Abstract
T cells recognizing poorly displayed self determinants escape tolerance mechanisms and persist in the adult repertoire. The process by which these T cells are primed is not clear, but once activated, they can cause autoimmunity. Here, we show that dendritic cells treated with interleukin 6 (IL-6) process and present determinants from a model native antigen in a qualitatively altered hierarchy, activating T cells in vitro and in vivo against determinants that were previously cryptic because of poor display. IL-6 does not induce conventional maturation of dendritic cells but alters the pH of peripheral, early endosomal compartments and renders the cells more susceptible to killing by chloroquine. Acidification of endosomes by ouabain mimics the effect of IL-6 and allows processing of the same cryptic determinant. These results suggest that cytokines such as IL-6 could initiate and help to propagate an autoimmune disease process by differentiating dendritic cells into a state distinct from that induced by normal maturation.
A characteristic of many human autoimmune diseases and rodent disease models is the spreading of the anti-self T cell response to include reactivity against cryptic determinants (1–4). Determinants can seem cryptic because of limited display by antigen-presenting cells (APC) after normal processing of native antigen or because of limitations in the T cell repertoire. Available T cells specific for cryptic determinants can be activated by molecular mimicry, whereby T cells are primed initially against a dominant determinant derived from an infectious pathogen, then continue to be stimulated by cross-reactive cryptic self-determinants during inflammation, thereby becoming autoreactive (5–8). Alternatively, inflammatory cytokines found in autoimmune lesions may alter processing by professional APC so that previously hidden self determinants are revealed to the T cell repertoire (9, 10).
Dendritic cells are APC that are specialized to prime naive T cells (11, 12) and to focus T cell responses on the determinants they present (13). Dendritic cells are matured into a state that activates T cells by inflammatory cytokines, lipopolysaccharide, or CD40L (14–17). The maturation process involves cell migration and alteration in intracellular major histocompatibility complex (MHC) class II transport and surface expression, causing antigen acquired in peripheral sites to be well presented to T cells in lymph nodes (11, 12, 17, 18). Maturation also decreases the ability of dendritic cells to take up and process antigen so that T cells are focused on those antigens previously derived from sites of inflammation (14, 15). However, the pleiotropic cytokine interleukin (IL)-6, although often present in inflammatory sites, does not induce similar maturation of dendritic cells (19). Recently, IL-6 has been shown to be essential for the development of autoimmunity in rodent disease models (20–22). It seemed likely to us that IL-6 might have an effect on dendritic cells without inducing their maturation and that this effect might have relevance to some disease situations. To test this hypothesis, we investigated the effect of IL-6 on the processing of the well characterized model antigen hen-egg lysozyme (HEL) by dendritic cells.
We found that dendritic cells treated with IL-6 processed native HEL for a panel of various T cell hybridomas specific for HEL determinants, so that a qualitatively different profile of hybridoma responses was observed compared with responses stimulated by control dendritic cells. In particular, dendritic cells treated with IL-6 and processing whole HEL activated a hybridoma specific for a determinant that was otherwise cryptic because of poor display. Dendritic cells exposed to IL-6 and HEL and injected into mouse footpads resulted in T cells being primed in vivo against cryptic determinants. When IL-6 itself was included in an immunization protocol designed to prime anti-HEL T cell responses, we found that splenocytes from mice that had been injected with HEL plus IL-6 in incomplete Freund’s adjuvant (IFA) responded to cryptic HEL determinants. Dendritic cells (but not B cells) isolated from lymph nodes draining HEL plus IL-6 injection sites stimulated T cell hybridoma cells specific for a cryptic determinant. We investigated the mechanistic effect of IL-6 on dendritic cells and found that peripheral endosomes were much more acidified therein than in control cells. This influence of IL-6 on dendritic cells represents a distinct differentiation pathway for this important cell type that, because it resulted in T cells being primed against cryptic determinants, could play a role in the generation of anti-self immunity.
MATERIALS AND METHODS
Media.
Iscove’s modified Dulbecco’s medium (GIBCO/BRL) supplemented with 10% fetal calf serum and 5 × 10−5 M 2-mercaptoethanol was used for dendritic cell culture (DC). RPMI medium 1640, 5% fetal calf serum, and 5 × 10−5 M 2-mercaptoethanol was used for hybridoma culture and hybridoma assays. HL-1 serum-free medium (BioWhittaker) was used for ex vivo lymphocyte proliferation assays.
DC.
Dendritic cells were grown from BALB/c or CBA female 8- to 12-week-old mouse bone-marrow precursor cells for 6 days in the presence of 10 ng/ml recombinant murine granulocyte–macrophage colony-stimulating factor (GM-CSF; Autogen Bioclear, Calne, U.K.) as described (23). On day 6, nonadherent cells and loosely adherent colonies were removed by gentle pipetting and subcultured overnight at 106 cells per ml with 10 ng/ml GM-CSF with 5 ng/ml recombinant murine IL-6 (DC/IL-6; Autogen Bioclear) or without IL-6 (DC); in some experiments 20 μM chloroquine (Sigma) also was added. The next day, nonadherent cells were washed three times and used for hybridoma assays, staining, or immunization. These cells were routinely >80% dendritic cells [MHC class II positive and CD11c positive as indicated by N418 mAb (a gift from A. Livingstone, Imperial College, London, biotinylated by C. Van Wely, Glaxo-Wellcome, Stevenage, U.K.)]. The remaining cells were MHC class II negative granulocytes. Therefore, this study assumed that the observed effects were caused by presentation by dendritic cells, although it is difficult to rule out the possibility of an influence of a very small contaminating population of other presenting cells on the overall response.
Hybridoma Assay.
DC or DC/IL-6 were cultured at 104 cells per well with hybridoma cells at 5 × 104 cells per well in the presence of whole HEL (Boehringer Mannheim) or HEL peptide (synthesized at the Imperial Cancer Research Fund) at graded doses, all in triplicate. After 24 h, supernatants were removed, and IL-2 levels were assayed by proliferation of the IL-2-dependent cell line CTLL measured by [3H]thymidine (Amersham) incorporation over the last 18 h of a 36-h culture. No supernatants from DC, DC/IL-6, or any hybridoma alone contained significant amounts of IL-2.
Immunizations and Proliferation Assays.
DC or DC/IL-6 from CBA or BALB/c mice were cultured with whole HEL at 50 μg/ml for 6 h, washed three times, and resuspended at 106 cells per ml in PBS, and 5 × 104 cells per foot were injected into opposite hind footpads of six CBA or BALB/c mice. Five days later, draining popliteal and inguinal lymph nodes from either side of the mice were pooled, and lymphocytes obtained were cultured at 5 × 105 cells per well in triplicate with HEL, ovalbumin (Sigma, grade VII), or HEL peptides at 60 μ/ml for 4 days. Proliferation was measured by [3H]thymidine incorporation over the last 18 h of culture. HEL (100 μg per mouse) with or without IL-6 (5 ng per mouse) in PBS was emulsified (1:1, vol/vol) in IFA (Sigma), then injected into five CBA or BALB/c mice at the base of the tail. After 10 days, spleens were harvested, and splenocytes were cultured at 106 cells per well for 5 days with antigens as above. Background values of proliferation without added antigen (≈2,000 cpm) were subtracted.
APC Separation.
HEL with or without IL-6 as above or ovalbumin (100 μg) with IL-6 were emulsified in IFA as above and injected into rear footpads of 10 BALB/c mice. After 5 days, draining nodes were harvested. The resulting lymphocytes were depleted of T cells with an anti-Thy 1.2 antibody (Serotec) and rabbit complement (Cedarlane Laboratories) and separated into B cells and N418-positive dendritic cells as described (24). The dendritic cell fraction was >85% pure (contaminants were B220-positive B cells), whereas the B cell fraction contained <2% dendritic cells. Cell fractions were cultured at graded concentrations with hybridoma cells specific for HEL 2–16 at 5 × 104 cells per well with or without HEL 2–16 peptide at 100 μg/ml for 24 h. IL-2 secreted by the hybridoma was measured by using CTLL cells as above.
Locating Acidic Vesicles with N-{3-[(2,4-Dinitrophenyl)amino]propyl}-N-(3-aminopropyl)methylamine Dihydrochloride (DAMP).
Acidic vesicles were located with DAMP as described (25). Briefly, DC and DC/IL-6 were incubated with 30 μM DAMP (Molecular Probes) for 30 min, then washed, fixed, made permeable, stained with a mouse anti-dinitrophenyl antibody (a gift from B. Askonas, Imperial College, London), washed again, and then stained with a fluorescein isothiocyanate-labeled second-layer goat anti-mouse antibody (Sigma). After further washing, these cells were mounted on poly-l-lysine-coated slides and visualized on a Zeiss Axiovert 100 microscope in conjunction with a Bio-Rad MRC 1024 laser scanning confocal imaging system.
Ouabain Treatment of Dendritic Cells.
DC (104 cells) were incubated with graded doses of ouabain (Sigma), 50 μM HEL 2–16 peptide, or whole HEL and 5 × 104 T cell hybridoma cells specific for HEL 2–16 for 24 h. The IL-2 concentrations in the supernatants from these cultures were determined by using a sandwich ELISA kit (Genzyme). CTLL cells could not be used in these experiments, because they are very sensitive to the ouabain in the supernatants.
RESULTS
IL-6 Qualitatively Alters Processing of HEL by in Vitro Cultured Dendritic Cells.
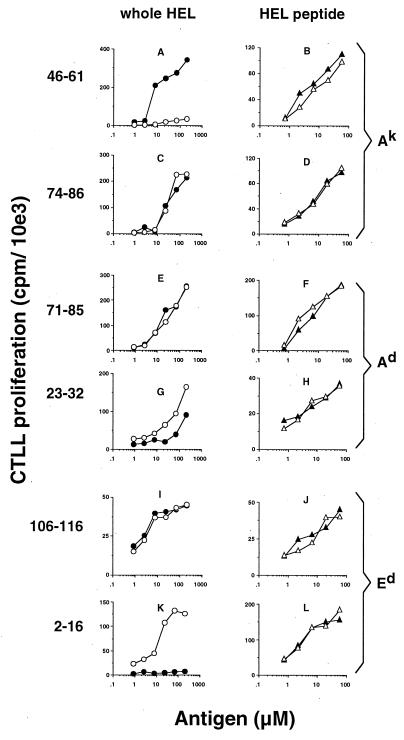
Dendritic cells were grown for 6 days from BALB/c (H-2d) or CBA (H-2k) mouse bone-marrow precursors and then subcultured overnight, with or without IL-6. These cells were then cocultured with whole HEL or an HEL-peptide determinant and an epitope-specific T cell hybridoma. We found that when cocultured with whole HEL, DC/IL-6 elicited a qualitatively different profile of T hybridoma responses compared with DC (Fig. 1 Left). When I-Ak-restricted determinants were studied, DC/IL-6 presented the codominant determinant 46–61 much less efficiently than DC (Fig. 1A), whereas display of the subdominant determinant 74–86 (Fig. 1C) was not significantly affected. When I-Ad-restricted determinants were studied, DC/IL-6 and DC presented the subdominant determinant 71–85 similarly (Fig. 1E), but 23–32 (Fig. 1G) was slightly better displayed by DC/IL-6. DC/IL-6 presented the dominant Ed determinant 106–116 as well as DC (Fig. 1I). Most notably, only DC/IL-6 efficiently processed and displayed the cryptic determinant 2–16 from whole HEL (Fig. 1K). DC and DC/IL-6 were able to present the respective peptide form of all determinants to the T cell hybridomas equivalently (Fig. 1 Right). This control shows that both DC and DC/IL-6 could present each determinant if it required minimal processing but that they processed native HEL differently; in several instances, they activated two distinct sets of determinant-specific hybridomas.
Figure 1.
DC/IL-6 process and present a different profile of determinants from whole HEL compared with DC. The immunogenicity of the determinants studied has been described (57). DC/IL-6 (open symbols) or DC (closed symbols) from CBA (A–D) or BALB/c (E–L) mice were cultured with hybridoma cells of the specificity indicated, as well as with whole HEL (circles, Left) or appropriate peptide determinants (triangles, Right). Dendritic cells were cultured from bone-marrow precursors and treated with IL-6, as described in Materials and Methods. Hybridoma responses are expressed as proliferation of CTLL cells as the mean of triplicates with all values within 10% of the mean. With no antigen, hybridoma responses to DC/IL-6 and DC were not significantly different, and cpm values were always less than 2,000. Each hybridoma was tested in at least three independent experiments, each producing similar results. 10e3 = 103.
We next investigated whether DC/IL-6 could prime T cells from the in vivo repertoire against cryptic and subdominant HEL determinants. HEL-exposed DC and DC/IL-6 were injected into both hind footpads of CBA and BALB/c mice. After 5 days, the recall specificities of draining node lymphocytes for HEL or HEL determinants were tested. In CBA mice (Fig. 2A), DC/IL-6 immunization caused a decrease in the response to the codominant determinant 46–61, and the response to whole HEL was decreased. In BALB/c mice (Fig. 2B), T cells from nodes draining DC/IL-6 immunization sites responded well to N-terminal HEL peptides, namely subdominant determinants 11–25 and 23–32 and particularly the cryptic determinant 2–16. Responses to other determinants were unchanged, and the response to whole HEL was increased relative to cells collected from nodes draining DC immunization sites. These in vivo responses are consistent with the in vitro responses of the T hybridomas to DC/IL-6 and DC.
Figure 2.
Lymphocytes from mice immunized with DC/IL-6 pulsed with HEL respond to subdominant and cryptic HEL determinants. Lymphocytes from nodes draining sites of injection of DC (black bars) or DC/IL-6 (white bars) in CBA (A) and BALB/c (B) mice were cultured with the proteins and peptides shown. Proliferation was measured by [3H]thymidine incorporation over the last 18 h of culture. Results are expressed as means of triplicates; all values fell within 10% of the mean. The experiment was repeated three times with similar results.
Injected IL-6 Alters the ex Vivo Response to HEL and Affects Processing of HEL by Dendritic Cells in Situ.
One index of the relative immunogenicity in vivo of MHC class II restricted determinants is defined by the T cell proliferative responses to these determinants from mice immunized with whole antigen; dominant determinants elicit the greatest responses (4). We investigated whether injecting IL-6 with antigen could affect this hierarchy of T cell responses. Mice were immunized at the base of the tail with HEL, with or without IL-6, emulsified in IFA. After 10 days, splenocytes were cultured with HEL or HEL determinants, and proliferation was measured for each individual mouse. In CBA mice (Fig. 3A), the response to the codominant determinant 46–61 was decreased when IL-6 was included. In BALB/c mice, the responses to whole HEL, 11–25, 23–32, and most dramatically 2–16 were up-regulated (Fig. 3B). Dendritic cells, not B cells, are the APC type that presents dominant determinants processed from injected antigen emulsified in adjuvant (24). However, because IL-6 is a growth/differentiation factor for B cells (26), we investigated which APC type was presenting the cryptic determinant in this case. BALB/c mice were injected with HEL in IFA (Fig. 3C) or HEL plus IL-6 in IFA (Fig. 3D) into opposing rear footpads. After 5 days, draining node lymphocytes were depleted of T cells, and dendritic cells or B cells were separated from the remaining population before coculture with a T cell hybridoma specific for the cryptic determinant HEL 2–16. Only dendritic cells from draining sites of HEL plus IL-6 immunization were able to stimulate the hybridoma efficiently and directly (Fig. 3D, open triangles), whereas all APC could present the 2–16 peptide determinant to the hybridoma. In a control experiment, neither B cells nor dendritic cells from lymph nodes draining sites of immunization with ovalbumin plus IL-6 stimulated the hybridoma specific for 2–16, unless peptide determinant was also present (Fig. 3E). These results show that the raised concentration of IL-6 affected the processing of HEL by dendritic cells in vivo, which then displayed the cryptic determinant in lymph nodes 5 days later. The altered specificities of the T cell responses to cryptic and other determinants shown in Fig. 3 A and B were therefore very likely caused by the action of IL-6 on dendritic cells.
Figure 3.
Splenocytes from mice immunized with HEL plus IL-6 emulsified in IFA respond to subdominant and cryptic determinants of HEL, and dendritic cells isolated from lymph nodes draining the immunization sites (of HEL plus IL-6 in IFA) present the cryptic determinant to specific T cell hybridomas. Splenocytes from CBA (A; one of two experiments) and BALB/c (B; one of three experiments) mice immunized with either HEL in IFA (closed symbols) or HEL plus IL-6 in IFA (open symbols) were cultured with the proteins and peptides shown, and proliferation was measured as for Fig. 2. Data points represent responses of individual mice. (C–E) B cells (circles) or dendritic cells (triangles) from BALB/c mouse lymph nodes draining immunization sites of either HEL in IFA (C), HEL plus IL-6 in IFA (D) or, from a separate experiment, ovalbumin plus IL-6 in IFA (E) were incubated at graded doses with cryptic-determinant (HEL 2–16) specific hybridoma cells at 5 × 104 cells per well with (closed symbols) or without (open symbols) HEL 2–16 peptide at 100 μg/ml for 24 h. Hybridoma responses are expressed as in Fig. 1.
IL-6 Renders Dendritic Cells Vulnerable to Chloroquine.
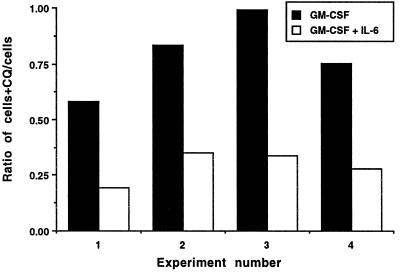
Inflammatory cytokines such as tumor necrosis factor α (TNF-α) mature dendritic cells in a well studied process involving up-regulation of surface MHC class II and a decrease in endocytosis (17, 18). However, dendritic cells treated with TNF-α did not present the cryptic HEL determinant to specific T cells (data not shown). Moreover, IL-6 does not mature dendritic cells in the conventional manner; levels of MHC class II, levels of other surface costimulatory and adhesion molecules, and the rate of endocytosis all remain unchanged (ref. 22; H.D., unpublished results). We therefore sought an alternative explanation of how IL-6 alters antigen processing. The acidotropic antigen-processing inhibitor chloroquine is used often to determine whether processing of particular determinants from an antigen requires acidic intracellular compartments. We attempted to use chloroquine to dissect the processing of HEL by dendritic cells treated with IL-6. However, as Fig. 4 shows, low levels of chloroquine were selectively toxic to dendritic cells treated with IL-6 and killed up to 80% of these cells overnight, whereas chloroquine killed only ≈15% of control cells. The remaining living cells treated with IL-6 were unable to present HEL or HEL peptide to any T cell hybridoma (data not shown). This result suggested a role for IL-6 in heightened vesicle acidification in dendritic cells, so that chloroquine might accumulate in and be toxic to IL-6 treated cells, but not to controls.
Figure 4.
Chloroquine selectively kills dendritic cells exposed to IL-6. Equal numbers of dendritic cells derived from day-6 bone marrow cultures were incubated overnight with GM-CSF or GM-CSF plus IL-6 with or without 20 μM chloroquine. The next day nonadherent dendritic cells were counted, and the ratios of the number of cells from cultures treated or untreated with chloroquine (CQ) from four experiments were calculated and plotted as shown.
IL-6 Acidifies Peripheral Early Endosomes of Dendritic Cells, and Acidification of Early Endosomes Results in the Processing of a Cryptic Determinant.
Perinuclear lysosomes are usually the most acidic intracellular vesicles. Fig. 5A shows that this localization was the case in control dendritic cells. However, in dendritic cells treated with IL-6, acidic vesicles were observed in peripheral areas of the cytoplasm just beneath the plasma membrane (Fig. 5B); these peripheral acidic compartments were accessed by Texas-Red dextran within 10 min of endocytosis (Fig. 5C) and hence were functionally part of the early endosome of these cells. Double labeling of these “early” Texas-Red-positive compartments confirmed that they contained the early endosome marker CD71 (the transferrin receptor), but the compartments were negative for the lysosomal marker LAMP-1 (not shown). We conclude that IL-6 acidifies peripheral compartments of dendritic cells, which form part of the early endosome.
Figure 5.
IL-6 acidifies peripheral compartments that are accessible to endocytosed antigen; independent of IL-6, acidification of early endosomes results in presentation of the cryptic determinant. The location of acidic compartments (shown in green) within DC (A) and DC/IL-6 (B) was visualized by using DAMP as described (25). (Bars = 10 μm.) (C) DC/IL-6 were allowed to internalize via endocytosis 1 mg/ml Texas-Red-conjugated dextran (Molecular Probes) by adding the dextran during the final 10 min of culture with DAMP. Cells were then washed, fixed, and stained for acidic compartments as in A and B. (Bar = 5 μm.) Colocalization of red dextran with green acidic vesicle is shown in yellow. Other markers such as HEL-fluorochrome conjugates also passed through acidic vesicles during the first 10 min of endocytosis (not shown). (D) Increasing ouabain concentration allows processing and presentation of HEL 2–16 from whole HEL (closed circles) but does not interfere with presentation of HEL 2–16 peptide (open circles) by DC to hybridoma cells specific for HEL 2–16. The hybridoma assay was carried out as in Fig. 1. Ouabain did not affect the rate of fluid-phase endocytosis (not shown). Experiments were carried out three times with similar results.
The pH of early (but not late) endosomes is controlled by the membrane Na+/K+ ATPase, and inhibition of this protein pump acidifies early endosomes (27, 28); IL-6 has been shown to inhibit this pump in hepatocytes (29), and mononuclear cells from rheumatoid arthritic patients (whose serum contains high levels of IL-6; ref. 30) also have dysfunctional Na+/K+ ATPase (31). The classical inhibitor of this pump is ouabain, and we found that, in the absence of IL-6, increasing doses of ouabain caused dendritic cells to process the cryptic determinant HEL 2–16 from whole HEL (Fig. 5D). We conclude that acidification of early endosomes independent of IL-6 is sufficient to allow processing of the cryptic determinant from whole HEL.
DISCUSSION
The major finding of this study was that exposure of dendritic cells to IL-6 dramatically altered the specificity of the T cell response to native HEL. Both in vitro and in vivo, dendritic cells treated with IL-6 were much less efficient at stimulating responses against a hitherto codominant determinant, HEL 46–61 in the H-2k haplotype; in contrast, they acquired the ability to activate T cells against other determinants, including the cryptic determinant HEL 2–16, which is not normally recognized after exposure to intact antigen. These changes in repertoire most likely reflect either the loading of different sets of peptides onto MHC class II molecules or, conceivably, changes in the orientation of peptides within the MHC class II groove (32). In either case, the net result was that dendritic cells treated with IL-6 activate a qualitatively different set of HEL-specific T cells than the set activated by untreated dendritic cells.
The molecular mechanisms mediating the effects of IL-6 are not defined completely. However, a key event seems to be that IL-6 increases the acidification of peripheral early endosomes, a process that may be mediated via inhibition of the endosomal Na+/K+ ATPase. A causal relationship between the activity of endosomal Na+/K+ ATPase and altered processing is supported by the observation that direct inhibition of the Na+/K+ ATPase with the inhibitor ouabain allowed presentation of the 2–16 cryptic determinant. Altered early endosome acidity could affect the generation of peptide/MHC complexes in several ways. The specificity of the endosomal proteinases (such as cathepsins D and E) in cleaving HEL are different at different pH (D.O. and B.C., unpublished results). Moreover, lower pH in the early endosome should favor an exchange of bound MHC-class-II-associated invariant chain peptide (CLIP) for antigenic peptide. A mildly acidic pH favors the association of HLA-DM with the MHC class II/invariant chain complex (33), enhances the spontaneous or HLA-DM-mediated removal of CLIP from MHC class II (34, 35), stabilizes empty forms of MHC class II generated after removal of CLIP (36), and enables peptide loading of MHC class II, possibly by inducing a conformational change in the MHC class II dimer (37). This means that a more acidic early endosome could allow cryptic determinants revealed by altered processing to be rescued by binding to MHC class II before they are destroyed by the full battery of late endosome proteinases. Further studies are needed to unravel the exact sequence of events in this complex pathway. Additionally, studies should investigate whether tumor antigens and autoantigens can be processed differently by dendritic cells treated with IL-6.
Mechanisms of antigen processing by dendritic cells are of critical importance, because these cells are initiators of adaptive immune responses. In fact, the immunological dominance of certain determinants in an immune response may be caused in great part by their preferential processing and presentation by dendritic cells. It is apparent that mechanisms of antigen processing by dendritic cells are different than those of other APC such as B cells. Dendritic cells undergo maturation in response to inflammatory stimuli such as TNF-α. During this process, the transport of MHC class II molecules to loading compartments and later to the surface of dendritic cells (38) is regulated differently than MHC class II transport in B cells, in which MHC class II molecules seem to scan most intracellular compartments for peptide to bind(39). Because of these differences, B cells and dendritic cells may process an antigen such that a slightly different profile of determinants is presented; dendritic cells may be more focused with only a few (dominant) determinants being selected, whereas B cell processing may maximize display of available MHC class II binding peptides. Because dendritic cells mediate both negative selection of thymocytes and priming of mature T cells, a major factor in determining the dominance or crypticity of a self or foreign determinant would be how efficiently a dendritic cell displays that determinant, with presentation by B cells being less important. This hypothesis could explain the results of Viner et al., who found that a particular determinant of HEL seemed to be cryptic, but was well processed and presented by B cells in vitro (40).
Our data also have implications for how dendritic cell differentiation should be viewed. It is becoming increasingly apparent that this process is a complex one, in which many factors (e.g., microbial products, local cytokines, and inflammatory mediators; ref. 41) contribute to the overall activation process. For example, dendritic cells exposed to CD40 on helper T cells can prime cytotoxic T cells (42–44), whereas dendritic cells treated with IL-10 can tolerize T cells (45). In inflammatory microenvironments in vivo, a number of proinflammatory cytokines, including IL-1, TNF-α, and IL-6, are produced simultaneously. However, with respect to dendritic cell regulation, their function is not overlapping. IL-1 and TNF-α, for example, play quite distinct roles in the initiation of dendritic cell migration (46). In this study, we document another aspect of this multilevel regulation. Both TNF-α and IL-6 regulate dendritic cell antigen processing, but, although TNF-α alters intracellular distribution of MHC class II molecules and down-regulates endocytosis, it does not result in presentation of the cryptic determinant HEL 2–16. In contrast, IL-6 alters endosome acidification and changes the peptides presented from HEL but does not alter distribution of MHC class II or overall rates of endocytosis (ref. 22; H.D., unpublished results). Thus, it seems that the response to IL-6 is just one of many potential pathways down which dendritic cells can differentiate after stimulation by exogenous signals. The flexibility of the dendritic cell in response to its microenvironment is a key aspect of its role in the orchestration of an adaptive immune response.
However, this flexibility itself could become dangerous if there were a chronic imbalance in the signals that dendritic cells receive. Thomas and Lipsky have proposed that dendritic cells may cause rheumatoid arthritis in humans, caused by the activation of dendritic cells by inflammatory signals such as TNF-α (47). Patients with rheumatoid and juvenile chronic arthritis have persistently high levels of IL-6 in their synovial fluid (30, 48), which also contains many dendritic cells (49, 50). It is noteworthy that IL-6 is absolutely essential in several experimental autoimmune encephalomyelitis and arthritis models (20–22), unlike interferon-γ and TNF-α, which are not required and can even confer resistance in reported disease models (51–55).
Therefore, we would refine the concept of Thomas and Lipsky, focusing not on dendritic cells conventionally activated by TNF-α, but on dendritic cells receiving an imbalance of signals—such as high levels of IL-6—that may differentiate the cells into a distinct functional state. The dendritic cell exposed to IL-6 may process self antigen qualitatively differently so that previously hidden (cryptic) determinants are revealed to the T cell repertoire; whether this phenomenon is an inevitable consequence of inflammation involving IL-6 remains to be learned. Our results point to IL-6 as a major pharmacological mediator of altered antigen processing in vivo and are consistent with the hypothesis that autoreactive T cells can be engaged to induce and propagate autoimmune disease as a result of imbalanced signals to dendritic cells. If this hypothesis proves to be correct, blocking antigen processing by dendritic cells exposed to IL-6 by using safe acidotropic drugs may ameliorate autoimmune disease; in fact chloroquine, although it has some undesired side effects, has been used to treat rheumatoid arthritis with some success (56).
Acknowledgments
We thank P. Fairchild for hybridomas, M. Keegan, L. Lopes, and V. Woodhead for help with experiments, and F. G. Drakesmith, J. F. Elliott, S. Kiani-Alikhan, P. Noble, H. Stauss, and A. R. M. Townsend for constructive criticism. This work was supported in part by the Arthritis and Rheumatism Council and by National Institutes of Health Grant AI-28419.
ABBREVIATIONS
- APC
antigen-presenting cells
- DAMP
N-{3-[(2,4-dinitrophenyl)amino]propyl}-N-(3-aminopropyl)methylamine dihydrochloride
- DC
dendritic cell culture
- DC/IL-6
dendritic cell culture with interleukin 6
- GM-CSF
granulocyte–macrophage colony-stimulating factor
- HEL
hen-egg lysozyme
- IFA
incomplete Freund’s adjuvant
- IL
interleukin
- MHC
major histocompatibility complex
- TNF
tumor necrosis factor
References
- 1.Lehmann P V, Forsthuber T, Miller A, Sercarz E E. Nature (London) 1992;358:155–157. doi: 10.1038/358155a0. [DOI] [PubMed] [Google Scholar]
- 2.Kaufman D L, Clare S M, Tian J, Forsthuber T, Ting G S, Robinson P, Atkinson M A, Sercarz E E, Tobin A J, Lehmann P V. Nature (London) 1993;366:69–72. doi: 10.1038/366069a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tisch R, Yang X D, Singer S M, Liblau R S, Fugger L, McDevitt H O. Nature (London) 1993;366:72–75. doi: 10.1038/366072a0. [DOI] [PubMed] [Google Scholar]
- 4.Sercarz E E, Lehmann P V, Ametani A, Benichou G, Miller A, Moudgil K. Annu Rev Immunol. 1993;11:729–766. doi: 10.1146/annurev.iy.11.040193.003501. [DOI] [PubMed] [Google Scholar]
- 5.Oldstone M B A. Cell. 1987;50:819–820. doi: 10.1016/0092-8674(87)90507-1. [DOI] [PubMed] [Google Scholar]
- 6.Bhardwaj V, Kumar V, Geysen H M, Sercarz E E. J Immunol. 1993;151:5000–5010. [PubMed] [Google Scholar]
- 7.Wucherpfennig K W, Strominger J L. Cell. 1995;80:695–705. doi: 10.1016/0092-8674(95)90348-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao Z S, Granucci F, Yeh L, Schaffer P A, Cantor H. Science. 1998;279:1344–1347. doi: 10.1126/science.279.5355.1344. [DOI] [PubMed] [Google Scholar]
- 9.Lehmann P V, Sercarz E E, Forsthuber T, Dayan C M, Gammon G. Immunol Today. 1993;14:203–208. doi: 10.1016/0167-5699(93)90163-F. [DOI] [PubMed] [Google Scholar]
- 10.Lanzavecchia A. J Exp Med. 1995;181:1945–1948. doi: 10.1084/jem.181.6.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Steinman R M. Annu Rev Immunol. 1991;9:271–296. doi: 10.1146/annurev.iy.09.040191.001415. [DOI] [PubMed] [Google Scholar]
- 12.Banchereau J, Steinman R M. Nature (London) 1998;392:245–252. doi: 10.1038/32588. [DOI] [PubMed] [Google Scholar]
- 13.Gapin L, Bravo d A Y, Casrouge A, Cabaniols J P, Kourilsky P, Kanellopoulos J. J Immunol. 1998;160:1555–1564. [PubMed] [Google Scholar]
- 14.Sallusto F, Lanzavecchia A. J Exp Med. 1994;179:1109–1118. doi: 10.1084/jem.179.4.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sallusto F, Cella M, Danieli C, Lanzavecchia A. J Exp Med. 1995;182:389–400. doi: 10.1084/jem.182.2.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamaguchi Y, Tsumura H, Miwa M, Inaba K. Stem Cells (Dayton) 1997;15:144–153. doi: 10.1002/stem.150144. [DOI] [PubMed] [Google Scholar]
- 17.Cella M, Engering A, Pinet V, Pieters J, Lanzavecchia A. Nature (London) 1997;388:782–787. doi: 10.1038/42030. [DOI] [PubMed] [Google Scholar]
- 18.Pierre P, Turley S J, Gatti E, Hull M, Meltzer J, Mirza A, Inaba K, Steinman R M, Mellman I. Nature (London) 1997;388:787–792. doi: 10.1038/42039. [DOI] [PubMed] [Google Scholar]
- 19.Winzler C, Rovere P, Rescigno M, Granucci F, Penna G, Adorini L, Zimmermann V S, Davoust J, Ricciardi C P. J Exp Med. 1997;185:317–328. doi: 10.1084/jem.185.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alonzi T, Fattori E, Lazzaro D, Costa P, Probret L, Kollias G, de Benedetti F, Poli V, Ciliberto G. J Exp Med. 1998;187:461–468. doi: 10.1084/jem.187.4.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mendel I, Katz A, Kozak N, Ben-Nun A, Revel M. Eur J Immunol. 1998;28:1727–1737. doi: 10.1002/(SICI)1521-4141(199805)28:05<1727::AID-IMMU1727>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 22.Okuda D, Sakoda S, Bernard C, Fujimura H, Saeki Y, Kishimoto T, Yanagihara T. Int Immunol. 1998;10:703–708. doi: 10.1093/intimm/10.5.703. [DOI] [PubMed] [Google Scholar]
- 23.Inaba K, Inaba M, Romani N, Aya H, Deguchi M, Ikehara S, Muramatsu S, Steinman R M. J Exp Med. 1992;176:1693–1702. doi: 10.1084/jem.176.6.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guery J C, Ria F, Adorini L. J Exp Med. 1996;183:751–757. doi: 10.1084/jem.183.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lutz M B, Rovere P, Kleijmeer M J, Rescigno M, Assmann C U, Oorschot V M, Geuze H J, Trucy J, Demandolx D, Davoust J, et al. J Immunol. 1997;159:3707–3716. [PubMed] [Google Scholar]
- 26.Muraguchi A, Hirano T, Tang B, Matsuda T, Horii Y, Nakajima K, Kishimoto T. J Exp Med. 1988;167:332–344. doi: 10.1084/jem.167.2.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cain C C, Sipe D M, Murphy R F. Proc Natl Acad Sci USA. 1989;86:544–548. doi: 10.1073/pnas.86.2.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fuchs R, Schmid S, Mellman I. Proc Natl Acad Sci USA. 1989;86:539–543. doi: 10.1073/pnas.86.2.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Green R M, Whiting J F, Rosenbluth A B, Beier D, Gollan J L. Am J Physiol. 1994;267:G1094–G1100. doi: 10.1152/ajpgi.1994.267.6.G1094. [DOI] [PubMed] [Google Scholar]
- 30.Houssiau F A, Devogelaer J P, Van Damme J, de Deuxchaisnes C N, Van Snick J. Arthritis Rheum. 1988;31:784–788. doi: 10.1002/art.1780310614. [DOI] [PubMed] [Google Scholar]
- 31.Maubach K, Foey A D, Hall N D. Agents Actions. 1993;39:C107–C109. doi: 10.1007/BF01972737. [DOI] [PubMed] [Google Scholar]
- 32.Viner N J, Nelson C A, Deck B, Unanue E R. J Immunol. 1996;156:2365–2368. [PubMed] [Google Scholar]
- 33.Watts C. Annu Rev Immunol. 1997;15:821–850. doi: 10.1146/annurev.immunol.15.1.821. [DOI] [PubMed] [Google Scholar]
- 34.Denzin L K, Cresswell P. Cell. 1995;82:155–165. doi: 10.1016/0092-8674(95)90061-6. [DOI] [PubMed] [Google Scholar]
- 35.Kropshofer H, Vogt A B, Stern L J, Hammerling G J. Science. 1995;270:1357–1359. doi: 10.1126/science.270.5240.1357. [DOI] [PubMed] [Google Scholar]
- 36.Vogt A B, Moldenhauer G, Hammerling G J, Kropshofer H. Immunol Lett. 1997;57:209–211. doi: 10.1016/s0165-2478(97)00061-8. [DOI] [PubMed] [Google Scholar]
- 37.Runnels H A, Moore J C, Jensen P E. J Exp Med. 1996;183:127–136. doi: 10.1084/jem.183.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pierre P, Mellman I. Cell. 1998;93:1135–1145. doi: 10.1016/s0092-8674(00)81458-0. [DOI] [PubMed] [Google Scholar]
- 39.German R N, Castellino F, Han R, Reis e Sousa C, Romagnoli P, Sadegh-Nasseri S, Zhong G-M. Immunol Rev. 1996;151:5–30. doi: 10.1111/j.1600-065x.1996.tb00701.x. [DOI] [PubMed] [Google Scholar]
- 40.Viner N J, Nelson C A, Unanue E R. Proc Natl Acad Sci USA. 1995;92:2214–2218. doi: 10.1073/pnas.92.6.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rutault, K., Alderman, C., Chain, B. M. & Katz, D. R. (1998) Free Radical Biol. Med., in press. [DOI] [PubMed]
- 42.Bennett S R M, Carbone F R, Karamalis F, Flavell R A, Miller J F A P, Heath W R. Nature (London) 1998;393:478–480. doi: 10.1038/30996. [DOI] [PubMed] [Google Scholar]
- 43.Ridge J P, Di Rosa F, Matzinger P. Nature (London) 1998;393:474–478. doi: 10.1038/30989. [DOI] [PubMed] [Google Scholar]
- 44.Schoenberger S P, Toes R E M, van der Voort E I H, Offringa R, Melief C J M. Nature (London) 1998;393:480–483. doi: 10.1038/31002. [DOI] [PubMed] [Google Scholar]
- 45.Steinbrink K, Wolfl M, Jonuleit H, Knop J, Enk A H. J Immunol. 1997;159:4772–4780. [PubMed] [Google Scholar]
- 46.Cumberbatch M, Dearman R J, Kimber I. Adv Exp Med Biol. 1997;417:125–128. doi: 10.1007/978-1-4757-9966-8_21. [DOI] [PubMed] [Google Scholar]
- 47.Thomas R, Lipsky P E. Immunol Today. 1996;17:559–564. doi: 10.1016/s0167-5699(96)20030-1. [DOI] [PubMed] [Google Scholar]
- 48.de Benedetti F, Massa M, Robbioni P, Ravelli A, Burgio G R, Martini A. Arthritis Rheum. 1991;34:1158–1163. doi: 10.1002/art.1780340912. [DOI] [PubMed] [Google Scholar]
- 49.de Vere Tyndall A, Knight S C, Edwards A J, Clarke J B. Lancet. 1983;i:472–473. doi: 10.1016/s0140-6736(83)91468-x. [DOI] [PubMed] [Google Scholar]
- 50.Harding B, Knight S C. Clin Exp Immunol. 1986;63:594–600. [PMC free article] [PubMed] [Google Scholar]
- 51.Billiau A, Heremans H, Vandekerckhove F, Dijkmans R, Sobis H, Meulpas E, Carton H. J Immunol. 1988;140:1506–1510. [PubMed] [Google Scholar]
- 52.Ferber I A, Brocke S, Taylor-Edwards C, Ridgway W, Dinisco C, Steinman L, Dalton D, Fathman C G. J Immunol. 1996;156:5–7. [PubMed] [Google Scholar]
- 53.Krakowski M, Owens T. Eur J Immunol. 1996;26:1641–1646. doi: 10.1002/eji.1830260735. [DOI] [PubMed] [Google Scholar]
- 54.Frei K, Eugster H P, Bopst M, Constantinescu C S, Lavi E, Fontana A. J Exp Med. 1997;185:2177–2182. doi: 10.1084/jem.185.12.2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu J, Marino M W, Wong G, Grail D, Dunn A, Bettadapura J, Slavin A J, Old L, Bernard C C. Nat Med. 1998;4:78–83. doi: 10.1038/nm0198-078. [DOI] [PubMed] [Google Scholar]
- 56.Percival S P, Meanock I. Br Med J. 1968;3:579–584. doi: 10.1136/bmj.3.5618.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moudgil K, Sekiguchi D, Kim S, Sercarz E. J Immunol. 1997;159:2574–2579. [PubMed] [Google Scholar]