Abstract
An intracellular protein termed CD2 binding protein 2 (CD2BP2), which binds to a site containing two PPPGHR segments within the cytoplasmic region of CD2, was identified. Mutagenesis and NMR analysis demonstrated that the CD2 binding region of CD2BP2 includes a 17-aa motif (GPY[orF]xxxxM[orV]xxWxxx GYF), also found in several yeast and Caenorhabditis elegans proteins of unknown function. In Jurkat T cells, over-expression of the isolated CD2BP2 domain binding to CD2 enhances the production of interleukin 2 on crosslinking of CD2 but not the T cell receptor. Hence, a proline-binding module distinct from SH3 and WW domains regulates protein–protein interactions.
Human CD2 functions to facilitate both adhesion and activation of T lymphocytes on binding to its ligand CD58 (1–3). Moreover, the binding of CD58 to CD2 augments interleukin (IL)-12-driven T cell responsiveness (4, 5) and initiates reversal of the anergic state (6). Although the important contribution of the cytoplasmic tail of CD2 to T cell activation has been studied in both rodents and humans (7–10), the mechanisms by which the CD2 tail mediates those functions are not clear. Interaction trap cloning was used to investigate the molecular basis of CD2-mediated signaling.
MATERIALS AND METHODS
Yeast Two-Hybrid System.
The yeast two-hybrid system interaction trap was used as described (11). In brief, as bait, a human CD2 cDNA encoding the cytoplasmic segment amino acids 221–327 was transferred into the pEG202 vector. cDNAs from an activated human T cell cDNA library, CD2 binding protein 2 (CD2BP2), or mutant cDNA variants of CD2BP2 were introduced into the pJG4–5 vector between EcoRI and XhoI restriction sites. Mutations were created by a previously described method (12) and were confirmed by DNA sequencing. Both of the yeast shuttle vectors were introduced into the yeast strain EGY48, were plated onto selection plates containing glucose, and were incubated at 30°C. After 48 h, >100 colonies were mixed, were transferred onto a plate containing galactose/raffinose and 5-bromo-4-chloro-3-indolyl-d-galactoside, and were incubated for 48 h at 30°C followed by 5 days at 8°C. The strength of the interaction was scored based on the time point when the blue color appeared in comparison with a negative control: ++++, <6 h; +++, <12 h; ++, <24 h; +, <48 h; ±, <72 h; and −, >72 h. CD2 mutations were generated and analyzed in a similar manner.
5′ Rapid Amplification of cDNA Ends (RACE) and PCR.
Standard 5′ RACE was performed by using human T cell total RNA as described (13). The primers corresponded to position bp 676–697 (for the reverse transcription primer) and bp 356–376 and 310–331 (for RACE primers), according to the numbering in Fig. 1. The RACE reaction produced a single product as detected by agarose gel electrophoresis. To introduce the Kozak consensus sequence (CCGCCACC) before the initiation ATG as well as FLAG epitope sequence (GACTACAAGGACGACGATGACAAG encoding DYKDDDDK) at the N terminus of the CD2BP2 sequence, two consecutive PCR reactions were performed in which the tagged primers extended the 5′ end of the product stepwise. As a downstream primer, an oligonucleotide corresponding to bp 676–697 was used. The PCR product then was cloned into the TA cloning vector pCR2.1 (Invitrogen), and the DNA sequence was verified. Subsequently, the 5′ DNA fragment (between the EcoRI site within the vector and an endogenous PstI site within the CD2BP2 sequence) was obtained as was a 3′ PstI-XhoI fragment isolated from the original pJG4–5 CD2BP2 clone. Both of the gel purified fragments were cloned into pCR2.1 (Invitrogen), which previously had been digested with EcoRI-XhoI, thus generating a full length FLAG-CD2BP2 construct.
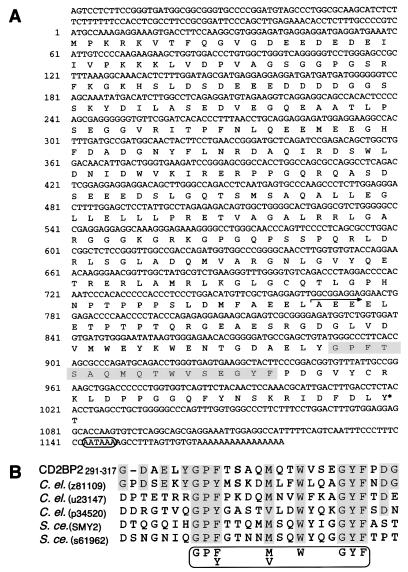
Figure 1.
CD2BP2 sequence and motif. (A) Nucleotide and predicted amino acid sequence of CD2BP2. The 17-residue segment showing homology with other proteins is shaded. The arrow defines the start of the smallest C-terminal CD2BP2 segment binding to CD2 as shown by mutation analysis using a yeast two-hybrid system. The asterisk indicates the tyrosine residue, which is a potential phosphorylation site. The polyadenylation signal is circled. The two clones obtained from the yeast two-hybrid system have an identical overlapping sequence of 768 bp with the longer of the two clones extending 408 bp further 5′. Seven independent RACE clones were sequenced to determine the upstream region from bp −24 to −120. (B) Alignment of the motif segment residing between amino acids 291–317 of CD2BP2 with sequences from several other proteins. To identify local homologies, segments of the CD2BP2 amino acid sequence were subjected to the standard blast search (30) against SWISSPROT and translated sequences of the GenBank data, using a sliding window of 50 residues over the sequence with a pitch of 20 residues. All of the sequences that meet the consensus 17-residue motif (GP[F, Y]-4x-[M, V]-2x-W-3x-GYF) were aligned with the use of megalign (DNAstar, Madison, WI). The origin (C.el., C. elegans; S.ce., S. cerevisiae) and the accession number or name of each locus is shown in parentheses. SMY2 was cloned as a suppressor of the defect in myosin (36).
Transfection, Immunoprecipitation, and Western Blotting.
As the vectors for transfection, pcDNA1.1 and pcDNA3.1 (Invitrogen) were used with COS7 and with Jurkat cells, respectively. For expression of CD2 and CD4 in COS7 cells, a pCDM8 plasmid containing each was used (14, 15). COS7 cells were transfected by the calcium phosphate precipitation method as described (14). For immunoblot analysis, 2 × 106 COS7 cells or 107 Jurkat transformants were washed with cold TBS (20 mM Tris, pH 7.4/0.15 M NaCl), were lysed in TBS containing 1% Triton X-100, 10 mM NaF, 10 mM sodium pyrophosphate, 0.2 trypin inhibitory units/ml aprotinin, 1 mM phenylmethylsulfonyl fluoride, 1 mM sodium orthovanadate, 5 μg/ml leupeptin, 1 mM MgCl2, and 1 mM ZnCl2, and were incubated for 2 h with 15 μl of CNBr Sepharose beads coupled with either 3T4–8B5 (anti-T111) anti-CD2 mAb, 19Thy5D7 anti-CD4, 2H11 anti-leucine zipper mAb, or M2 anti-FLAG mAb agarose beads (Kodak). The bead-bound immune complexes were washed with lysis buffer and TBS and were eluted by boiling in Laemmli SDS sample buffer. Western blotting was performed as described (16). For detection of the FLAG epitope, M2 mAb was used. For CD2 detection, M32 rabbit serum was used at 1:2,000, followed by detection with protein A-HRP (Bio-Rad) and ECL (Amersham) according to the manufacturer’s protocol.
Protein Expression and NMR Analysis.
The CD2BP2 protein used for the NMR studies comprises amino acid residues 256–341 with an additional glycine as well as six histidines placed at the N terminus. The cDNA encoding this sequence was cloned into the T7-based expression vector pTFT74 (17) and was expressed in the Escherichia coli strain BL21(DE3) (18). Purification of the soluble, cytoplasmic protein was achieved with a single-step purification on Ni-Sepharose (Qiagen). Protein samples were concentrated and buffer exchanged against 50 mM sodium-phosphate buffer (pH 6.3) with Centricon-3-concentrators (Amicon). The glutathione S-transferase (GST)-CD2 construct comprised residues 221–282 of the CD2 molecule fused to the GST gene of the pGEX-4T-1 vector (Pharmacia). The fusion protein was expressed in E. coli BL21(DE3) and was purified by glutathione-Sepharose (Pharmacia) in a single step. Except for prolines, NMR backbone assignments of the isolated CD2BP2 binding domain were achieved by the use of an HNCA experiment (19) applied to a 15N-13C-labeled sample of the CD2BP2 domain. Starting points for the sequential assignment were obtained by the acquisition of a gradient enhanced version of the heteronuclear single quantum coherence experiment (20) by using the WATERGATE sequence for water suppression (21) for selectively labeled samples (15N-leucine, 15N-valine and 15N-phenylalanine). NMR experiments were performed either on a Unity 500 machine (Varian) or a Bruker AM500 spectrometer (Bruker, Billerica, MA). Sample concentrations were 1.5 mM for the double-labeled sample and 0.4 mM for the selectively labeled samples. Spectra were measured at 298 K. The data were processed and analyzed with the programs prosa (22) and xeasy (23), respectively.
Regulated CD2BP2 Expression and Cell Sorting.
Jurkat cells expressing the tetracycline responsive transcription activator (Jurkat-tTA) were generated according to the manufacturer’s protocol (Clontech). For expression and sorting analysis, typically, 106 cells were electroporated with 20 μg pBI-G CD2BP2 cDNA by using a cell porator (Bio-Rad) set at 800 μF and 250 V, incubated in standard B′ medium [RPMI medium 1640 with 10% fetal calf serum, 2 mM l-glutamine, 1 mM sodium pyruvate, and 50 units/ml penicillin-streptomycin (Gibco/BRL)] for 24 h and were stained by fluorescence-2-galactopyranoside as described (24). In brief, 107 cells were loaded with fluorescence-2-galactopyranoside for 2 min at 37°C in hypoosmotic medium (0.5× RPMI medium 1640/2% fetal calf serum/10 mM Hepes, pH 7.6/0.5 mM fluorescence-2-galactopyranoside) and were returned to normal osmolarity (B′ medium) for incubation for 1 h at 4°C. Cell sorting was performed by using either a Vantage (Becton Dickinson) or MoFlo (Cytomation, Fort Collins, CO) cell sorter.
Ca2+ Flux and IL-2 Assays.
Ca2+ influx was analyzed as reported (25, 26). For IL-2 production assays, sorted cells were plated onto U-bottom 96-well plates at 105 cells per well in 200 μl of B′ medium, which was supplied with 0.5 nM phorbol myristate acetate and, when necessary, either a combination of anti-T112 plus anti-T113 anti-CD2 mAbs or anti-CD3 mAb 2Ad2 (1:100 dilution of ascites). After 24 h, the amount of IL-2 in the medium was assayed by ELISA (Endogen).
RESULTS AND DISCUSSION
To define proteins interacting with the CD2 cytoplasmic region, we used a yeast two-hybrid system (11) and isolated eight clones from an activated human T cell cDNA library, which demonstrated strong interaction with the CD2 cytoplasmic tail. DNA sequencing showed that five contained SH3 domains (27, 28), one contained a WW domain (29), and the remaining clones lacked both of these known domains. The latter two clones were derived from the same gene but differed in size. 5′ RACE analysis (13) showed that the complete coding region of the gene product termed CD2BP2 specifies a 341-aa polypeptide (Fig. 1A). Although standard blast homology search (30) with the entire sequence did not reveal any significant homology with entries in SWISSPROT and translated sequences of the GenBank database, the C-terminal region showed local homology with some Caenorhabditis elegans and yeast (Saccharomyces cerevisiae) proteins of unknown function. Sequence alignment identified 17 residues that are particularly conserved, comprising the following motif: GPF[or Y]xxxxM[orV]xxWxxxGYF (Fig. 1B). We note that a segment near the C terminus contains a consensus site for potential tyrosine phosphorylation. RNA blot analysis indicates that CD2BP2 is expressed in a wide variety of organs as ≈1.35- and 4-kilobase RNAs (data not shown).
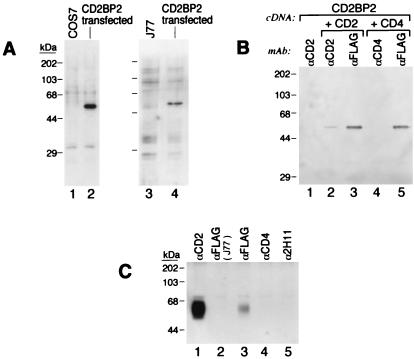
To characterize the nature and associations of the CD2BP2 protein, we transfected Jurkat cells (J77) and COS7 cells with the CD2BP2 cDNA tagged with a FLAG epitope. The apparent molecular size (≈50 kDa) detected by anti-FLAG mAb Western blotting is larger in both cell types than that expected based on the theoretical molecular weight (molecular weight = 37.6) (Fig. 2A). This anomalous mobility is likely a consequence of the acidic nature of the CD2BP2 protein (pI = ≈4.49). To determine whether CD2BP2 associates specifically with CD2, we cotransfected COS7 cells with FLAG-CD2BP2 and CD2 cDNAs or, as a control, CD4 cDNA. Subsequently, the CD2 protein was immunoprecipitated with anti-CD2 mAb and association with CD2BP2 examined by Western blotting with anti-FLAG mAb. As shown in Fig. 2B, FLAG-CD2BP2 specifically associates with CD2 (Fig. 2B, lane 2) but not with CD4 (Fig. 2B, lane 4). The amount of CD2BP2 associated with CD2, however, is only a fraction of the CD2BP2 protein expressed in the COS7 cells.
Figure 2.
In vivo binding of CD2 to CD2BP2. (A) Western blot analysis of lysates from parental cells or cells transfected with the cDNA encoding the CD2BP2 protein tagged with an N-terminal FLAG epitope. The FLAG epitope was detected on a protein migrating with apparent MW of 50KD. (Left) Lanes: 1, control untransfected COS7 cells; 2, transiently transfected COS7 cells. (Right) Lanes 3, control untransfected Jurkat (J77) cells; 4, a representative stable Jurkat FLAG-tagged CD2BP2 cDNA transformant. (B) Interaction between CD2BP2 and CD2 in COS7 cells. COS7 cells were transfected with FLAG-CD2BP2 cDNA (lane 1), with FLAG-CD2BP2 and a human CD2 cDNA (lanes 2 and 3), or with FLAG-CD2BP2 and a human CD4 cDNA (lanes 4 and 5) then were lysed and immunoprecipitated with the anti-FLAG mAb M2 (lanes 3 and 5), with anti-CD2 mAb (anti-T111) (lanes 1 and 2), or with anti-CD4 mAb (19Thy5D7) (lane 4). Subsequently, the amount of FLAG-CD2BP2 in the immunoprecipitates was assayed by anti-FLAG mAb M2 Western blotting. (C) Binding of CD2BP2 to CD2 in Jurkat transformants. Lysates from control Jurkat cells (lane 2) and Jurkat cells stably transfected with FLAG-CD2BP2 cDNA (lanes 1 and 3–5) were immunoprecipitated with anti-CD2 mAb anti-T111 (lane 1), anti-FLAG M2 mAb (lanes 2 and 3), anti-CD4 mAb (lane 4), or the irrelevant leucine zipper mAb 2H11 (lane5), and the associated CD2 protein was detected by using the M32 anti-CD2 rabbit heteroantisera. ECL was used as described.
To determine next whether the interaction between CD2 and CD2BP2 could be detected in lymphoid cells, lysates of J77 FLAG-CD2BP2 transfectants were immunoprecipitated with anti-FLAG mAb followed by Western blotting with anti-CD2 heteroantisera (Fig. 2C). Although anti-FLAG mAb precipitated CD2 from the Jurkat cells transfected with FLAG-CD2BP2 (Fig. 2C, lane3), anti-CD4 mAb (Fig. 2C, lane4), and an irrelevant control mAb (anti-leucine zipper mAb 2H11) (Fig. 2C, lane 5), anti-CD8 (21Thy2D3) and anti-HLA (W6–32) (unpublished results) showed no significant CD2 coprecipitation. Moreover, as expected, in untransfected J77 cells, no CD2 was detected in the anti-FLAG mAb immunoprecipitate (Fig. 2C, lane 2).
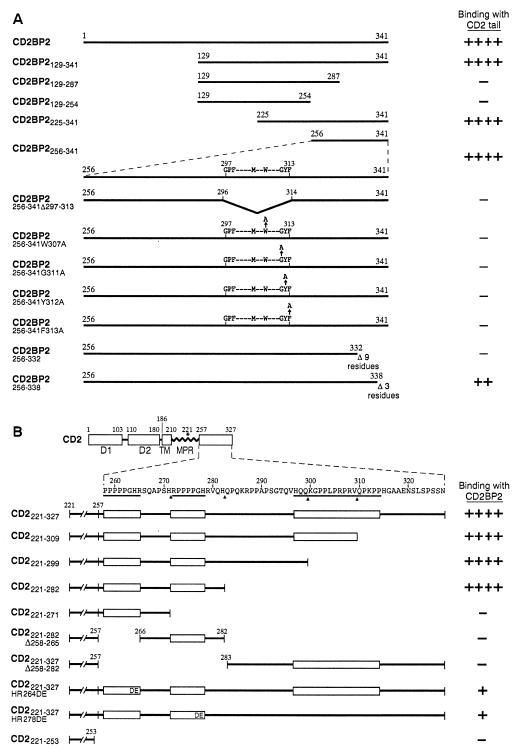
Molecules such as lck and fyn, reported to bind to CD2, each contain SH3 domains (27, 28, 31, 32). Because CD2BP2 possessed no known protein–protein interaction binding motif, we first generated CD2BP2 deletion mutants and then determined their ability to bind to CD2 in the yeast two-hybrid system. All clones with N-terminal deletions (CD2BP2129–341, CD2BP2225–341, CD2BP2256–341) showed binding activity whereas two C-terminal deletion mutants tested (CD2BP2129–287, CD2BP2129–254) did not (Fig. 3A). This finding is of note because the shortest functional clone, namely, CD2BP2256–341, contains the motif identified by sequence homology (amino acids 297–317) (Fig. 1). Consistent with the notion that the identified motif may be functionally relevant, a deletion clone lacking those 17 residues, CD2BP2256–341Δ297–313, failed to bind to CD2 as detected in complementation analysis.
Figure 3.
Mutational analysis of CD2 and CD2BP2 interaction sites. For each construct, the strength of interaction was scored by yeast two-hybrid analysis. (A) CD2BP2 and its derivatives were tested for their binding to the CD2 cytoplasmic region (CD2221–327). Note that the 17 residues highlighted in Fig. 1B are residues 297–313. The amino acid numbers corresponding to the boundaries of each fragment are shown. The nomenclature denotes the residues that were contained or modified in each construct. (B) Human CD2 and its derivatives tested for binding with CD2BP2. In the schematic diagram, D1 and D2 represent domains 1 and 2 of the extracellular region of CD2, respectively. TM, transmembrane segment; MPR, membrane proximal region. The three proline rich regions conserved among all species characterized to date are underlined. Binding is scored as described in Materials and Methods.
To test further the functional importance of this segment, four alanine scanning mutants involving the most conserved residues were generated (W307A, G311A, Y312A, and F313A). Each mutation abolished detectable binding, suggesting that the 17-residue motif is critical for CD2 binding. To further delineate any other region of CD2BP2 responsible for the CD2 binding, two additional constructs whose C termini were truncated also were generated. Of note, with the three-residue C-terminal truncation (CD2BP2256–338), interaction was weakened whereas, with the nine-residue truncation (CD2BP2256–332), binding was no longer detectable. Collectively, these results support the idea that C-terminal residues of CD2BP2 may fold in such a way that they interact directly or indirectly with the 17-residue motif to mediate binding to CD2.
To map the CD2BP2 binding site on CD2, several CD2 cytoplasmic tail truncation mutants were generated for yeast two-hybrid analysis (Fig. 3B). Two deletion mutants, CD2221–299 and CD2221–282, lacking the conserved 18-aa C-terminal segment that binds to the SH3 domains of fyn (37), and CD2BP1 (unpublished results) still maintained strong binding to CD2BP2. By contrast, when either of the two PPPGHR sequences was deleted (CD2221–271 and CD2221–282Δ258–265), binding was abolished. Moreover, mutation within either sequence (CD2221–327HR264DE or CD2221–327HR278DE) markedly attenuated binding. We conclude that the two PPPGHR sequences within the CD2257–282 cytoplasmic tail segment mediate the binding to CD2BP2.
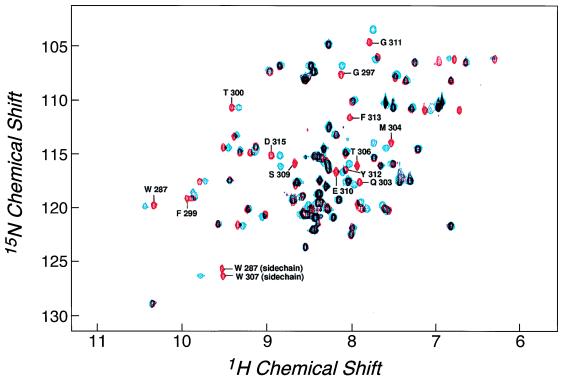
To define further the binding between CD2BP2 and the cytoplasmic tail of CD2 in vitro, NMR titration studies were performed. Fig. 4 shows the 15N-1H correlation spectrum of the purified binding domain of CD2BP2 comprising amino acids 256–341 of the protein, where each NH group within the protein results in a signal in the NMR spectrum. The chemical shift dispersion is in agreement with a folded, native protein domain. Backbone resonance assignments were performed to define the in vitro binding at the level of individual amino acids. Fig. 4 also shows the 15N-1H correlation spectrum of the CD2BP2 protein domain after the addition of equimolar amounts of a GST-fusion of the truncated CD2 tail (CD2221–282). This truncated CD2 tail segment contains both PPPGHR regions and showed full affinity in the yeast two-hybrid assay (Fig. 3B). Because the GST-CD2 tail fusion protein was not enriched isotopically with 15N, only the resonances arising from the CD2BP2 domain are seen in this spectrum. Compared with the unligated spectrum, certain of the resonances display significant chemical shift differences indicative of a binding event. This chemical shift difference indicates either that the respective residue directly participates in complex formation or is involved in a ligand-induced conformational change at a distance from the binding site. Because only a subset of residues show a significant chemical shift difference, it is most probable that this difference is caused by direct involvement in binding rather than a large conformational change of the protein. The latter would be expected to affect most of the residues within such a small protein domain. No chemical shift differences can be seen when the GST protein alone is used in the titration experiments (unpublished results). The latter result indicates that the truncated CD2 tail is responsible for the binding.
Figure 4.
HSQC spectra of the isolated and complexed CD2BP2 binding domain. The comparison shows that some NH resonances denoted by their sequence number in the uncomplexed spectrum (red) shift on the addition of equimolar amounts of a construct comprising a CD2-tail segment (amino acids 248–310 of CD2) fused to GST (blue).
Resonances demonstrating chemical shift differences of >0.1 ppm (1H) and >0.5 ppm (15N) on binding are W287, G297, F299, Q303, M304, T306, W307, S309, G311, Y312, F313, and D315. Except for W287 and D315, each of these residues lies within the 17-aa segment suggested from the mutational studies to be essential for binding. The chemical shifts of the CD2BP2 cross peaks change in a manner dependent on the concentration of the ligand (unpublished results). This observation indicates that CD2BP2 is in fast exchange between free and bound forms, and, hence, the averaged signals are observed. Because there is no significant line broadening (<2Hz) for proton signals that shift by ≈50 Hz, we estimate that the off rates are faster than 1,000 s−1 because the koff is πΔ2/(4 δν) for a two-site exchange where Δ is the chemical shift difference between the two states and δν is the line broadening.
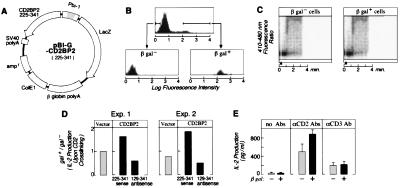
Next, the functional role of the CD2BP2 in T lymphocytes was investigated. To this end, we used a transient transfection assay in Jurkat cells by using a reporter protein expression system based on a bidirectional promoter in combination with FACS-gal sorting as described (Fig. 5 A and B) (24, 33). Jurkat cells transfected with CD2BP2225–341 were separated by FACS into plasmid expressing and nonexpressing subpopulations based on β-galactosidase (β-gal) activity and then were examined in functional assays. As shown in Fig. 5C, regardless of CD2BP2225–341 (i.e., β-gal) expression, there was no difference in terms of Ca2+ influx induced by CD2 crosslinking. However, although the pBI-G vector alone did not alter the CD2-triggered IL-2 production, the pBI-G-CD2BP2225–341 sense construct reproducibly enhanced IL-2 production 150–200%. By contrast, expression of the anti-sense strand decreased IL-2 production to ≈50% (Fig. 5D). To test whether CD2BP2 is also involved in T cell receptor-stimulated IL-2 production, we performed additional experiments in which transfected cells either were left unstimulated or were triggered by anti-CD2 or anti-CD3 mAbs. As shown in Fig. 5E, expression of CD2BP2 selectively enhances IL-2 production on CD2 crosslinking (clustering by a combination of anti-T112 plus anti-T113 mAbs). This selectivity of CD2BP2 for CD2-mediated activation is consistent with data showing differences in CD2 and CD3 signaling pathways (4, 5, 34, 35).
Figure 5.
Regulation of CD2-stimulated T cell activation by CD2BP2. Shown is the effect of the expression of the CD2 binding segment (amino acids 225–341) of CD2BP2 on Ca2+ influx and IL-2 production in Jurkat cells on CD2 crosslinking. (A) pBI-G vector system. This vector has a bidirectional promoter (Pbi-1) that allows the expression of two genes on either side of the promoter. Thus, both CD2BP2225–341 and β-gal are expressed, with the latter functioning as the reporter. (B) Sorting of β-gal positive and negative populations of Jurkat cells after transient CD2BP2225–341 transfection. Presorted and sorted populations are shown as florescence histograms of cell number on the y axis versus log fluorescence of the cleaved β-gal substrate fluorescence-2-galactopyranoside on the x axis. (C) CD2-triggered Ca2+ flux in β-gal− and β-gal+ cells. Ca2+ influx was measured as described (7, 8). Filled arrowhead indicates the time point when cells were stimulated with anti-T112 and anti-T113 (ascites 1:200 dilution). D) Effect of the expression vector pBI-G, pBI-G-CD2BP2225–341 sense and pBI-G-CD2BP2129–341 antisense on IL-2 production following CD2 crosslinking. The height of the bars shows the ratio of IL-2 produced from β-gal+ versus β-gal− subpopulations isolated by cell sorting from the same transient transfection. Results from two representative experiments are shown. For statistical analysis, a t test was used to examine the difference between the pairwise data (such as those from galactosidase [gal] positive [+] and negative [−] cells from each experiment set). This test showed clearly significant differences between gal+ and gal− cells on transfection of CD2BP2225–341 sense (P < 0.001) and CD2BP2129–341 antisense (P < 0.01) but no significant difference on transfection with the vector pBI-G (P > 0.1). (E) Effect of the expression of pBI-G-CD2BP2225–341 sense on IL-2 production of unstimulated Jurkat-tTA or Jurkat-tTA triggered by CD2 crosslinking or CD3 crosslinking. β-gal+ (solid bar) and β-gal− (hatched bar) cells were stimulated, as indicated in parallel. One of two representative experiments is shown in which IL-2 levels were measured in triplicate, and the mean ± SD is displayed. The mean and SD of the data regarding the amount of IL-2 produced by β-gal negative (positive) cells are as follows: with no stimulation, 25 ± 18 (28 ± 10); CD2 stimulation, 480 ± 159 (870 ± 83.5); and CD3 stimulation, 201 ± 58.1 (215 ± 54.0).
The CD2BP2 molecule has several unique features that distinguish it from SH3 and WW domain-containing proteins. Although there is some difference in their detailed specificity, SH3 and WW domains both bind to PxxP-containing sequences. Consistent with this specificity, we and several other groups have identified CD2-binding proteins with SH3 domains and determined that they mostly bind to the PPLP sequence (amino acids 302–305 in Fig. 3B), which is an SH3 ligand consensus site (refs. 31 and 32; unpublished results) within the most highly conserved portion of the CD2 tail segment. By contrast, CD2BP2 binds to a site containing the two tandem PPPGHR motifs but not to the SH3 ligand consensus sites (Fig. 3B). Moreover, unlike SH3 domains whose ligands require only eight residues for binding (27, 28), CD2BP2 requires a 21-residue segment. We anticipate that this CD2BP2 binding segment transiently assumes a configuration necessary for interaction, perhaps regulated by divalent cations. Conservation of the dibasic residue within the two tandem motifs including the histidine in human, mouse, rat, and horse CD2 is noteworthy. We previously have shown that the PPPGHR-containing region of the CD2 tail is essential for CD2 ectodomain-stimulated IL-2 production (7, 8). Although the mechanism by which the tandem PPPGHR sequences trigger IL-2 gene activation on CD2 clustering is still unclear, it is possible that this region is needed for the proper orientation and/or function of the downstream SH3 ligand binding motif. Consistent with this notion, replacement of the dibasic HR residues of the PPPGHR segment with DE residues weakens not only the binding of CD2BP2 to this region but also that of the SH3 domain of p59fyn to the downstream SH3 consensus site (unpublished results). Thus, we speculate that CD2BP2 may play a biologic role in coordinating the binding of other interactors to the more C-terminal region of the CD2 tail.
SH3 domains are made up of 5–6 antiparallel β-strands forming a compact, barrel-like structure. As shown by analysis of complexes of SH3 domains and their ligands, the ligand for a given SH3 domain forms a left-handed polyproline-type II helix whose interactions with the SH3 domain are mediated primarily by hydrophobic residues within the binding site (27, 28). WW domains form a three-stranded antiparallel β-strand with one of the two conserved tryptophan residues crucially involved in the interaction with the proline-rich ligand (38). Our in vivo binding assays show that a number of aromatic residues of CD2BP2 probably are involved directly in the interaction with the proline-rich sequence motif of the CD2 cytoplasmic domain. However, structure-prediction methods and initial nuclear overhauser effect analysis (unpublished results) indicate the presence of a central α-helix within the binding domain of CD2BP2 (residues 301–311). This helix is predicted to reside within the conserved 17-aa sequence shown herein to be necessary for the binding of the proline-rich ligand. It therefore seems probable that the binding domain of CD2BP2 defines a class of proline-rich recognition domains. In this fold, an α-helical rather than a β-strand structure displays those aromatic and hydrophobic residues necessary for the binding to the proline-rich ligand. Given that the CD2BP2 protein involved in binding to the PPPGHR motif is expressed in different tissues and that there is conservation of the GPY[or F]xxxxM[or V]xxWxxxGYF sequence in other unrelated proteins derived from different species, it is likely that this interaction is not restricted to lymphocytes. Rather, it may represent a basis for protein–protein interaction.
Acknowledgments
We acknowledge the important early contributions of Dr. W. An to the cDNA cloning effort. K.N. is gratefully indebted to Dr. S. Okinaga and Teikyo Univ. for financial support. C.F. is supported by the Schweizerische Nationalfonds. This work was supported by National Institutes of Health Grants AI21226 to E.L.R. and AI27581 to G.W.
ABBREVIATIONS
- IL
interleukin
- CD2BP2
CD2 binding protein 2
- RACE
rapid amplification of cDNA ends
- GST
glutathione S-transferase
- β-gal
β-galactosidase
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF104222).
References
- 1.Moingeon P, Chang H-C, Sayre P H, Clayton L K, Alcover A, Gardner P, Reinherz E L. Immunol Rev. 1989;111:111–144. doi: 10.1111/j.1600-065x.1989.tb00544.x. [DOI] [PubMed] [Google Scholar]
- 2.Bierer B E, Sleckman B P, Ratnofsky S E, Burakoff S J. Annu Rev Immunol. 1989;7:579–599. doi: 10.1146/annurev.iy.07.040189.003051. [DOI] [PubMed] [Google Scholar]
- 3.Springer T A. Nature (London) 1990;346:425–434. doi: 10.1038/346425a0. [DOI] [PubMed] [Google Scholar]
- 4.Gollob J A, Li J, Reinherz E L, Ritz J. J Exp Med. 1995;182:721–731. doi: 10.1084/jem.182.3.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gollob J A, Li J, Kawasaki H, Daley J F, Groves C, Reinherz E L, Ritz J. J Immunol. 1996;157:1886–1893. [PubMed] [Google Scholar]
- 6.Boussiotis V A, Freeman G J, Griffin J D, Gray G S, Gribben J G, Nadler L M. J Exp Med. 1994;180:1665–1673. doi: 10.1084/jem.180.5.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang H-C, Moingeon P, Lopez P, Krasnow H, Stebbins C, Reinherz E L. J Exp Med. 1989;169:2073–2083. doi: 10.1084/jem.169.6.2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang H-C, Moingeon P, Pedersen R, Lucich J, Stebbins C, Reinherz E L. J Exp Med. 1990;172:351–355. doi: 10.1084/jem.172.1.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bierer B E, Bogart R E, Burakoff S J. J Immunol. 1990;144:785–789. [PubMed] [Google Scholar]
- 10.He Q, Beyers A D, Barclay A N, Williams A F. Cell. 1988;54:979–984. doi: 10.1016/0092-8674(88)90112-2. [DOI] [PubMed] [Google Scholar]
- 11.Finley R L, Brent R. Interaction Trap Cloning with Yeast. New York: Oxford Univ. Press; 1995. [Google Scholar]
- 12.Higuchi R, Krummel B, Saiki R K. Nucleic Acids Res. 1988;16:7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frohman M A. Methods Enzymol. 1993;218:340–356. doi: 10.1016/0076-6879(93)18026-9. [DOI] [PubMed] [Google Scholar]
- 14.Sakihama T, Smolyar A, Reinherz E L. Proc Natl Acad Sci USA. 1995;92:6444–6448. doi: 10.1073/pnas.92.14.6444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li J, Smolyar A, Sunder-Plassmann R, Reinherz E L. J Mol Biol. 1996;263:209–226. doi: 10.1006/jmbi.1996.0570. [DOI] [PubMed] [Google Scholar]
- 16.Johnstone A, Thorpe R. Immunochemistry in Practice. Oxford: Blackwell Scientific; 1996. pp. 211–225. [Google Scholar]
- 17.Freund C, Ross A, Guth G, Plückthun A, Holak T A. FEBS Lett. 1993;320:97–100. doi: 10.1016/0014-5793(93)80070-b. [DOI] [PubMed] [Google Scholar]
- 18.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 19.Kay L E, Ikura M, Tschudin R, Bax A. J Magn Reson. 1990;89:496–514. doi: 10.1016/j.jmr.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 20.Mori S, Abeygunawardara C, Johnson M O, van Zijl P C. J Magn Reson B. 1995;108:94–98. doi: 10.1006/jmrb.1995.1109. [DOI] [PubMed] [Google Scholar]
- 21.Sklenar V, Piotto M, Leppik R, Saudek V. J Magn Reson. 1993;102:241–245. [Google Scholar]
- 22.Güntert P, Dötsch V, Wider G, Wüthrich K. J Biomol NMR. 1992;2:619–630. [Google Scholar]
- 23.Eccles C, Güntert P, Billeter M, Wüthrich K. J Biomol NMR. 1991;1:111–130. doi: 10.1007/BF01877224. [DOI] [PubMed] [Google Scholar]
- 24.Nolan G P, Herzenberg L A. Proc Natl Acad Sci USA. 1988;85:2603–2607. doi: 10.1073/pnas.85.8.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reem G H, Carding S, Reinherz E L. J Immunol. 1987;139:130–134. [PubMed] [Google Scholar]
- 26.Targan S R, Deem R L, Liu M, Wang S, Nel A. J Immunol. 1995;154:664–675. [PubMed] [Google Scholar]
- 27.Ren R, Mayer B J, Cicchetti P, Baltimore D. Science. 1993;259:1157–1161. doi: 10.1126/science.8438166. [DOI] [PubMed] [Google Scholar]
- 28.Musacchio A, Wilmannis M, Saraste M. Prog Biophys Mol Biol. 1994;61:283–297. doi: 10.1016/0079-6107(94)90003-5. [DOI] [PubMed] [Google Scholar]
- 29.Bork P, Sudol M. Trends Biochem Sci. 1994;19:531–533. doi: 10.1016/0968-0004(94)90053-1. [DOI] [PubMed] [Google Scholar]
- 30.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bell G M, Fargnoli J, Bolen J B, Kish L, Imboden J B. J Exp Med. 1996;183:169–178. doi: 10.1084/jem.183.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gassmann M, Amrein K E, Flint N A, Schraven B, Burn P. Eur J Immunol. 1994;24:139–144. doi: 10.1002/eji.1830240121. [DOI] [PubMed] [Google Scholar]
- 33.Baron V, Feundlieb S, Gossen M, Bujard H. Nucleic Acids Res. 1995;23:3605–3606. doi: 10.1093/nar/23.17.3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Semnani R T, Svoboda K, Khoshnood B, Seventer G A. Scand J Immunol. 1998;47:436–443. doi: 10.1046/j.1365-3083.1998.00312.x. [DOI] [PubMed] [Google Scholar]
- 35.Sunder-Plassmann R, Reinherz E L. J Biol Chem. 1998;273:24249–24257. doi: 10.1074/jbc.273.37.24249. [DOI] [PubMed] [Google Scholar]
- 36.Lillie S H, Brown S S. Nature (London) 1992;356:358–361. doi: 10.1038/356358a0. [DOI] [PubMed] [Google Scholar]
- 37.Li, J., Nishizawa, K., An, W., Hussey, R. E., Lialios, F. E., Salgia, R., Sunder-Plassmann, R. & Reinherz, E. L. EMBO J., in press. [DOI] [PMC free article] [PubMed]
- 38.Macias M J, Hyvonen M, Baraldi E, Schultz J, Sudol M, Saraste M, Oschkinat H. Nature (London) 1996;382:646–649. doi: 10.1038/382646a0. [DOI] [PubMed] [Google Scholar]