Abstract
Nucleolar dominance is an epigenetic phenomenon in which one parental set of ribosomal RNA (rRNA) genes is silenced in an interspecific hybrid. In natural Arabidopsis suecica, an allotetraploid (amphidiploid) hybrid of Arabidopsis thaliana and Cardaminopsis arenosa, the A. thaliana rRNA genes are repressed. Interestingly, A. thaliana rRNA gene silencing is variable in synthetic Arabidopsis suecica F1 hybrids. Two generations are needed for A. thaliana rRNA genes to be silenced in all lines, revealing a species-biased direction but stochastic onset to nucleolar dominance. Backcrossing synthetic A. suecica to tetraploid A. thaliana yielded progeny with active A. thaliana rRNA genes and, in some cases, silenced C. arenosa rRNA genes, showing that the direction of dominance can be switched. The hypothesis that naturally dominant rRNA genes have a superior binding affinity for a limiting transcription factor is inconsistent with dominance switching. Inactivation of a species-specific transcription factor is argued against by showing that A. thaliana and C. arenosa rRNA genes can be expressed transiently in the other species. Transfected A. thaliana genes are also active in A. suecica protoplasts in which chromosomal A. thaliana genes are repressed. Collectively, these data suggest that nucleolar dominance is a chromosomal phenomenon that results in coordinate or cooperative silencing of rRNA genes.
Nucleolar dominance was among the first recognized epigenetic phenomena, discovered in interspecific hybrids in the plant genus Crepis. Navashin noted secondary constrictions at metaphase on D chromosomes inherited from one Crepis species, but not on the D chromosome from the other species (1, 2). Navashin’s contemporary, McClintock, showed that these secondary constrictions are sites where nucleoli were organized in the preceding interphase (3). Decades later, nucleolus organizer regions (NORs) were identified as loci where genes encoding the precursor of the 18S, 5.8S, and 25–28S ribosomal RNAs are tandemly arrayed (4–7). Thus, nucleolar dominance was shown ultimately to result from uniparental rRNA gene expression (8).
Nucleolar dominance occurs throughout the plant and animal kingdoms (9, 10) via mechanisms that remain unclear. Navashin noted that dominant NOR-bearing chromosomes could be contributed through the pollen or egg (2), ruling out maternal or paternal effects. A dominant D chromosome could also be contributed as part of an incomplete chromosome set. In Drosophila melanogaster × Drosophila simulans hybrids, the D. melanogaster NORs on the X and Y chromosomes are dominant over the single NOR on the D. simulans X (11). In hybrid XO males lacking a D. melanogaster sex chromosome, but containing all autosomes, the D. simulans NOR is active. These results indicate that NOR-bearing chromosomes must be present for nucleolar dominance to occur. Evidence from Xenopus suggested that the rRNA genes themselves might be responsible. In hybrids of X. laevis and X. borealis, only X. laevis rRNA genes are expressed in early development (12, 13). Using X. laevis and X. borealis rRNA minigenes coinjected into oocytes, Reeder and Roan found that sequences in the X. laevis intergenic spacer conferred transcriptional dominance (14). They hypothesized that X. laevis rRNA genes compete better than X. borealis genes for one or more limiting transcription factors because of an increase in enhancer number or strength (14). A similar hypothesis was proposed for nucleolar dominance in wheat (15, 16).
Nucleolar dominance has been thought to be independent of rRNA gene dosage or ploidy. Navashin observed dominant D chromosomes suppressing secondary constrictions on as many as three underdominant D chromosomes (in a 1:3 allotetraploid). Likewise, dominant Brassica rRNA genes can be outnumbered in a 4:2 allohexaploid (17) and the dominant NOR in hexaploid wheat has only half as many rRNA genes as the second most active NOR (18, 19). The favored, although unproved, explanation has been that rRNA genes are present in excess over transcription factors such that dominant genes titrate these factors even when outnumbered.
We report the occurrence and molecular analysis of nucleolar dominance in Arabidopsis suecica, an allotetraploid of Arabidopsis thaliana and Cardaminopsis arenosa (20). Surprisingly, A. thaliana rRNA gene silencing in newly formed A. suecica hybrids is highly variable, with two generations needed to establish dominance in some lines. Also, unexpectedly, the direction of dominance can be switched (C. arenosa genes silenced) in 3:1 A. thaliana to C. arenosa allotetraploids, an observation that challenges the hypothesis that dominant genes have inherently higher binding affinities for transcription factors. The possibility that uniparental rRNA gene expression results from silencing a species-specific transcription factor is argued against by transient expression experiments. These results suggest that nucleolar dominance is a chromosomal phenomenon that is enforced independent of transcription factor availability.
MATERIALS AND METHODS
Plant Material.
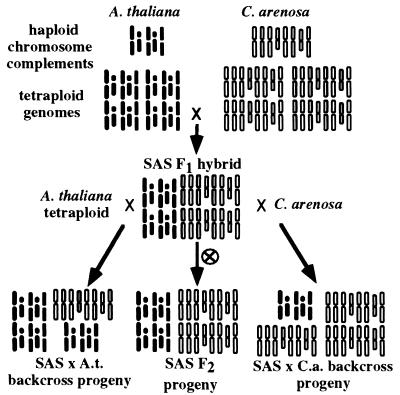
A. suecica race 90.10.085 was provided by Dr. Steven O’Kane (University of Northern Iowa, Cedar Falls, IA). Tetraploid A. thaliana Landsberg erecta plants were regenerated from cultured roots (21) treated with 0.5% (vol/vol) colchicine. A single C. arenosa plant from race 9509 (designated Care-1 in the Comai laboratory) was the pollen donor onto emasculated flowers of tetraploid A. thaliana to create synthetic allotetraploid A. suecica-like hybrids [designated synthetic A. suecica (SAS)]. Four self-fertile SAS F1 plants were obtained: SAS 1-4 (original strain designations are 605A, 605B, 49-2B, and 49-2A). F1 plants were self-pollinated to produce F2 seeds. SAS-2 was the female parent for backcrosses to tetraploid A. thaliana or C. arenosa. Chromosome analyses confirmed that SAS plants were allotetraploid; biparental inheritance of multiple molecular markers was also confirmed (L.C. et al., unpublished work).
Nucleic Acid Isolation.
Nucleic acids were generally purified from pooled tissues of 5–10 plants. For SAS F1 and F2 plants and backcross progeny, leaves from single plants were used. Tissues frozen in liquid nitrogen were ground to a powder, then mixed with three volumes (wt/vol) of extraction buffer (250 mM Tris⋅HCl, pH 8.5/375 mM NaCl/25 mM EDTA, pH 8.0/1% SDS/1% β-mercaptoethanol/0.5 mg/ml heparin). The homogenate was extracted twice with phenol/chloroform, and total nucleic acids were ethanol precipitated (22). Following centrifugation, pellets were resuspended in diethylpyrocarbonate-treated sterile water and total RNA was precipitated with 3 M LiCl. Genomic DNA in the supernatant was recovered by ethanol precipitation.
PCR Amplification.
PCR was used to amplify rRNA gene sequences from −265 (relative to the A. thaliana transcription start site, +1) to the 18S rRNA coding region using genomic DNA from A. thaliana, A. suecica, and C. arenosa. The upstream primer (see Fig. 1A) was 5′-TCGGTACCGAGTTTAGGATGTCAAGT-3′; the 18S primer was 5′-GCATATGACTACTGGCAGGATCAACC-3′. PCR products were cloned in pBluescript plasmids (Stratagene). Multiple clones were sequenced.
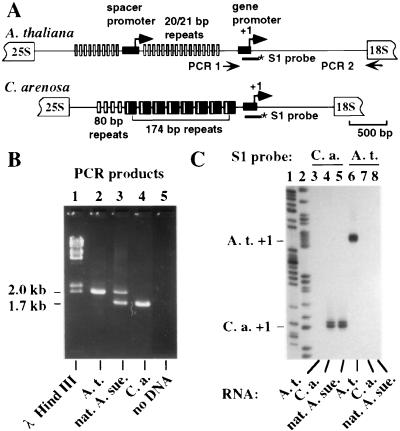
Figure 1.
Nucleolar dominance in A. suecica. (A) Organization of the intergenic spacers separating 25S and 18S rRNA coding sequences of adjacent rRNA genes. A. thaliana intergenic spacers are longer and contain repetitive elements smaller than those located in C. arenosa spacers. (B) Ethidium bromide-stained agarose gel of PCR products using the primers depicted in A and A. suecica, A. thaliana, and C. arenosa genomic DNA. HindIII-digested λDNA served as size markers in lane 1. The PCR reaction in lane 5 was performed with both primers but no genomic DNA. (C) Detection of rRNA transcripts by using the S1 nuclease protection assay. A. thaliana, C. arenosa, and natural A. suecica rRNA transcripts were detected with C. arenosa- (lanes 3–5) or A. thaliana-specific (lanes 6–8) DNA probes. Dideoxynucleotide sequencing reactions served as size markers in lanes 1 and 2.
Southern Blot Hybridization.
Restriction endonuclease-treated genomic DNA was subjected to agarose gel electrophoresis and blotted (23) to ZetaProbe membranes (Bio-Rad). Filters were hybridized (24) to radioactive probes (see figure legends) labeled by random hexamer priming (25).
S1 Protection Assay.
rRNA transcripts were detected by S1 nuclease protection as described previously (26). Briefly, 5 μg of total RNA was hybridized to 5′ end-labeled DNA fragments spanning the transcription start site. The A. thaliana probe was an SphI-EcoRV (−115 to +96) fragment labeled at +96; the C. arenosa probe was an SphI-BspE I fragment (−111 to +59) labeled at +59. RNA/DNA probe hybrids were treated with S1 nuclease (150 units/ml, 37°C, 30 min). Digestion products were resolved on a sequencing gel and exposed to x-ray film. The size of protected products corresponds to the distance from the labeled nucleotide to the transcription start site, +1. S1 probes were used in excess over RNA such that the amount of protected product was proportional to the amount of RNA transcript.
Transient Expression.
Protoplasts of sterile-grown 14- to 21-day-old A. thaliana and C. arenosa plants were transfected as described previously (26, 27). Briefly, 5 × 106 protoplasts were transfected with 50 pmols of supercoiled plasmid containing an A. thaliana or a C. arenosa rRNA minigene. Following transfection, protoplasts were incubated ≈24 hr. RNA was then purified and analyzed by S1 nuclease protection. A. thaliana S1 probes were end-labeled in plasmid sequences such that only minigene transcripts would be detected (27). For pAt1 5′Δ−520 (27), the probe was the SphI–BamHI fragment labeled at the BamHI site; for pAt1 5′Δ-2590/3′Δ+6 (27), the SphI–BssH II fragment was labeled at the BssH II site. The C. arenosa probe was the SphI–BspE I fragment described previously.
RESULTS
A. thaliana rRNA Genes Are Silent in A. suecica.
A. suecica is an allotetraploid derived from A. thaliana and C. arenosa. A. thaliana rRNA genes have been sequenced (28–31) and the intergenic spacer sequence of C. arenosa was determined recently (Hayworth and Schaal, personal communication). Based on these data, we designed PCR primers to amplify A. thaliana and C. arenosa rRNA gene sequences (see Fig. 1A). PCR amplification of A. thaliana genomic DNA yielded a ≈2.0-kb product (Fig. 1B, lane 2) whereas a ≈1.7-kb product was obtained by using C. arenosa DNA (Fig. 1B, lane 4). PCR amplification of A. suecica genomic DNA yielded both 1.7- and 2.0-kb products (lane 3) in similar amounts.
A. suecica PCR products were cloned and sequenced from −265 to ≈+150. A. thaliana- and C. arenosa-type clones were obtained in approximately equal numbers. Their sequences perfectly matched A. thaliana or C. arenosa rRNA gene sequences downstream of the transcription start site (+1). This homogeneity allowed the same S1 nuclease protection probes to detect rRNA transcripts in A. suecica or the appropriate progenitor species (Fig. 1C). By using the C. arenosa probe (lanes 3–5), a 59-nucleotide product corresponding to accurately initiated transcripts was detected with C. arenosa (lane 4) and A. suecica (lane 5) RNA, but not A. thaliana RNA (lane 3), verifying the specificity of the probe. Aliquots of the same RNA samples were tested using the A. thaliana probe (lanes 6–8). A 96-nucleotide-protected probe fragment was detected with A. thaliana RNA (lane 6), but not with C. arenosa RNA (lane 7), as expected. Using A. suecica RNA, no A. thaliana rRNA gene transcripts were detected (lane 8). We conclude that C. arenosa rRNA genes are dominant in A. suecica.
Stochastic Onset of Nucleolar Dominance in Synthetic A. suecica.
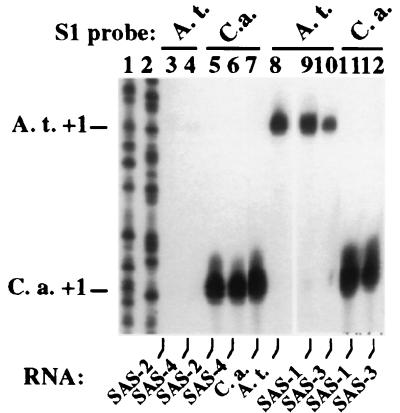
We have shown that nucleolar dominance occurs in natural and synthetic strains (32) of Brassica napus, B. juncea and B. carinata allotetraploids (17) but F3–F5 generations of synthetic lines were the earliest available for testing (32). Using the Arabidopsis system, we asked when dominance is first established. Chromosome counts in meiotic cells of C. arenosa 9509 indicated that this and several other races tested are tetraploid (L.C. et al., unpublished work), most likely autotetraploids, because only one type of rRNA gene was detected on sequencing multiple PCR clones (Z.J.C., unpublished work). Autotetraploid A. thaliana were generated by colchicine treatment and crossed with C. arenosa to recreate synthetic allotetraploid A. suecica (Fig. 2). Four self-fertile SAS plants were obtained, designated SAS-1 through SAS-4. C. arenosa transcripts were detected in all SAS plants (Fig. 3, lanes 5, 6, 11, 12) at levels similar to the C. arenosa control (lane 7). In contrast, expression of A. thaliana rRNA genes was variable. In SAS-2 and SAS-4, only trace amounts of A. thaliana transcripts could be detected in long exposures (lanes 3 and 4). However, in SAS-1, A. thaliana transcripts were as abundant as in the A. thaliana control (compare lanes 9 and 8). A. thaliana transcripts were expressed at moderate levels (≈40%) in SAS-3 (lane 10). Although the trend is toward dominance of C. arenosa genes in SAS plants, as in natural A. suecica, at least three epigenetic states for the rRNA genes are revealed in these newly formed allotetraploids: complete dominance, partial dominance, and codominance. PCR and Southern blot analyses of SAS plants confirmed the presence of A. thaliana and C. arenosa rRNA genes in similar abundance (Z.J.C., data not shown).
Figure 2.
Creation of synthetic A. suecica allotetraploids and backcross progeny. A. thaliana is typically diploid, with a haploid complement of five chromosomes. C. arenosa is a natural tetraploid with a haploid complement of eight chromosomes. Tetraploid A. thaliana was used as the maternal parent in a cross with C. arenosa, yielding SAS plants. SAS F2 progeny were obtained from self-pollinated flowers. Alternatively, flowers were emasculated and manually pollinated with tetraploid A. thaliana or C. arenosa pollen. Thus progeny with 3:1, 2:2 (F2), or 1:3 A. thaliana to C. arenosa genome complements were obtained from the same mother plant.
Figure 3.
Variable severity of nucleolar dominance in SAS F1 hybrids. Total RNA from SAS, A. thaliana, and C. arenosa plants (lanes 7 and 8) was assayed by using S1 nuclease protection with A. thaliana- (lanes 3, 4, 8–10) or C. arenosa-specific probes (lanes 5–7, 11, 12). DNA sequencing reactions (lanes 1 and 2) served as size markers.
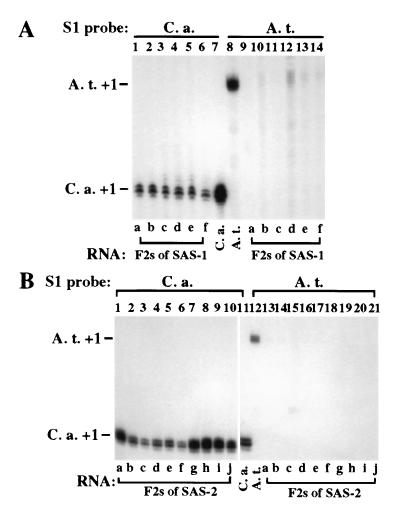
To determine whether the variable rRNA gene expression states observed in F1 plants persist in subsequent generations, we examined F2 siblings of self-pollinated SAS-1 and SAS-2 plants (Fig. 4). In SAS-1, A. thaliana and C. arenosa were codominant (see Fig. 3). However, in all F2 sibs of SAS-1, C. arenosa rRNA genes were now dominant, with only trace amounts of A. thaliana transcripts detected (Fig. 4A, compare lanes 1–6 with lanes 9–14). In SAS-2, C. arenosa rRNA genes were already dominant in the F1 generation, with only a trace of A. thaliana rRNA transcription detectable (see Fig. 3). In all F2 sibs of SAS-2, A. thaliana transcripts were not detectable even in trace amounts (Fig. 4B). A. thaliana and C. arenosa rRNA gene PCR products were obtained in similar abundance in SAS-1 and all F2 progeny, indicating that A. thaliana rRNA genes were not lost or underrepresented in F2 plants (data not shown). Collectively, these data show that nucleolar dominance occurs in newly formed A. suecica but two generations are necessary for its complete establishment.
Figure 4.
Two generations are needed for establishment of nucleolar dominance in some SAS lines. (A) SAS-1, in which A. thaliana and C. arenosa rRNA genes were codominant, was self-pollinated to generate F2 plants. rRNA transcripts in six F2 siblings and C. arenosa and A. thaliana controls (lanes 7, 8) were detected using C. arenosa- (lanes 1–7) or A. thaliana-specific (lanes 8–14) S1 probes. (B) F2 progeny of SAS-2 were analyzed in the same way.
The Direction of Nucleolar Dominance Is Subject to Gene or Genome Dosage Effects.
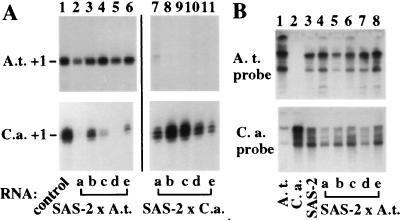
To investigate parental gene dosage effects on nucleolar dominance, SAS plants were backcrossed to C. arenosa or tetraploid A. thaliana. In five siblings derived from SAS-2 backcrossed to A. thaliana, A. thaliana transcripts were detected at levels (Fig. 5A, lanes 2–6, top autoradiogram) comparable to the A. thaliana control (lane 1, top), whereas in five siblings from a backcross to C. arenosa, A. thaliana transcripts were not detected (lanes 7–11, top; the weak signal in lane 7 does not correspond to the A. thaliana start site and was because of incomplete probe digestion). Examination of C. arenosa rRNA gene expression in the SAS-2 × A. thaliana backcross progeny yielded interesting results (Fig. 5A, bottom left autoradiogram). Only trace amounts of C. arenosa transcripts were detected in sibs a and d (lanes 2 and 5). C. arenosa transcripts were detected at low levels in sibs c and e (lanes 4 and 6). In sib b, arenosa transcripts were abundant (lane 3) but below control levels (lane 1). Overall, the data suggest that in A. suecica × A. thaliana backcross progeny, the trend is reversed toward dominance of A. thaliana genes, although the extent of dominance is variable, reminiscent of the results with the initial synthetic A. suecica plants (see Fig. 3). Ethidium-stained RNA gels showed that the variable C. arenosa transcript levels in Fig. 5A could not be explained by differences in RNA quantity or quality (data not shown). Southern blotting confirmed that both parental sets of rRNA genes were present (Fig. 5B). The presence of A. thaliana rRNA genes in SAS-2 and its backcross progeny (Top, lanes 3–8) was confirmed by using a A. thaliana-specific intergenic spacer probe (compare lanes 1 and 2). The equivalent C. arenosa probe crossreacted weakly with A. thaliana rRNA gene fragments (Bottom, compare lanes 1 and 2), but fragment size differences allowed confirmation of C. arenosa rRNA genes in SAS-2 and its backcross progeny (lanes 3–8). PCR analyses supported the Southern blot analysis results (not shown). Collectively, these data suggest that the differential expression of C. arenosa rRNA genes in SAS-2 × A. thaliana backcross progeny cannot be explained by defects in the transmission of C. arenosa rRNA genes.
Figure 5.
The direction of nucleolar dominance can be switched. SAS-2, which displayed complete dominance of C. arenosa over A. thaliana rRNA genes (see Fig. 3) was the maternal parent for backcrosses to tetraploid A. thaliana or C. arenosa. (A) Five backcross sibs from each cross were assayed for rRNA gene expression using A. thaliana- (Top) or C. arenosa-specific (Bottom) S1 probes. A. thaliana or C. arenosa controls are in lane 1 of the relevant autoradiograms. (B) Southern blot analysis of BssH II-digested DNA of A. thaliana (lane 1), C. arenosa (lane 2), SAS-2 (lane 3), or the backcross sibs (lanes 4–8) tested in A. The filter was first hybridized to an A. thaliana-specific rRNA gene intergenic spacer probe (Top), then was stripped and reprobed (49) with a similar C. arenosa probe (Bottom). The former corresponded to −2,590 to +6 of plasmid pAt1 5′Δ−2,590/3′Δ+6 (27); the latter spanned −2,316 to +33 of pCa1.
Transient Expression Argues Against Species-Specific Transcription Factors.
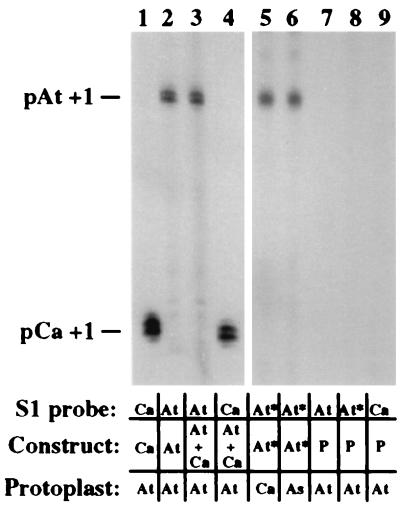
Coordinate silencing of parental sets of rRNA genes could be accomplished by silencing a species-specific RNA polymerase I transcription factor (9). To test this possibility, we performed reciprocal transfections of cloned A. thaliana and C. arenosa rRNA minigenes into protoplasts of each species (Fig. 6). The C. arenosa minigene was expressed in A. thaliana (lane 1) and did not outcompete a A. thaliana minigene when both were cotransfected (compare lanes 3 and 4). Likewise, a A. thaliana minigene was transcribed both in C. arenosa (lane 5) and A. suecica (lane 6), despite the fact that the endogenous A. thaliana genes are silenced in A. suecica.
Figure 6.
A. thaliana and C. arenosa rRNA minigenes are transcribed in the other species. A. thaliana (At), C. arenosa (Ca) or A. suecica (As) protoplasts were transfected with 50 pmols of pBluescript plasmid containing C. arenosa rRNA gene promoter sequences (pCa; sequences −265 to ≈+1,700) or one of two A. thaliana minigenes, pAt or pAt*. Constructs pAt and pAt* correspond to minigenes pAt1 5′Δ−520 (sequences −520 to +92) and pAt1 5′Δ−2,590/3′Δ+6 (−2,590 to +6), respectively (27). Transcripts were detected with minigene-specific S1 probes. Transcripts in lanes 3 and 4 derive from an experiment in which 50 pmols each of pAt and pCa were cotransfected and half of the isolated RNA was hybridized with the At (lane 3) or Ca (lane 4) probe. Data in lanes 1–4 and 5–9 are derived from the same exposure of a single autoradiogram. Note that no transcripts are detected in protoplasts transfected only with pBluescript (P) DNA (lanes 7–9).
Cytosine Methylation Enforces rRNA Gene Silencing, as in Brassica.
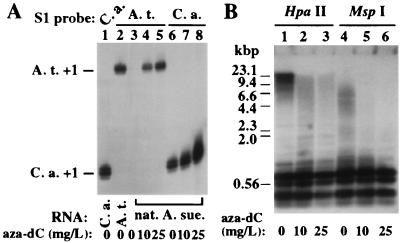
In our previous work with Brassica, a genus closely related to Arabidopsis, inhibiting cytosine methylation (with aza-dC) or histone deacetylation (with trichostatin A or sodium butyrate) derepressed silenced rRNA genes subjected to nucleolar dominance (33). In agreement with these results, A. thaliana rRNA transcripts were induced to 40% or 100% of control levels in natural A. suecica plants germinated on media containing aza-dC (Fig. 7, compare lanes 4 and 5 to lane 2). C. arenosa transcripts were also up-regulated 2- to 3-fold at the highest level of aza-dC (compare lane 8 to lanes 1 and 6). These results do not provide mechanistic insights beyond our previous study but confirm that A. thaliana genes in A. suecica are not defective, but are silenced.
Figure 7.
Derepression of A. thaliana rRNA genes in natural A. suecica using 5-aza-2′ deoxycytidine (aza-dC). Approximately 30 A. suecica seeds were surface-sterilized and germinated on medium containing 0, 10, or 25 mg/L, as described previously (33). (A) Total RNA from 3-week-old plantlets was purified and analyzed by S1 nuclease protection with A. thaliana- (lanes 2–5) or C. arenosa-specific (lanes 1, 6–8) probes. C. arenosa and A. thaliana control reactions are in lanes 1 and 2. (B) Southern blot analysis of DNA from control and treated plants following digestion with HpaII (lanes 1–3) or MspI (lanes 4–6) and hybridization to an A. thaliana probe spanning the ≈2-kb region from −365 to the beginning of the 18S rRNA coding sequences. This probe detects both A. thaliana and C. arenosa rRNA genes.
Verification that aza-dC altered cytosine methylation levels in A. suecica was obtained using the restriction endonucleases HpaII or MspI, both of which cut CCGG motifs. HpaII will not cut if the inner C is methylated and cuts very inefficiently if the outer C is methylated. MspI will cut if the inner, but not the outer, C is methylated. Southern blot hybridization using a probe that hybridizes to all A. suecica rRNA genes in the conserved ≈2-kb region from the promoter to the 18S rRNA coding sequences showed that without aza-dC treatment, ≈50% of the crosshybridizing signal following HpaII digestion ranged in size from 6- to 23-kb (Fig. 7B lane 1). This signal corresponds to A. thaliana and C. arenosa genes methylated at most or all CCGGs (each gene is ≈10 kb in length). The DNA is more susceptible to MspI cleavage (lane 4), as expected. Aza-dC treatment caused increased susceptibility to HpaII and MspI cleavage, confirming loss of cytosine methylation (lanes 2, 3, 5, 6).
Using Southern blotting and a variety of probes and methylation-sensitive restriction endonucleases, we have been unable to identify specific restriction sites in Arabidopsis (or Brassica) rRNA gene promoters or intergenic spacers whose methylation state correlates with gene activity or silencing. This observation contrasts with studies of nucleolar dominance in wheat, triticale, and maize (16, 34, 49). The overall degree of A. thaliana rRNA gene methylation, estimated by resistance to HpaII digestion, is also not different in A. thaliana, natural or synthetic A. suecica, or SAS F2 or backcross progeny, despite their varying degrees of A. thaliana rRNA gene expression (data not shown). It is unknown currently whether demethylation of the rRNA genes or a distinct regulatory locus is responsible for the derepression of silenced rRNA genes by aza-dC (see Discussion and ref. 33).
DISCUSSION
Despite a long history of cytological and cytogenetic descriptions, few studies have examined nucleolar dominance at a transcriptional level (8, 17, 33). The variable severity of nucleolar dominance we observed in newly formed hybrids contrasts with the longstanding idea that nucleolar dominance is the same in all individuals for a particular cross (2). Furthermore, our study shows that establishment of nucleolar dominance can require multiple generations. This requirement might have gone unnoticed in previous studies using diploid species because their dihaploid F1 progeny are often sterile due to defects in chromosome pairing and segregation. For allotetraploids (amphidiploids) such as A. suecica, which contain diploid chromosome complements from both progenitors, infertility is less problematic.
Our most important finding is that gene or genome dosage effects can switch the direction of nucleolar dominance, negating the truism that dominance is independent of rRNA gene dosage or ploidy (9, 10). An effect of genome dosage on the number of nucleoli visible at interphase in allopolyploid Ribes (gooseberry and black currant) hybrids was reported, suggesting that dominance could be overcome by increasing the number of underdominant NORs (35). Studies in wheat have shown also that NOR activity is variable in different chromosome addition lines, suggesting that genes unlinked to the NORs can affect their activity (18, 36). However, the demonstration that normally dominant rRNA genes can be made underdominant is unprecedented.
Dominance switching is also at odds with the prevailing hypothesis that dominant rRNA genes outcompete underdominant genes for transcription factors because of higher protein binding affinities (9, 14, 19). Such affinities, described by equilibrium binding constants, should be invariant. Indeed, the presumed insensitivity of nucleolar dominance to gene or genome dosage effects has been interpreted as evidence that rRNA genes are always in excess over transcription factors, such that the genes with highest binding affinities are always dominant. It follows that decreasing the dosage of dominant genes might allow transcription factors to become available to underdominant rRNA genes, but dominant genes should never fall silent. Our demonstration of dominance switching argues that rRNA genes are not independently regulated based on their intrinsic affinities for transcription factors. Consistent with this conclusion from genetic experiments, dominant and underdominant Brassica rRNA genes compete equally for transcription factors (Frieman and C.S.P., unpublished data) in the in vitro transcription system we have developed (37).
Coordinate control of parental sets of rRNA genes might be accomplished by controlling the expression of a species-specific transcription factor. However, our transient expression results argue against this possibility. The transient expression results suggest further that nucleolar dominance is a chromosomal phenomenon that is enforced, and possibly established, independent of transcription factor availability. Supporting evidence is that in Drosophila hybrids, chromosomal rearrangements adjacent to the NORs of D. melanogaster eliminate suppression of D. simulans nucleolus formation in trans without reducing D. melanogaster rRNA gene or nucleolus expression (38). Therefore, expression of rRNA genes at dominant NORs is not sufficient to cause repression of an underdominant NOR, as predicted by the limiting transcription factor hypothesis (38). Durica and Krider’s observations also show that loci flanking the NORs play a role in their regulation (38). Being closely linked to the NORs, it is possible that these flanking loci are the true determinants of nucleolar dominance rather than structural features of the rRNA genes within the dominant NORs.
Variable rRNA gene silencing in F1 hybrids suggests a stochastic component to the establishment of nucleolar dominance. Furthermore, completion of nucleolar dominance by the F2 generation implies that silencing is cumulative and is not erased at meiosis. De novo cytosine methylation is a candidate DNA modification that might explain the stochastic onset and progressive establishment of nucleolar dominance (39–43) and is consistent with the derepression of silenced rRNA genes by aza-dC. However, we have been unable to correlate rRNA gene activity with overall rRNA gene methylation levels or methylation of specific restriction sites in either Brassica (33) or Arabidopsis (this study). Perhaps critical methylated cytosines have gone unnoticed using this approach. However, we have also found that rRNA gene transcription in vitro is insensitive to cytosine methylation (Frieman and C.S.P., unpublished data). Furthermore, histone deacetylase inhibitors derepress silenced rRNA genes without detectably altering rRNA gene methylation in vivo (Z.J.C. and C.S.P., unpublished work). These results argue against models in which hypermethylation of underdominant rRNA gene promoters directly blocks transcription factor binding (44). However, it is possible that methylation acts indirectly via recruitment of methylcytosine-binding protein complexes that include histone deacetylase activity (45, 46). Histone deacetylation, in turn, might render the chromatin structure of rRNA gene promoters inaccessible to transcription factors. The latter model is consistent with the derepression of silenced rRNA genes caused by histone deacetylase inhibitors (33), the lack of synergy between these inhibitors and aza-dC (33), and the fact that histone deacetylase inhibitors have no detectable effect on rRNA gene methylation levels (Z.J.C. and C.S.P., unpublished work).
Another possibility is that aza-dC and histone deacetylase inhibitors derepress silenced rRNA genes by activating a regulatory locus that controls the transcriptional competence of the NOR. Involvement of other loci is consistent with studies showing loss of nucleolar dominance because of chromosome substitution in wheat and triticale (18, 36) or because of chromosome rearrangements in Drosophila (38) and barley (47). Our current study suggests that such hypothetical regulatory loci are unlikely to encode species-specific transcription factors. Beyond this conclusion, our current understanding falls short of considering other candidate loci. The availability of A. thaliana hypomethylation mutants (48) and other Arabidopsis genetic resources should make A. suecica a promising model system for further study.
Acknowledgments
We thank Douglas Hayworth and Barbara Schaal for sharing unpublished C. arenosa rRNA gene sequence data and Steve O’Kane for providing C. arenosa and A. suecica seeds. We thank Ronald Reeder (Fred Hutchinson Cancer Center, Seattle, WA) and Eric Richards (Washington University, St. Louis, MO) for helpful discussions. This work was supported by National Science Foundation Grant MCB-9617471 to C.S.P. and by a Monsanto Postdoctoral Fellowship in Plant Biology and a National Institutes of Health National Research Service award (1 F32 GM19072) to Z.J.C.
ABBREVIATIONS
- NOR
nucleolus organizer region
- rRNA
ribosomal RNA
- SAS
synthetic A. suecica.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Navashin M S. Proc Int Conf Genet Eng. 1928;5:1148–1152. [Google Scholar]
- 2.Navashin M. Cytologia. 1934;5:169–203. [Google Scholar]
- 3.McClintock B. Z Zellforsch Mikrosk Anat. 1934;21:294–328. [Google Scholar]
- 4.Ritossa F M, Spiegelman S. Proc Natl Acad Sci USA. 1965;53:737–745. doi: 10.1073/pnas.53.4.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wallace H, Birnstiel M L. Biochem Biophys Acta. 1966;114:296–310. doi: 10.1016/0005-2787(66)90311-x. [DOI] [PubMed] [Google Scholar]
- 6.Phillips R L, Kleese R A, Wang S S. Chromosoma. 1971;36:79–88. [Google Scholar]
- 7.Givens J F, Phillips R L. Chromosoma. 1976;57:103–117. [Google Scholar]
- 8.Honjo T, Reeder R H. J Mol Biol. 1973;80:217–228. doi: 10.1016/0022-2836(73)90168-x. [DOI] [PubMed] [Google Scholar]
- 9.Reeder R H. J Cell Biol. 1985;101:2013–2016. doi: 10.1083/jcb.101.5.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pikaard C S, Chen Z J. In: RNA Polymerase I: Transcription of Eukaryotic Ribosomal RNA. Paule M R, editor. Austin, TX: R. G. Landes; 1998. pp. 275–293. [Google Scholar]
- 11.Durica D S, Krider H M. Dev Biol. 1977;59:62–74. doi: 10.1016/0012-1606(77)90240-8. [DOI] [PubMed] [Google Scholar]
- 12.Blackler A W, Gecking C A. Dev Biol. 1972;27:385–394. doi: 10.1016/0012-1606(72)90177-7. [DOI] [PubMed] [Google Scholar]
- 13.Cassidy D M, Blackler A W. Dev Biol. 1974;41:84–96. doi: 10.1016/0012-1606(74)90285-1. [DOI] [PubMed] [Google Scholar]
- 14.Reeder R H, Roan J G. Cell. 1984;38:39–44. doi: 10.1016/0092-8674(84)90524-5. [DOI] [PubMed] [Google Scholar]
- 15.Martini G, O’Dell M, Flavell R B. Chromosoma. 1982;84:687–700. [Google Scholar]
- 16.Sardana R, O’Dell M, Flavell R. Mol Gen Genet. 1993;236:155–162. doi: 10.1007/BF00277107. [DOI] [PubMed] [Google Scholar]
- 17.Chen Z J, Pikaard C S. Proc Natl Acad Sci USA. 1997;94:3442–3447. doi: 10.1073/pnas.94.7.3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flavell R B, O’Dell M. Chromosoma. 1979;71:135–152. [Google Scholar]
- 19.Flavell R B. Oxford Surv Plant Mol Cell Biol. 1986;3:252–274. [Google Scholar]
- 20.O’Kane S, Schaal B, Al-Shehbaz I. Syst Bot. 1995;21:559–566. [Google Scholar]
- 21.Valvekens D, Van Montagu M, Van Lijsebettens M. Proc Natl Acad Sci USA. 1988;85:5536–5540. doi: 10.1073/pnas.85.15.5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, New York: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 23.Chomczynski P. Anal Biochem. 1992;201:134–139. doi: 10.1016/0003-2697(92)90185-a. [DOI] [PubMed] [Google Scholar]
- 24.Church G M, Gilbert W. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feinberg A P, Vogelstein B. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 26.Doelling J H, Gaudino R J, Pikaard C S. Proc Natl Acad Sci USA. 1993;90:7528–7532. doi: 10.1073/pnas.90.16.7528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Doelling J H, Pikaard C S. Plant J. 1995;8:683–692. doi: 10.1046/j.1365-313x.1995.08050683.x. [DOI] [PubMed] [Google Scholar]
- 28.Gruendler P, Unfried I, Pointner R, Schweizer D. Nucleic Acids Res. 1989;17:6395–6396. doi: 10.1093/nar/17.15.6395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gruendler P, Unfried I, Pascher K, Schweizer D. J Mol Biol. 1991;221:1209–1222. doi: 10.1016/0022-2836(91)90929-z. [DOI] [PubMed] [Google Scholar]
- 30.Unfried I, Stocker U, Gruendler P. Nucleic Acids Res. 1989;17:7513. doi: 10.1093/nar/17.18.7513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Unfried I, Gruendler P. Nucleic Acids Res. 1990;18:4011. doi: 10.1093/nar/18.13.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song K, Tang K, Osborn T C. Theor Appl Genet. 1993;86:811–821. doi: 10.1007/BF00212606. [DOI] [PubMed] [Google Scholar]
- 33.Chen Z J, Pikaard C S. Genes Dev. 1997;11:2124–2136. doi: 10.1101/gad.11.16.2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Flavell R B, O’Dell M, Thompson W F. J Mol Biol. 1988;204:523–534. doi: 10.1016/0022-2836(88)90352-x. [DOI] [PubMed] [Google Scholar]
- 35.Keep E. Can J Genet Cytol. 1962;4:206–218. [Google Scholar]
- 36.Neves N, Silva M, Heslop-Harrison J S, Viegas W. Chromosome Res. 1997;5:125–131. doi: 10.1023/a:1018470208730. [DOI] [PubMed] [Google Scholar]
- 37.Saez-Vasquez J, Pikaard C S. Proc Natl Acad Sci USA. 1997;94:11869–11874. doi: 10.1073/pnas.94.22.11869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Durica D S, Krider H M. Genetics. 1978;89:37–64. doi: 10.1093/genetics/89.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bestor T H, Chandler V L, Feinberg A P. Dev Genet (Amsterdam) 1994;15:458–462. doi: 10.1002/dvg.1020150603. [DOI] [PubMed] [Google Scholar]
- 40.Bird A. Cell. 1992;70:5–8. doi: 10.1016/0092-8674(92)90526-i. [DOI] [PubMed] [Google Scholar]
- 41.Eden S, Cedar H. Curr Opin Genet Dev. 1994;4:255–259. doi: 10.1016/s0959-437x(05)80052-8. [DOI] [PubMed] [Google Scholar]
- 42.Razin A, Cedar H. Cell. 1994;77:473–476. doi: 10.1016/0092-8674(94)90208-9. [DOI] [PubMed] [Google Scholar]
- 43.Martienssen R A, Richards E J. Curr Opin Genet Dev. 1995;5:234–242. doi: 10.1016/0959-437x(95)80014-x. [DOI] [PubMed] [Google Scholar]
- 44.Houchins K, O’Dell M, Flavell R B, Gustafson J P. Mol Gen Genet. 1997;255:294–301. doi: 10.1007/s004380050500. [DOI] [PubMed] [Google Scholar]
- 45.Nan X, Ng H H, Johnson C A, Laherty C D, Turner B M, Eisenman R N, Bird A. Nature (London) 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 46.Jones P L, Veenstra G J, Wade P A, Vermaak D, Kass S U, Landsberger N, Strouboulis J, Wolffe A P. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 47.Schubert I, Kunzel G. Chromosoma. 1990;99:352–359. [Google Scholar]
- 48.Vongs A, Kakutani T, Martienssen R A, Richards E J. Science. 1993;260:1926–1928. doi: 10.1126/science.8316832. [DOI] [PubMed] [Google Scholar]
- 49.Jupe E R, Zimmer E A. Plant Mol Biol. 1993;21:805–821. doi: 10.1007/BF00027113. [DOI] [PubMed] [Google Scholar]