Abstract
T cell antigen receptor (TCR) and pre-TCR complexes are composed of clonotypic heterodimers in association with dimers of signal transducing invariant subunits (CD3γ, -δ, -ɛ, and ζ). The role of individual invariant subunits in T cell development has been investigated by generating gene-specific mutations in mice. Mutation of CD3γ, -δ, or ζ results in an incomplete block in development, characterized by reduced numbers of mature T cells that express low levels of TCR. In contrast, mature T cells are absent from CD3ɛ−/− mice, and thymocyte development is arrested at the early CD4−CD8− stage. Although these results suggest that CD3ɛ is essential for pre-TCR and TCR expression/function, their interpretation is complicated by the fact that expression of the CD3γ and CD3δ genes also is reduced in CD3ɛ−/− mice. Thus, it is unclear whether the phenotype of CD3ɛ−/− mice reflects the collective effects of CD3γ, -δ, and -ɛ deficiency. By removing the selectable marker (PGK-NEO) from the targeted CD3ɛ gene via Cre/loxP-mediated recombination, we generated mice that lack CD3ɛ yet retain normal expression of the closely linked CD3γ and CD3δ genes. These (CD3ɛΔ/Δ) mice exhibited an early arrest in T cell development, similar to that of CD3ɛ−/− mice. Moreover, the developmental defect could be rescued by expression of a CD3ɛ transgene. These results identify an essential role for CD3ɛ in T cell development not shared by the CD3γ, CD3δ, or ζ-family proteins and provide further evidence that PGK-NEO can influence the expression of genes in its proximity.
Differentiation of thymocytes into mature, functional T cells requires the input of signals delivered through the T cell antigen receptor (TCR) and a precursor form of the TCR, the pre-TCR (1, 2). The TCR complexes expressed on mature T cells are composed of subunits originating from six different genes: TCRα and TCRβ (or TCRγ and TCRδ), CD3γ, CD3δ, CD3ɛ, and ζ (or a related family member) (3). The clonotypic (TCRα/β or TCRγ/δ) chains, which specify distinct lineages of T cells, are responsible for ligand recognition and are generated during development by programmed rearrangement of germline [V-(D)-J] gene segments. TCRα/β and TCRγ/δ heterodimers lack inherent signaling activity but associate noncovalently with dimers composed of the invariant signal transducing CD3 and ζ-family subunits. The generally accepted stoichiometry for the TCR complex is TCRαβ or TCRγδ/CD3γɛ/CD3δɛ/ζζ.
Rearrangement of genes encoding the lineage-specific αβ- and γδ-TCR chains is initiated in immature CD4− CD8− [or double negative (DN)] thymocytes that constitutively express the CD3γ, CD3δ, CD3ɛ, and ζ subunits (4). Productive rearrangement of both the TCRγ and TCRδ genes results in surface expression of γδTCR complexes and commitment of DN thymocytes to the γδ-T cell lineage (5). On the other hand, productive rearrangement of the TCRβ locus results in surface expression of a “pre-TCR” complex composed of TCRβ chain paired with an invariant, pre-Tα chain in association with CD3 and ζ-chain dimers (2). Signals transduced by the pre-TCR induce commitment of DN thymocytes to the αβ-T cell lineage by triggering cell proliferation (resulting in “β-selection”) and transition to the CD4+CD8+ [or double positive (DP)] stage, in which TCRα gene rearrangement is initiated (2, 6).
The role of each of the TCR subunits in T cell development has been examined by generating gene-specific mutations in mice. Mice rendered deficient in [V-(D)-J] recombination (Rag1−/− or Rag2−/−) are devoid of T cells and contain only immature DN thymocytes (7, 8) whereas mutations that selectively block expression of the TCRα/β or TCRγ/δ chain heterodimers result in lineage-restricted defects (i.e., absence of α/β or γ/δ T cells, respectively) (6, 9, 10). Moreover, α/β-T cell development is arrested at the DN stage in TCRβ−/− mice and at the DP stage in TCRα−/− mice (6, 9), consistent with a requirement for TCRβ but not TCRα for pre-TCR assembly and function (2).
Mice lacking all members of the ζ-family (ζ, η, FcɛRIγ) exhibit a partial block in T cell development at both the DN→DP and DP→CD4+CD8− or CD4−CD8+ transitions (11). Small numbers of peripheral T cells are generated in ζ/η/FcɛRIγ−/− mice despite the fact that they express extremely low levels of TCR (11). Thus, pre-TCR and TCR complexes can still be expressed and are capable of transducing developmental signals, albeit much less efficiently, in the absence of ζ-family dimers (11).
The phenotype of CD3ɛ−/− mice has shown that, unlike the ζ-family dimers, CD3 dimers are absolutely required for pre-TCR and TCR assembly and/or signal transduction (12). Both αβTCR+ and γδTCR+ T cells are absent from CD3ɛ−/− mice, and, although rearrangement of TCRβ occurs, thymocyte development is arrested at the immature DN stage (12). In contrast, mutation of either CD3γ or CD3δ results in a less severe developmental impairment (13, 14). As in ζ/η/FcɛRIγ−/− mice, both CD3γ−/− and CD3δ−/− mice exhibit an incomplete block in development and contain low numbers of peripheral αβTCRlo T cells (11, 13, 14). In addition, CD3δ−/− mice contain normal numbers of DP thymocytes, and γδ-T cell development appears unaffected (13). One interpretation of these results has been that CD3γ and CD3δ are functionally redundant such that CD3γɛ dimers or CD3δɛ dimers alone can, though with different efficiency, promote pre-TCR and TCR assembly. On the other hand, CD3ɛ is absolutely required because, in its absence, neither heterodimer can be formed. However, a potentially confounding factor in the interpretation of these data has been that thymocytes from CD3ɛ−/− mice were found to express abnormally low levels of CD3γ and CD3δ transcripts (12). Thus, it is has remained unclear whether the phenotype observed in CD3ɛ−/− mice reflects a specific requirement for CD3ɛ in early T cell development or the collective effects of CD3γ, CD3δ, and CD3ɛ deficiency. In this study, we generated mice that lack expression of CD3ɛ yet retain normal expression of the closely linked CD3γ and CD3δ genes. Significantly, the developmental defect in these mice was found to be identical to that of the previously reported CD3ɛ−/− mice and could be rescued by expression of a CD3ɛ transgene. Collectively, these results identify an absolute requirement for CD3ɛ in early T cell development not shared by the CD3γ, CD3δ, or ζ-family proteins.
MATERIALS AND METHODS
Generation of CD3ɛN/N, CD3ɛΔ/Δ, and CD3ɛtg Mice.
The CD3ɛ targeting vector pɛNEO-loxP (Fig. 1A) was constructed by subcloning genomic DNA fragments that contained CD3ɛ exons 4 and 5 (5′ flank; 3.5 kilobases) and exons 7 and 8 (3′ flank; 3.0 kilobases) into plasmid pZINIMn, which contains both the loxP-PGK-NEO-loxP and PGK-HSV-TK expression cassettes. Embryonic stem (ES) cell electroporation and selection for homologous recombinants was performed essentially as described (15). CD3ɛΔ/Δ mice (see allele designations below) were generated by mating CD3ɛN/N mice with EIIa-cre transgenic mice (16) and then screening offspring for loss of the PGK-NEO cassette. The CD3ɛ transgene was generated by substituting the murine CD3ɛ coding sequences for the ζ cDNA sequences in construct ζ-CT108 (17). Four CD3ɛ transgenic founder lines were generated by zygote injection, and CD3ɛ protein expression was quantitated by Western blotting. Two founder lines [C7530 (2× expression relative to wild-type), and C7705 (0.25× expression relative to wild-type)] were used in the present experiments.
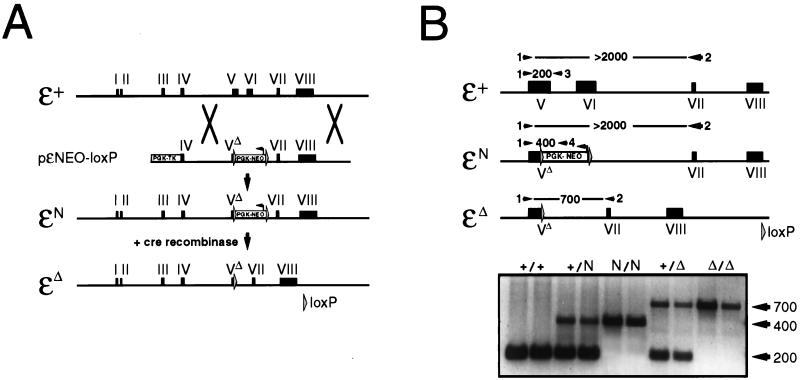
Figure 1.
Generation of CD3ɛN/N and CD3ɛΔ/Δ mice. (A) Representations of the CD3ɛ+ (wild-type), CD3ɛN (PGK-NEO+ mutant), and CD3ɛΔ (PGK-NEO− mutant) alleles. The CD3ɛN mutation was generated in ES cells by homologous recombination with the vector pɛNEO-loxP as shown. The CD3ɛΔ allele subsequently was generated from the CD3ɛN allele by Cre/loxP-mediated recombination as described in Materials and Methods. (B) Competitive PCR strategy for distinguishing between the CD3ɛ+, CD3ɛN, and CD3ɛΔ alleles. Mouse tail DNA was amplified with a mixture of oligonucleotides 1–4, and the products were visualized by gel electrophoresis. Allele-specific products were distinguished on the basis of size: CD3ɛ+ (200-bp product of oligos 1+3); CD3ɛN (400-bp product of oligos 1+4); and CD3ɛΔ (700-bp product of oligos 1+2). Although oligos 1+2 also can amplify a product from both the CD3ɛ+ and CD3ɛN alleles, the large size of the product (>2,000 bp) precludes its accumulation under the competitive PCR conditions. Results shown are from two independent DNA samples obtained from mice of the indicated genotypes.
PCR Screening.
Mice were genotyped initially by Southern blotting then subsequently by using a competitive PCR reaction that distinguishes between the CD3ɛ+ (wild-type), CD3ɛN (mutant CD3ɛ with PGK-NEO insertion), and CD3ɛΔ(mutant CD3ɛ without PGK-NEO) alleles (Fig. 2). PCR was performed with a mixture of oligonucleotide 1 (CD3ɛ-exonV/5′-3′; TACAAAGTCTCCATCTCAGG), 2 (CD3ɛ-exonVII/3′-5′; TGGCCGCTCCTTGTTTTG), 3 (CD3ɛ-exonV/3′-5′; CTCGAGCTTTCAGGTACAA), and 4 (NEO; GGATTAGATAAATG CCTGCT). PCR parameters were 95°C, 20′′; 55°C, 20′′; 72°C, 90′′ × 35 cycles. Products were resolved on a 2% agarose gel and were visualized by staining with ethidium bromide.
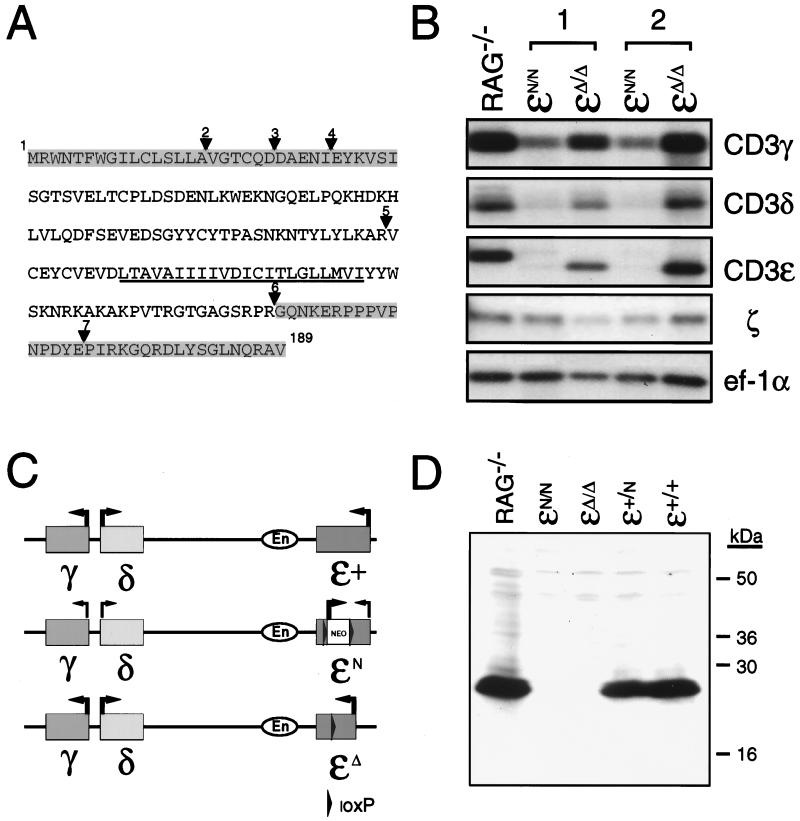
Figure 2.
(A) Predicted amino acid sequence of CD3ɛΔ. Amino acid sequence of the fully processed CD3ɛ protein is shown. The transmembrane domain is underlined. Triangles indicate the location of introns (2–7) within the CD3ɛ gene. Shaded areas represent the protein that potentially could be expressed in CD3ɛΔ/Δ mice based on the coding sequences that are retained within thymocyte transcripts from these mice. (B) Northern blot analysis of thymocyte total RNA from Rag−/− mice and from CD3ɛN/N and CD3ɛΔ/Δ mice that had been generated from two independently derived ES cell clones (1 and 2). Blots were hybridized with probes corresponding to CD3ɛ, CD3γ, CD3δ, and ζ and then were hybridized with a probe corresponding to the ubiquitously expressed ef-1α to assess the integrity and quantity of RNA in each lane. (C) Representation of the CD3-γ,-δ,-ɛ cluster on mouse chromosome 9. Location of the CD3 enhancer (En) element 3-prime of CD3ɛ is shown. Arrows indicate the direction of gene transcription. Thin lines indicate reduced gene expression. Depicted are the relative transcription levels observed from the (CD3)ɛ+, (CD3)ɛN, and (CD3)ɛΔ alleles. (D) Western blot of thymocyte protein from Rag−/−, (CD3)ɛN/N, (CD3)ɛΔ/Δ, (CD3)ɛ+/N, and (CD3)ɛ+/+ mice. Extracts from 7 × 106 thymocytes were run under reducing conditions on a 15% SDS-polyacrylamide gel, were transferred to poly(vinylidene difluoride) membrane, and were analyzed for CD3ɛ expression by using a rabbit antiserum raised against a peptide corresponding to amino acids within the C terminus of the CD3ɛ. Positions of the molecular mass standards are indicated in kilodaltons.
RNA and Protein Analysis.
For Northern blot analysis, RNA was purified from total thymocytes, was electrophoresed, was transferred to membranes, and was hybridized with CD3γ-, CD3δ-, CD3ɛ-, and ζ-specific probes as described (17). The ef-1α probe was generated by PCR of total embryo (fetal day 9.5) cDNA with oligos ef-1αA and ef-1αB (18). For Western blot analysis, thymocytes were enumerated, were lysed at a concentration of 1 × 108/ml in SDS-loading buffer plus β-mercaptoethanol, and were resolved by 15% SDS/PAGE. Separated proteins were transferred to poly(vinylidene difluoride) membranes, were blotted with a 1:100 dilution of polyclonal anti-CD3ɛ (A452; Dako), and were detected by chemiluminescence (ECL, Amersham).
Multicolor Flow Cytometry.
Single-cell suspensions of thymocytes or lymph node cells were processed, stained, and analyzed as described (17). mAbs used for flow cytometric analysis (purchased from PharMingen unless noted otherwise) included unlabeled anti-CD16/CD32 (2.4G2; blocking antibody), and fluorescein isothiocyanate- or phycoerythrin conjugated anti-CD4 (RM4.5), anti-CD8α (53–6.7), anti-TCRβ (H57–597), anti-TCRδ (GL-3), anti-CD3ɛ (145–2C11), anti-CD25 (7D4), anti-B220 (RA3–6B2), and anti-CD44 (IM7). Quantum-red conjugated anti-CD4 and anti-CD8 were purchased from Sigma.
RESULTS AND DISCUSSION
Generation of CD3ɛN/N and CD3ɛΔ/Δ Mice. The vector pɛNEO-loxP, which contains a PGK-NEO expression cassette flanked by loxP sites situated within a portion of the CD3ɛ gene, was used to generate targeted mutations of CD3ɛ in mouse ES cells (Fig. 1A). ES cell clones in which the targeting construct had integrated by homologous recombination contained a mutated CD3ɛ allele (ɛN) in which PGK-NEO was substituted for most of exon V and all of exon VI, which encode most of the extracellular domain and the entire transmembrane domain of CD3ɛ, respectively (Figs. 1A and 2A). Two positive ES clones were used to generate chimeric mice, which subsequently transmitted the mutated allele to their offspring.
Because it previously had been reported that mutation of CD3ɛ alters the expression of the CD3γ and CD3δ genes (12), thymocytes from mice homozygous for the CD3ɛ mutant allele (CD3ɛN/N) were screened for expression of CD3γ and CD3δ. Indeed, CD3ɛN/N mice generated from both of the independently derived ES clones exhibited a phenotype identical to that previously described [i.e., a reduction in the level of CD3γ transcripts and a near absence of CD3δ transcripts (ref. 12 and Fig. 2B)]. CD3ɛ transcripts, which were of aberrant size, were also nearly undetectable in CD3ɛN/N mice whereas expression of the unlinked ζ gene was unaffected (Fig. 2B).
It has been shown that the PGK-NEO cassette can negatively influence the expression of genes in its proximity, presumably by competing for regulatory sequences (19). Because the CD3γ, CD3δ, and CD3ɛ genes are linked closely, residing within 50 kilobases on mouse chromosome 9 (20), and are thought to be coregulated by an enhancer element located directly 3′ of CD3ɛ (ref. 21 and Fig. 2C), it was conceivable that such a mechanism could account for the phenotype of CD3ɛN/N mice. Consistent with this idea, multiple thymocyte-specific NEO transcripts were detectable in both CD3ɛ+/N and CD3ɛN/N mice (data not shown). To ascertain whether the presence of PGK-NEO was in fact responsible for the decrease in CD3γ and CD3δ expression, the loxP-PGK-NEO-loxP cassette was removed from CD3ɛN/N mice by Cre/loxP-mediated recombination. CD3ɛN/N mice were mated with EIIa-cre transgenic mice, which express Cre recombinase in all cells at the preimplantation stage of embryogenesis (16). Offspring of these matings initially were screened for loss of PGK-NEO (designated CD3ɛΔ) by Southern blotting and subsequently were tracked by a competitive PCR reaction that scores for the reduction in length of a product generated by primers corresponding to sequences within exon V and exon VII (Fig. 1B).
Analysis of thymocyte mRNA from CD3ɛΔ/Δ mice by Northern blotting revealed a single abundant CD3ɛ transcript of reduced length compared with the wild-type CD3ɛ transcript (Fig. 2B). Sequencing of cDNAs generated from CD3Δ transcripts by RT-PCR demonstrated that these mRNAs contained the intact coding sequences from exons I-IV and VII-VIII of CD3ɛ and thus represented a precise deletion of the exon V and exon VI coding sequences (Fig. 2A). Because these transcripts could conceivably direct the synthesis of a mutant CD3ɛ protein, thymocyte extracts from CD3Δ/Δ mice were screened by Western blotting with an antibody directed against a peptide corresponding to amino acids 156–168 of CD3ɛ, which should be retained within the intact CD3Δ product (Fig. 2A). Immunoreactive protein was undetectable in CD3Δ/Δ mice, indicating that either the protein encoded by the CD3Δ transcripts is highly unstable or that the epitope recognized by the antibody has been deleted (Fig. 2D). Significantly, in contrast to CD3ɛN/N mice, thymocytes from CD3ɛΔ/Δ mice contained normal levels of CD3γ and CD3δ transcripts, which were of the predicted size (Fig. 2B). Thus, the generation of CD3ɛΔ/Δ mice enabled a specific examination of the requirement for CD3ɛ in development.
Phenotype of CD3ɛΔ/Δ Mice.
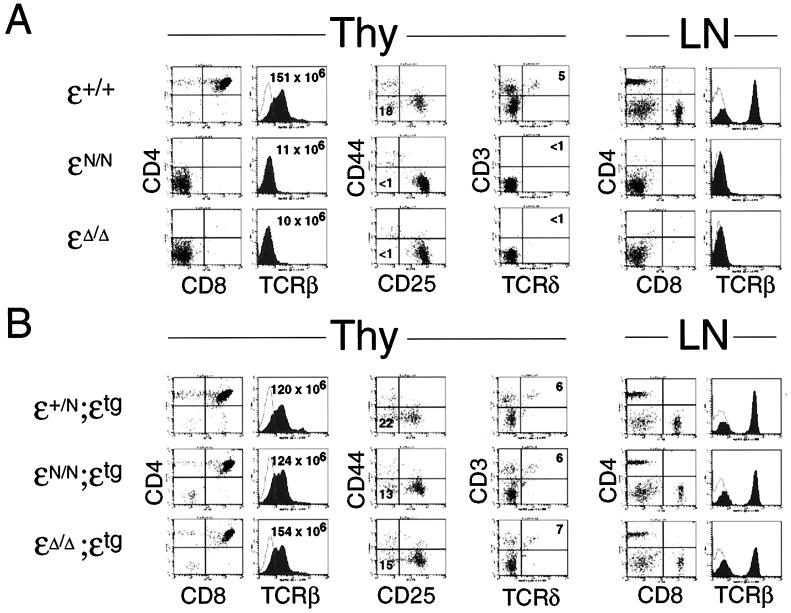
Analysis of T cell development in CD3ɛΔ/Δ mice revealed a phenotype virtually indistinguishable from that of CD3ɛN/N mice (ref. 12 and Fig. 3). Total thymocyte numbers in both CD3ɛΔ/Δ and CD3ɛN/N mice were <10% of control (CD3ɛ+/+), and the thymocytes that were present consisted entirely of the DN cells (i.e., both DP and CD4+CD8− or CD4−CD8+ thymocytes were absent) (Fig. 3A). DN thymocytes progress through four stages of maturation before their transition to the DP stage: CD44+ CD25− → CD44+ CD25+ → CD44−/lo CD25+ → CD44− CD25− (22). Rearrangement of the TCRβ locus is first detected in CD44−/lo CD25+ thymocytes, and progression of these cells to the CD44− CD25− stage is thought to require expression of the pre-TCR (or TCR) complex (2, 22). Consistent with this idea, thymocyte development is arrested at the CD44−/lo CD25+ stage in both Rag−/− and CD3ɛN/N mice (refs. 7, 8, and 12 and Fig. 3A). Significantly, thymocyte development was arrested at the identical (CD44−/lo CD25+) stage in CD3ɛΔ/Δ mice (Fig. 3A). As observed previously in CD3ɛN/N mice (12), TCRβ, TCRγ, and TCRδ gene rearrangements were readily detectable in thymocytes from CD3ɛΔ/Δ mice (data not shown). Thus, these results demonstrate that CD3ɛ specifically is required for maturation of CD44−/lo CD25+ thymocytes to the CD44− CD25− stage. In addition, the fact that T cell development is blocked at the identical stage in CD3ɛN/N and CD3ɛΔ/Δ mice indicates that CD3γ and CD3δ are incapable of compensating for the loss of CD3ɛ.
Figure 3.
(A) Phenotype of CD3ɛN/N and CD3ɛΔ/Δmice. Thy, thymocytes; LN, lymph node. The left two panels show two-color (CD4 vs. CD8) and single-color (TCRβ) analysis of thymocytes from (CD3)ɛ+/+, (CD3)ɛN/N, and (CD3)ɛΔ/Δmice. Total thymocyte numbers from representative mice are provided in the two color plots. The central panels show two-color (CD44 vs. CD25) and two-color (CD3 vs. TCRδ) staining of gated (CD4−CD8−B220−) thymocytes. Numbers in quadrants indicate percentage of gated cells within the quadrant. The right two panels show two-color (CD4 vs. CD8) and single-color (TCRβ) analysis of total lymph node cells. (B) Phenotype of CD3ɛN/N;ɛtg and CD3ɛΔ/Δ;ɛtg mice. The panels show representative data from (CD3)ɛ+/N, (CD3)ɛN/N, and (CD3)ɛΔ/Δmice that contain a CD3ɛ transgene under the control of the human CD2 promoter and enhancer sequences (founder line C7530). Analysis is identical to that described in A.
As predicted from the early block in thymocyte development, mature CD4+CD8− or CD4−CD8+ T cells were absent from the periphery of CD3ɛΔ/Δ mice (Fig. 3A). Moreover, surface αβTCR and γδTCR complexes were undetectable in both the thymus and peripheral lymphoid organs as assessed by staining for TCRβ and TCRδ chains (Fig. 3A). As previously reported for CD3ɛN/N mice (12), the development of intestinal intraepithelial T cells also was arrested in CD3ɛΔ/Δ mice whereas B and NK cell development appeared unaffected (data not shown).
Expression of a CD3ɛ Transgene Rescues T Cell Development in CD3ɛΔ/Δ Mice.
To confirm that the phenotype exhibited by CD3ɛΔ/Δ mice could be attributed entirely to the absence of CD3ɛ, we reconstituted CD3ɛ expression in CD3ɛΔ/Δ mice by the introduction of a CD3ɛ transgene. Our own investigations have shown that transgenes placed under the control of the T-lineage-specific human CD2 promoter and enhancer begin to be expressed in thymocytes at the DN, CD44−/lo CD25+ stage and continue to be expressed at all subsequent stages of development (ref. 23 and data not shown). Thus, the CD3ɛ transgene construct should provide expression of CD3ɛ at the stage in development at which an arrest is first observed in CD3Δ/Δ mice (Fig. 3A). Because overexpression of CD3ɛ has been shown to block T cell development (24), we used transgenic founder lines that express relatively low levels of CD3ɛ for reconstitution experiments (0.25- and 2× wild-type). These transgenes did not inhibit thymocyte development when introduced into CD3+/+ or CD3+/N mice (Fig. 3B and data not shown).
As depicted in Fig. 3B, reexpression of CD3ɛ in CD3ɛΔ/Δ mice effectively reversed the developmental defects as assessed by the presence of DN, CD44− CD25− thymocytes, as well as DP and CD4+CD8− and CD4−CD8+ thymocytes in CD3ɛΔ/Δ CD3ɛtg mice. In addition, total thymocyte cellularity was increased markedly (to normal levels) in CD3ɛΔ/Δ; CD3ɛtg mice, and mature T cells were present in the periphery (Fig. 3). Finally, reexpression of CD3ɛ also restored surface expression of αβ- and γδ-TCR complexes on both immature and mature T cells (Fig. 3 A and B). Thus, these results demonstrate that the defects in T cell development observed in CD3ɛΔ/Δ mice are caused specifically by CD3ɛ deficiency. Moreover, because the CD3ɛ transgene would not be expected to restore expression of CD3ɛ at stages in development that precede the DN, CD44−/lo CD25+ stage, these findings lend support to the idea that CD3ɛ first is required at the point at which expression of the pre-TCR begins (12).
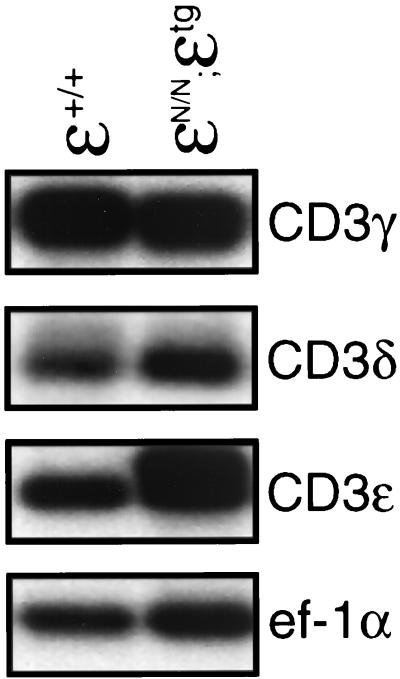
Of interest, introduction of the CD3ɛ transgene also fully corrected the developmental defect in CD3ɛN/N mice (Fig. 3B). This result was unexpected because DN thymocytes from CD3ɛN/N mice contain abnormally low levels of CD3γ and CD3δ transcripts (ref. 12 and Fig. 2B), and thymocyte development is compromised in the absence of either CD3γ or CD3δ (13, 14). Analysis of CD3γ and CD3δ gene expression in total thymocytes from CD3ɛN/N;CD3ɛtg mice (which consisted mostly of DP cells) revealed normal levels of CD3γ or CD3δ transcripts (Fig. 4). Thus, the restoration of thymocyte development in CD3ɛN/N;CD3ɛtg mice could be attributed to normalization of CD3γ and CD3δ expression as well as reconstitution of CD3ɛ. There are several possible explanations for these results. For example, the inhibitory effect of PGK-NEO mice may be variable between individual cells, such that some DN thymocytes express relatively normal levels of CD3γ and CD3δ and these cells are expanded selectively when CD3ɛ is reexpressed. Alternatively, the transcriptional suppression of CD3γ and CD3δ by PGK-NEO may be cell cycle- or stage-dependent. Regardless of the mechanism, these results demonstrate that the inhibitory effect of PGK-NEO on CD3γ and CD3δ gene expression is not developmentally limiting if CD3ɛ expression is restored.
Figure 4.
Northern blot analysis of total thymocyte RNA from CD3ɛ+/+ and CD3ɛN/N;ɛtg mice. Blots were hybridized with probes corresponding to CD3ɛ, CD3γ, CD3δ, and ζ and then were hybridized with the a probe corresponding to the ubiquitously expressed ef-1α to assess the integrity and quantity of RNA in each lane.
In summary, these results reveal a specific requirement for CD3ɛ in T cell development not exhibited by any of the other invariant subunits of the TCR complex (i.e., CD3γ, CD3δ, and ζ) (11, 13, 14). The present data are consistent with previous results obtained in T cell lines demonstrating that αβTCR complexes are still expressed, though at reduced levels, in the absence of CD3γ, CD3δ, or ζ but that CD3ɛ is absolutely required for surface expression of TCRα/β heterodimers (25–27). The absence of DP thymocytes and γδTCR+ cells in CD3ɛΔ/Δ mice demonstrates that CD3ɛ is also required for the expression and/or function of the pre-TCR and γδTCR complexes. That this need not necessarily have been the case is suggested by the discovery that the pre-TCR and γδTCR differ from the αβTCR in their requirement for CD3δ (13, 28). Taken together, current data indicate that a minimal requirement for pre-TCR/TCR expression and/or function is the expression of either a CD3γɛ or CD3δɛ heterodimer, although the former are much more efficient at promoting the assembly and surface expression of pre-TCR and γδTCR complexes. Finally, although these experiments demonstrate an obligate role for CD3ɛ in T cell development, it is important to note that the defect in CD3ɛΔ/Δ mice results in loss of both the structural and signal transducing functions of CD3ɛ. Therefore, additional studies will be required to determine whether CD3ɛ-mediated signal transduction is required at specific stages of development.
Acknowledgments
We thank B. J. Fowlkes for critical review of the manuscript. K.J.W. is a Howard Hughes Medical Institute–National Institutes of Health Research Scholar.
ABBREVIATIONS
- DN
double negative (CD4−CD8−)
- DP
double positive (CD4+CD8+)
- TCR
T cell antigen receptor
- ES cell
embryonic stem cell
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Jamison S C, Hogquist K A, Bevan M J. Annu Rev Immunol. 1995;13:93–126. doi: 10.1146/annurev.iy.13.040195.000521. [DOI] [PubMed] [Google Scholar]
- 2.Von Boehmer H, Fehling H J. Annu Rev Immunol. 1997;15:432–452. doi: 10.1146/annurev.immunol.15.1.433. [DOI] [PubMed] [Google Scholar]
- 3.Klausner R D, Lippincott-Schwartz J, Bonifacino J S. Annu Rev Cell Biol. 1990;6:403–431. doi: 10.1146/annurev.cb.06.110190.002155. [DOI] [PubMed] [Google Scholar]
- 4.Wiest D L, Kearse K P, Shores E W, Singer A. J Exp Med. 1994;180:1375–1382. doi: 10.1084/jem.180.4.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kang J, Raulet D H. Semin Immunol. 1997;9:171–179. doi: 10.1006/smim.1997.0069. [DOI] [PubMed] [Google Scholar]
- 6.Mombaerts P, Clarke A R, Rudnicki M A, Iacomini J, Itohara S, Lafille J J, Wang L, Ichikawa Y, Jaenisch R, Hooper M L, et al. Nature (London) 1992;360:225–231. doi: 10.1038/360225a0. [DOI] [PubMed] [Google Scholar]
- 7.Mombaerts P, Iacomini J, Johnson R S, Herrup K, Tonegawa S, Papaioannou B E. Cell. 1992;68:869–877. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- 8.Shinkai Y, Rathbun G, Lam K-P, Oltz E M, Stewart V, Mendelsohn M, Charon J, Datta M, Young F, Stall A M, et al. Cell. 1992;68:855–867. doi: 10.1016/0092-8674(92)90029-c. [DOI] [PubMed] [Google Scholar]
- 9.Philpott K L, Viney J L, Kay G, Rastan S, Gardiner E M, Chae S, Hayday H, Owen M J. Science. 1992;256:1448–1452. doi: 10.1126/science.1604321. [DOI] [PubMed] [Google Scholar]
- 10.Itohara S, Mombaerts P, Lafaille J, Iacomini J, Nelson A, Clarke A R, Hooper M L, Farr A, Tonegawa S. Cell. 1993;72:337–348. doi: 10.1016/0092-8674(93)90112-4. [DOI] [PubMed] [Google Scholar]
- 11.Shores E W, Ono M, Kawabe T, Sommers C L, Tran T, Lui K, Udey M C, Ravetch J, Love P E. J Exp Med. 1998;187:1093–1101. doi: 10.1084/jem.187.7.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Malissen M, Gillet A, Ardouin L, Bouvier G, Trucy J, Ferrier P, Vivier E, Malissen B. EMBO J. 1995;14:4641–4653. doi: 10.1002/j.1460-2075.1995.tb00146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dave V P, Cao Z, Browne C, Alarcon B, Fernandez-Miguel G, Lafaille J, de la Hera A, Tonegawa S, Kappes D J. EMBO J. 1997;16:1360–1370. doi: 10.1093/emboj/16.6.1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haks M C, Krimpenfort P, Borst J, Kruisbeek A. EMBO J. 1998;17:1871–1882. doi: 10.1093/emboj/17.7.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Love P E, Shores E W, Johnson M D, Tremblay M L, Lee E J, Grinberg A, Huang S P, Singer A, Westphal H. Science. 1993;261:918–921. doi: 10.1126/science.7688481. [DOI] [PubMed] [Google Scholar]
- 16.Lakso M, Pichel J G, Gorman J R, Sauer B, Okamoto Y, Lee E, Alt F W, Westphal H. Proc Natl Acad Sci USA. 1996;93:5860–5865. doi: 10.1073/pnas.93.12.5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Love P E, Shores E W, Lee E J, Grinberg A, Munitz T I, Westphal H, Singer A. J Exp Med. 1994;179:1485–1494. doi: 10.1084/jem.179.5.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anderson S J, Abraham K M, Nakayama T, Singer A, Perlmutter R. EMBO J. 1992;11:4877–4886. doi: 10.1002/j.1460-2075.1992.tb05594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fiering S, Epner E, Robinson K, Zhuang Y, Telling A, Hu M, Martin D I K, Enver T, Ley T J, Groudine M. Genes Dev. 1995;9:2203–2213. doi: 10.1101/gad.9.18.2203. [DOI] [PubMed] [Google Scholar]
- 20.Letourneur F, Mattei M-G, Malissen B. Immunogenetics. 1989;29:265–268. doi: 10.1007/BF00717911. [DOI] [PubMed] [Google Scholar]
- 21.Clevers H, Lonberg N, Dunlap S, Lacy E, Terhorst C. EMBO J. 1989;8:2527–2535. doi: 10.1002/j.1460-2075.1989.tb08390.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Godfrey D I, Zlotnik A. Immunol Today. 1993;14:547–553. doi: 10.1016/0167-5699(93)90186-O. [DOI] [PubMed] [Google Scholar]
- 23.Greaves D R, Wilson F D, Lang G, Kioussis D. Cell. 1989;56:979–986. doi: 10.1016/0092-8674(89)90631-4. [DOI] [PubMed] [Google Scholar]
- 24.Wang B, Levelt C, Salio M, Zheng D, Sancho J, Liu C P, She J, Huang M, Higgins K, Sunshine M J, et al. Int Immunol. 1995;7:435–448. doi: 10.1093/intimm/7.3.435. [DOI] [PubMed] [Google Scholar]
- 25.Sussman J J, Bonificino J S, Lippincott-Schwartz J, Weissman A M, Saito T, Klausner R D, Ashwell J D. Cell. 1988;52:85–95. doi: 10.1016/0092-8674(88)90533-8. [DOI] [PubMed] [Google Scholar]
- 26.Kappes D J, Tonegawa S. Proc Natl Acad Sci USA. 1991;88:10619–10623. doi: 10.1073/pnas.88.23.10619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hall C, Berkhout B, Alarcon B, Sancho J, Wileman T, Terhorst C. Int Immunol. 1991;3:359–368. doi: 10.1093/intimm/3.4.359. [DOI] [PubMed] [Google Scholar]
- 28.Berger M A, Davé V, Rhodes M R, Bosma G C, Bosma M J, Kappes D J, Wiest D L. J Exp Med. 1997;186:1461–1467. doi: 10.1084/jem.186.9.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]