was confirmed in all families, and nonparametric multipoint linkage analysis was performed on the collected marker data by using genehunter (13), with extensions to include calculation of appropriate logarithm of odds (lod) scores and support intervals (12).
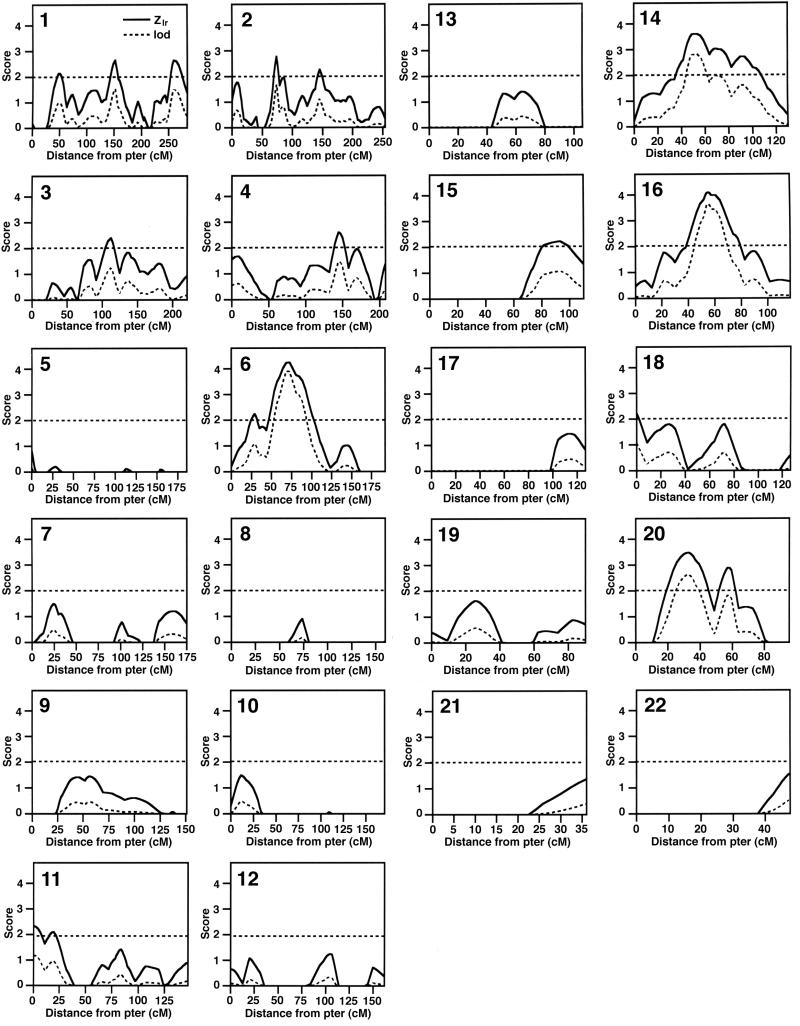
Results of the multipoint analysis are shown in Fig. 1, with Zlr, the test statistic generated by genehunter plus, and the calculated lod scores for each marker plotted for each chromosome. We considered any region with a Zlr ≥ 2.0 as “potentially interesting,” and applied the criteria of Lander and Kruglyak (14) to further define regions of significant (Z ≥ 4.1, lod ≥ 3.6, P ≤ 0.00002) or suggestive (Z ≥ 3.2, lod ≥ 2.2, P ≤ 0.0007) linkage. Overall, 25 of the 341 markers tested (7%) gave Zlr scores >2.1 and lod scores >1.00. Strikingly, 16 of the 25 positive markers clustered into 4 distinct genomic intervals (Fig. 1 and Table 2). Two of these intervals contained a marker that met criteria for significant linkage: 6p11-p21, mapping near the HLA region (D6S257, Zlr = 4.24, lod = 3.90, P = 0.000011), and 16q13 (D16S415, Zlr = 4.09, lod = 3.64, P = 0.000022). Two other regions fulfilled criteria for suggestive linkage: 14q21-q23 (D14S276, Zlr = 3.60, lod = 2.81, P = 0.00016) and 20p12 (D20S186, Zlr = 3.48, lod = 2.62, P = 0.00025). Nine additional chromosomal regions were identified by single markers with Zlr scores >2.1 and accompanying lod scores ≥1.00 (Table 2). In all of the most suggestive areas, these multipoint results were confirmed by single point analyses (data not shown).
Figure 1.
Nonparametric multipoint linkage analysis in 105 SLE sib-pair families. Shown are the analyses of chromosomes 1–22, plotting both Zlr (solid line) and lod (dotted line) scores. Subjects were genotyped with 341 polymorphic markers at an average interval of 9.7 cM (longest, 26.4 cM; shortest, 0.5 cM). The average value of the information statistic, which is a measure of the percent inheritance information extracted at each marker (13), was 0.78, indicating that, overall, the markers were highly informative.
Table 2.
Summary of potential SLE susceptibility loci
| Zlr range | Interval* | Zlr | P value | lod score† | Positive markers | Map position, cM‡ | IC§ |
|---|---|---|---|---|---|---|---|
| >4.0 | 6p11-p21 | 3.69 | 0.00011 | 2.96 | D6S426 | 60.44 | 0.77 |
| 4.24 | 0.000011 | 3.90 | D6S257 | 79.92 | 0.79 | ||
| 2.19 | 0.014 | 1.04 | D6S462 | 99.01 | 0.63 | ||
| 16q13 | 3.81 | 0.000069 | 3.15 | D16S3136 | 62.11 | 0.77 | |
| 4.09 | 0.000022 | 3.64 | D16S415 | 67.62 | 0.73 | ||
| 2.42 | 0.0077 | 1.27 | D16S503 | 83.55 | 0.69 | ||
| 3.0–4.0 | 14q21-23 | 2.94 | 0.0016 | 1.87 | D14S288 | 47.51 | 0.70 |
| 3.60 | 0.00016 | 2.81 | D14S276 | 56.36 | 0.77 | ||
| 2.80 | 0.0025 | 1.71 | D14S63 | 69.18 | 0.81 | ||
| 2.99 | 0.0014 | 1.94 | D14S258 | 76.28 | 0.78 | ||
| 2.54 | 0.0055 | 1.40 | D14S74 | 87.36 | 0.50 | ||
| 2.29 | 0.011 | 1.14 | D14S280 | 105.00 | 0.67 | ||
| 20p12 | 3.48 | 0.00025 | 2.62 | D20A186 | 32.30 | 0.80 | |
| 3.21 | 0.00066 | 2.23 | D20S112 | 39.25 | 0.80 | ||
| 2.38 | 0.0087 | 1.23 | D20S107 | 55.74 | 0.87 | ||
| 2.77 | 0.0028 | 1.67 | D20S119 | 61.77 | 0.78 | ||
| 2.0–3.0 | 2p15 | 2.78 | 0.0027 | 1.68 | D2S337 | 80.69 | 0.85 |
| 1p13 | 2.66 | 0.0039 | 1.53 | D1S252 | 150.27 | 0.76 | |
| 1q42 | 2.64 | 0.0041 | 1.51 | D1S235 | 254.64 | 0.67 | |
| 4q28 | 2.60 | 0.0047 | 1.46 | D4S424 | 144.46 | 0.74 | |
| 3cent-q11 | 2.39 | 0.0084 | 1.24 | D3S1271 | 117.76 | 0.57 | |
| 11p15 | 2.34 | 0.0096 | 1.19 | D11S922 | 2.11 | 0.86 | |
| 2q21–33 | 2.27 | 0.012 | 1.12 | D2S151 | 152.04 | 0.76 | |
| 15q26 | 2.22 | 0.013 | 1.07 | D15S127 | 86.81 | 0.86 | |
| 1p36 | 2.15 | 0.016 | 1.00 | D1S234 | 55.10 | 0.74 |
Chromosome locations were determined from the maps available at www.gdb.org/and http://cedar.genetics.soton.ac.uk/pub/.
lod = Zlr2/2ln10 (12).
Map positions obtained from the sex-averaged maps compiled by J. Weber (www.marshmed.org/genetics/).
Information content (13) for the marker designated in each interval.
Because of sample size considerations, we stratified the data by ethnic group only for the 84 Caucasian families in the overall sample of 105 families. The results of this analysis are shown in Table 3. Lod scores dropped in 10 of the top 13 identified potential susceptibility intervals when non-Caucasian families were eliminated from the analysis, suggesting that families of all ethnic groups contributed to the evidence for genetic linkage in these regions. In contrast, three intervals (4q28, 11p15, and 15q26) were characterized by an improvement in lod scores when only the Caucasian families were considered.
Table 3.
Comparison of lod scores obtained for Caucasian SLE sib-pair families versus entire family cohort
| Loci | Caucasian families* | All families† | Difference |
|---|---|---|---|
| lod score‡ | |||
| 6p11-p21 | 2.50 | 3.90 | −1.40 |
| 16q13 | 3.36 | 3.64 | −0.28 |
| 14q21–23 | 2.56 | 2.81 | −0.25 |
| 20p12 | 2.09 | 2.62 | −0.53 |
| 2p15 | 1.50 | 1.68 | −0.18 |
| 1p13 | 1.27 | 1.53 | −0.26 |
| 1q42 | 0.46 | 1.51 | −1.05 |
| 4q28 | 2.00 | 1.46 | +0.54 |
| 3cent-q11 | 0.87 | 1.24 | −0.37 |
| 11p15 | 1.41 | 1.19 | +0.22 |
| 2q21–33 | 0.61 | 1.12 | −0.51 |
| 15q26 | 2.09 | 1.07 | +1.02 |
| 1p36 | 0.41 | 1.00 | −0.59 |
The strongest evidence for linkage was found at 6p11-p21, with the best markers (D6S426 and D6S257) mapping just centromeric to the HLA region (located at 6p21.3). There is a long history of interest in the role of the major histocompatibility complex (MHC) in many autoimmune diseases, including IDDM, familial psoriasis, multiple sclerosis, rheumatoid arthritis, and SLE. Genetic associations have been shown for HLA-DR2 or HLA-DR3 alleles in SLE patients from several ethnic groups (relative risks ranging from 2.0 to 3.4) (15). However, in large lupus-prone families, HLA alleles do not necessarily segregate with disease (16). A stronger association of HLA alleles (DQ in particular) has been demonstrated for specific autoantibody subsets in SLE (17). Non-HLA genes within the MHC, notably the tumor necrosis factor α gene and the complement components C2 and C4, also have been implicated in SLE (17). Further work will be necessary to determine whether this 6p locus represents a polymorphic HLA locus or a linked gene within or near HLA.
A recent comparison of the linkage results from 23 independent genome screens in various human and experimental animal autoimmune or inflammatory diseases identified 18 “clusters” of non-MHC candidate human autoimmune loci (18). Of interest, two of the non-MHC intervals identified in this screen of SLE families (16q13 and 11p15) are located within clusters implicated in other human autoimmune diseases. The 16q cluster is a large interval of ≈35 cM that includes potential susceptibility loci for Crohn’s disease (19), Blau syndrome (20), psoriasis (21), IDDM (22), and asthma (23). Cluster 11p is a narrow interval (≈2–3 cM) identified in asthma (23), multiple sclerosis (24), and IDDM (22). SLE is not known to associate strongly with any of the diseases that define these clusters, but these data suggest the possibility that there is a sharing of genetic predisposition to multiple autoimmune diseases or, alternatively, that each of these intervals may contain closely linked autoimmune susceptibility genes.
The results of an independent genome scan of SLE families performed by Moser et al. (25) at the Oklahoma Medical Research Foundation support many of the findings of this screen. Of the top 13 intervals identified in this sib-pair family screen, 4 (20p12, 1p13, 1q42, and 4q28 with lod scores ranging from 2.62 to 1.46) also were identified in the Oklahoma Medical Research Foundation screen of mostly larger families multiplex for SLE. The 20p12 region shows suggestive evidence for linkage in this study (D20S186, Zlr = 3.48, P = 0.00025, lod = 2.62) and was also one of the strongest loci identified in the Oklahoma screen (25). This interval maps centromeric to a recently identified susceptibility interval for psoriasis (21). The locus in the 1q42 region, originally identified by Tsao et al. in a candidate approach in 52 SLE sib-pairs (26), now has been identified in three independent SLE screens [refs. 25 and 26 and this study (D1S235, Zlr = 2.64, P = 0.0041, lod = 1.51)]. Despite these similarities, several of the strong intervals identified in this screen (6p, 14q and 16q) did not show evidence for linkage in the Oklahoma Medical Research Foundation study. Also, in our primarily Caucasian sample, we find little evidence for linkage at 1q22–23. This region showed evidence for significant linkage in the Oklahoma Medical Research Foundation study and harbors the various Fc receptors, which are associated with renal disease in African-American SLE patients (27). These differences in the genome screen results may reflect “false positive” evidence for linkage at some loci (14) or may result from ethnic and/or genetic heterogeneity in the samples.
In summary, using an affected sib-pair family approach, we have identified four potential SLE loci with lod scores >2.6 and an additional nine loci with lod scores ranging from 1.00 to 1.68. The four most interesting regions (6p11-p21-HLA, 14q21–23, 16q13, and 20p12) show surprisingly strong evidence for linkage given the sample size of 114 affected sib pairs and a total of 127 affected relative pairs. Two of the non-MHC intervals identified in this screen (16q13 and 11p15) have been implicated in other human autoimmune diseases. These data support results from genetic studies in lupus-prone mouse strains suggesting that multiple genes are responsible for conferring susceptibility to SLE (28–30). It will now be important to confirm these findings in additional cohorts of sib-pair families and to initiate efforts to further narrow these intervals in preparation for gene identification.
Acknowledgments
We are grateful to all of the patients and their family members who participated in this study and the many physicians who referred families and verified diagnoses. Thanks to J. Harley and K. Moser for sharing data before publication. Supported by grants from the Minnesota chapters of the Arthritis Foundation and the Lupus Foundation of America, the Texas Gulf Coast Chapter of the Lupus Foundation of America, and National Institutes of Health Grants R01 AR/AI43271, SLE SCOR P50 AR45231, and KO8-AR01986. P.M.G. is supported by a Physician-Scientist Award from the University of Minnesota Department of Medicine.
ABBREVIATIONS
- sib
sibling
- SLE
systemic lupus erythematosus
- lod
logarithm of odds
- MHC
major histocompatibility complex
- cM
centimorgan
References
- 1.Rothfield N F. In: Arthritis and Allied Conditions. McCarty D J, editor. Philadelphia: Lea & Febiger; 1985. pp. 911–935. [Google Scholar]
- 2.Hochberg M C. In: Dubois’ Lupus Erythematosus. Wallace D J, Hahn B H, editors. Baltimore: Williams & Wilkins; 1997. pp. 49–65. [Google Scholar]
- 3.Lawrence J S, Martins C L, Drake G L. J Rheumatol. 1987;14:913–921. [PubMed] [Google Scholar]
- 4.Hochberg M C. J Rheumatol. 1987;14:867–869. [PubMed] [Google Scholar]
- 5.Deapen D M, Escalante A, Weinrib L, Horwitz D A, Mack T M. Arthritis Rheum. 1992;35:311–318. doi: 10.1002/art.1780350310. [DOI] [PubMed] [Google Scholar]
- 6.Block S R, Winfield J B, Lockshin M D, D’Angelo W A, Christian C L. Am J Med. 1975;59:533–552. doi: 10.1016/0002-9343(75)90261-2. [DOI] [PubMed] [Google Scholar]
- 7.Tan E M, Cohen A S, Fries J F, Masi A T, McShane D J, Rothfield N F, Schaller J G, Talal N, Winchester R J. Arthritis Rheum. 1982;25:1271–1277. doi: 10.1002/art.1780251101. [DOI] [PubMed] [Google Scholar]
- 8.Hochberg M C. Arthritis Rheum. 1997;40:1725. doi: 10.1002/art.1780400928. [DOI] [PubMed] [Google Scholar]
- 9.Risch N. Am J Hum Genet. 1990;46:229–241. [PMC free article] [PubMed] [Google Scholar]
- 10.Vyse T J, Kotzin B L. Annu Rev Immunol. 1998;16:261–292. doi: 10.1146/annurev.immunol.16.1.261. [DOI] [PubMed] [Google Scholar]
- 11.Kearns G M, Messner R P, Behrens T W. J Rheumatol. 1998;25:482–485. [PubMed] [Google Scholar]
- 12.Kong A, Cox N J. Am J Hum Genet. 1997;61:1179–1188. doi: 10.1086/301592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kruglyak L, Daly M J, Reeve-Daly M P, Lander E S. Am J Hum Genet. 1996;58:1347–1363. [PMC free article] [PubMed] [Google Scholar]
- 14.Lander E, Kruglyak L. Nat Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- 15.Arnett F C, Reveille J D. Rheum Dis Clin North Am. 1992;18:865–892. [PubMed] [Google Scholar]
- 16.Reveille J D, Bias W B, Winkelstein J A, Provost T T, Dorsch C A, Arnett F C. Medicine (Baltimore) 1983;62:21–35. doi: 10.1097/00005792-198301000-00002. [DOI] [PubMed] [Google Scholar]
- 17.Arnett F C. In: Dubois’ Lupus Erythematosus. Wallace D J, Hahn B H, editors. Baltimore: Williams & Wilkins; 1997. pp. 77–117. [Google Scholar]
- 18.Becker K G, Simon R M, Bailey-Wilson J E, Freidlin B, Biddison W E, McFarland H F, Trent J M. Proc Natl Acad Sci USA. 1998;95:9979–9984. doi: 10.1073/pnas.95.17.9979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hugot J P, Laurent P P, Gower R C, Olson J M, Lee J C, Beaugerie L, Naom I, Dupas J L, Van G A, Orholm M, et al. Nature (London) 1996;379:821–823. doi: 10.1038/379821a0. [DOI] [PubMed] [Google Scholar]
- 20.Tromp G, Kuivaniemi H, Raphael S, Ala-Kokko L, Christiano A, Considine E, Dhulipala R, Hyland J, Jokinen A, Kivirikko S, et al. Am J Hum Genet. 1996;59:1097–1107. [PMC free article] [PubMed] [Google Scholar]
- 21.Nair R P, Henseler T, Jenisch S, Stuart P, Bichakjian C K, Lenk W, Westphal E, Guo S-W, Christophers E, Voorhees J J, Elder J T. Hum Mol Genet. 1997;6:1349–1356. doi: 10.1093/hmg/6.8.1349. [DOI] [PubMed] [Google Scholar]
- 22.Davies J L, Kawaguchi Y, Bennett S T, Copeman J B, Cordell H J, Pritchard L E, Reed P W, Gough S C L, Jenkins S C, Palmer S M, et al. Nature (London) 1994;371:130–136. doi: 10.1038/371130a0. [DOI] [PubMed] [Google Scholar]
- 23.Daniels S E, Bhattacharrya S, James A, Leaves N I, Young A, Hill M R, Faux J A, Ryan G F, le Souef P N, Lathrop G M, et al. Nature (London) 1996;383:247–250. doi: 10.1038/383247a0. [DOI] [PubMed] [Google Scholar]
- 24.Haines J L, Ter-Minassian M, Bazyk A, Gusella J F, Kim D J, Terwedow H, Pericak-Vance M A, Rimmler J B, Haynes C S, Roses A D, et al. Nat Genet. 1996;13:469–471. doi: 10.1038/ng0896-469. [DOI] [PubMed] [Google Scholar]
- 25.Moser, K. L., Neas, B. R., Salmon, J. E., Yu, H., Gray-McGuire, C., Asundi, N., Bruner, G. R., Fox, J., Kelly, J., Henshall, S., et al. Proc. Natl. Acad. Sci. USA 95, 14869–14874. [DOI] [PMC free article] [PubMed]
- 26.Tsao B P, Cantor R M, Kalunian K C, Chen C J, Badsha H, Singh R, Wallace D J, Kitridoou R C, Chen S, Shen N, et al. J Clin Invest. 1997;99:725–731. doi: 10.1172/JCI119217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salmon J E, Millard S, Schachter L A, Arnett F C, Ginzler E M, Gourley M F, Ramsey-Goldman R, Peterson M G, Kimberly R P. J Clin Invest. 1996;97:1348–1354. doi: 10.1172/JCI118552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morel L, Rudofsky U H, Longmate J A, Schiffenbauer J, Wakeland E K. Immunity. 1994;1:219–229. [PubMed] [Google Scholar]
- 29.Drake C G, Babcock S K, Palmer E, Kotzin B L. Proc Natl Acad Sci USA. 1994;91:4062–4066. doi: 10.1073/pnas.91.9.4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vidal S, Kono D H, Theofilopoulos A N. J Clin Invest. 1998;101:696–702. doi: 10.1172/JCI1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wallace D J. In: Dubois’ Lupus Erythematosus. Wallace D J, Hahn B H, editors. Baltimore: Williams & Wilkins; 1997. pp. 627–633. [Google Scholar]