Abstract
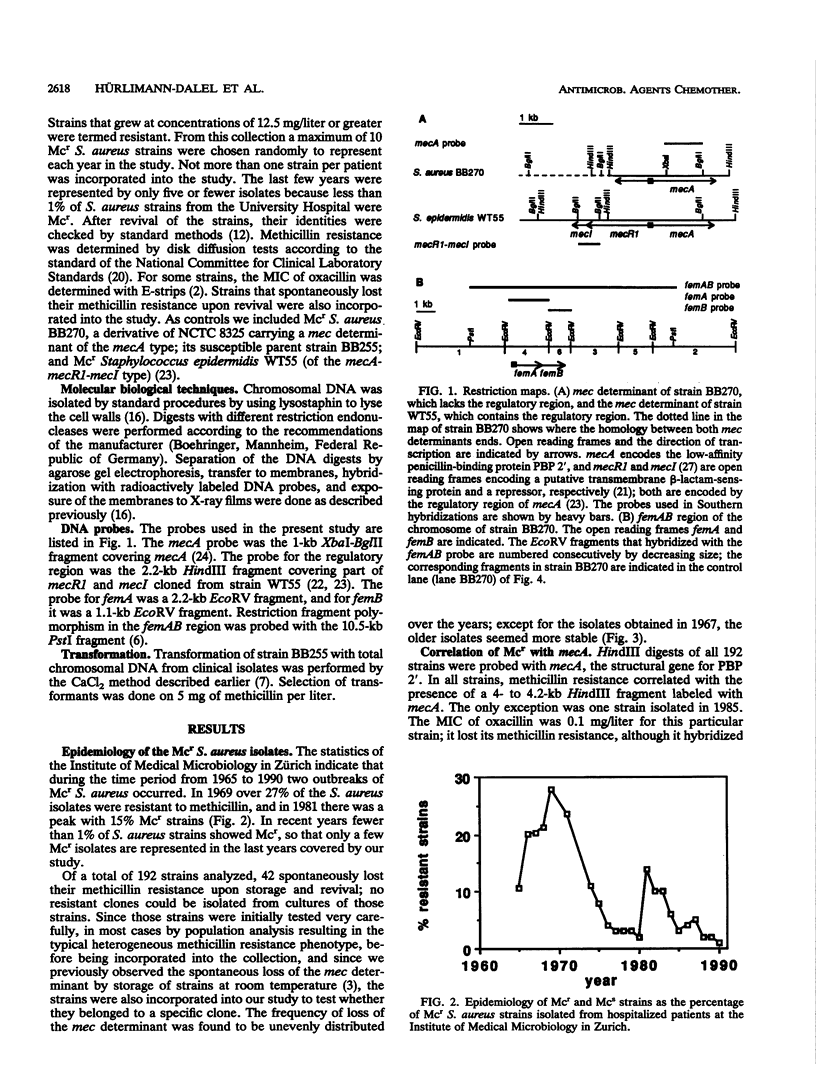
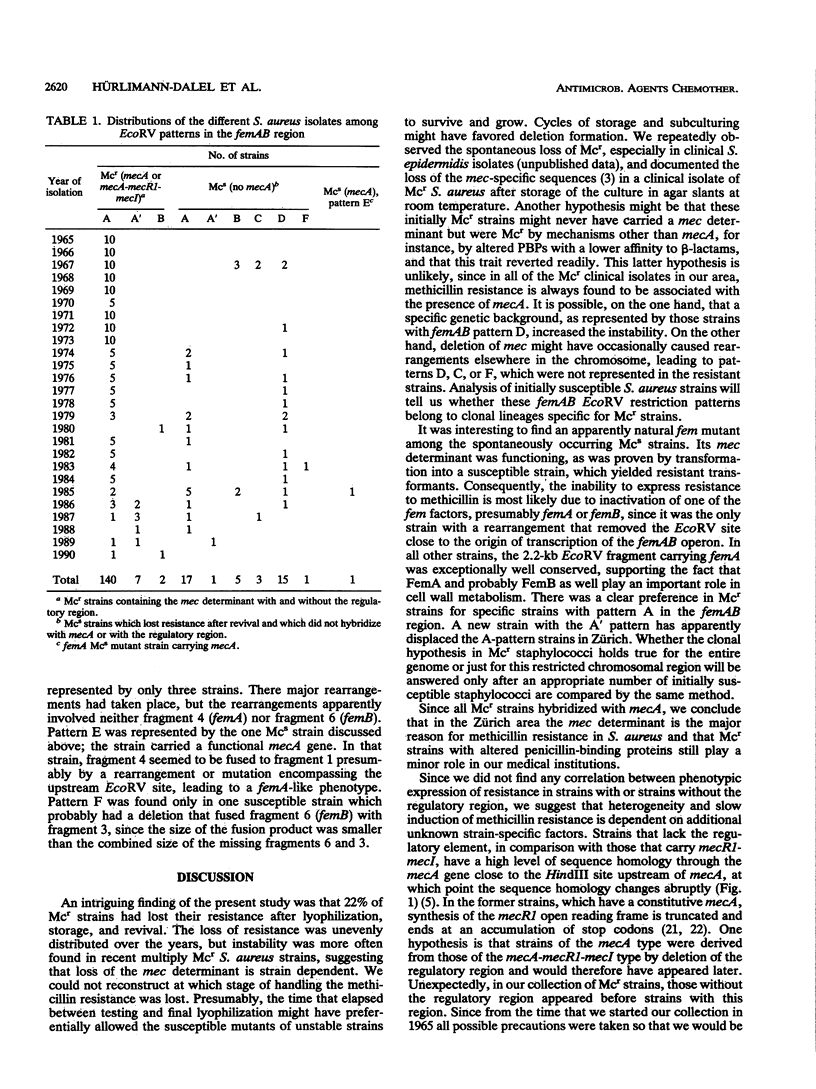
The restriction site polymorphism of the chromosomal femAB region and the first appearance of the regulatory element mecR1-mecI associated with the methicillin resistance determinant (mec) were analyzed in 192 initially methicillin resistant (Mcr) Staphylococcus aureus clinical isolates collected between 1965 and 1990 in the Zurich area. Forty-three of the strains lost the resistance spontaneously. All isolates that were still Mcr hybridized with mecA, the gene for the low-affinity penicillin-binding protein PBP 2'. Mcr strains isolated before 1977 lacked sequences that hybridized with mecR1-mecI, a regulatory element controlling the expression of mecA; exceptions to this were one strain isolated in 1966 and one strain isolated in 1972. The size of the EcoRV fragment carrying femA, a chromosomally encoded factor involved in pentaglycine side chain formation of the peptidoglycan and essential for the expression of methicillin resistance, was conserved in all strains but one, which was susceptible to methicillin even though it carried a functional mecA gene. The methicillin susceptibility of this particular strain was presumably due to a spontaneous femA-like mutation. The 192 strains belonged to seven different EcoRV restriction fragment patterns recognizable with a 10.5-kb probe covering the femAB region. Some 93% of the 149 Mcr strains belonged to pattern A, and the remaining Mcr strains shared patterns A' and B. The 42 isolates which spontaneously lost their resistance upon storage and revival represented all seven different patterns. This strong conservation of femA suggests an important role for femA in cell wall metabolism and methicillin resistance.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Archer G. L., Pennell E. Detection of methicillin resistance in staphylococci by using a DNA probe. Antimicrob Agents Chemother. 1990 Sep;34(9):1720–1724. doi: 10.1128/aac.34.9.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker C. N., Stocker S. A., Culver D. H., Thornsberry C. Comparison of the E Test to agar dilution, broth microdilution, and agar diffusion susceptibility testing techniques by using a special challenge set of bacteria. J Clin Microbiol. 1991 Mar;29(3):533–538. doi: 10.1128/jcm.29.3.533-538.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck W. D., Berger-Bächi B., Kayser F. H. Additional DNA in methicillin-resistant Staphylococcus aureus and molecular cloning of mec-specific DNA. J Bacteriol. 1986 Feb;165(2):373–378. doi: 10.1128/jb.165.2.373-378.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger-Bächi B., Barberis-Maino L., Strässle A., Kayser F. H. FemA, a host-mediated factor essential for methicillin resistance in Staphylococcus aureus: molecular cloning and characterization. Mol Gen Genet. 1989 Oct;219(1-2):263–269. doi: 10.1007/BF00261186. [DOI] [PubMed] [Google Scholar]
- Berger-Bächi B. Insertional inactivation of staphylococcal methicillin resistance by Tn551. J Bacteriol. 1983 Apr;154(1):479–487. doi: 10.1128/jb.154.1.479-487.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger-Bächi B., Strässle A., Gustafson J. E., Kayser F. H. Mapping and characterization of multiple chromosomal factors involved in methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1992 Jul;36(7):1367–1373. doi: 10.1128/aac.36.7.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiramatsu K., Asada K., Suzuki E., Okonogi K., Yokota T. Molecular cloning and nucleotide sequence determination of the regulator region of mecA gene in methicillin-resistant Staphylococcus aureus (MRSA). FEBS Lett. 1992 Feb 24;298(2-3):133–136. doi: 10.1016/0014-5793(92)80039-j. [DOI] [PubMed] [Google Scholar]
- Inglis B., Matthews P. R., Stewart P. R. The expression in Staphylococcus aureus of cloned DNA encoding methicillin resistance. J Gen Microbiol. 1988 Jun;134(6):1465–1469. doi: 10.1099/00221287-134-6-1465. [DOI] [PubMed] [Google Scholar]
- Kornblum J., Hartman B. J., Novick R. P., Tomasz A. Conversion of a homogeneously methicillin-resistant strain of Staphylococcus aureus to heterogeneous resistance by Tn551-mediated insertional inactivation. Eur J Clin Microbiol. 1986 Dec;5(6):714–718. doi: 10.1007/BF02013311. [DOI] [PubMed] [Google Scholar]
- Lacey R. W., Grinsted J. Genetic analysis of methicillin-resistant strains of Staphylococcus aureus; evidence for their evolution from a single clone. J Med Microbiol. 1973 Nov;6(4):511–526. doi: 10.1099/00222615-6-4-511. [DOI] [PubMed] [Google Scholar]
- Maidhof H., Reinicke B., Blümel P., Berger-Bächi B., Labischinski H. femA, which encodes a factor essential for expression of methicillin resistance, affects glycine content of peptidoglycan in methicillin-resistant and methicillin-susceptible Staphylococcus aureus strains. J Bacteriol. 1991 Jun;173(11):3507–3513. doi: 10.1128/jb.173.11.3507-3513.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami K., Tomasz A. Involvement of multiple genetic determinants in high-level methicillin resistance in Staphylococcus aureus. J Bacteriol. 1989 Feb;171(2):874–879. doi: 10.1128/jb.171.2.874-879.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryffel C., Kayser F. H., Berger-Bächi B. Correlation between regulation of mecA transcription and expression of methicillin resistance in staphylococci. Antimicrob Agents Chemother. 1992 Jan;36(1):25–31. doi: 10.1128/aac.36.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryffel C., Tesch W., Birch-Machin I., Reynolds P. E., Barberis-Maino L., Kayser F. H., Berger-Bächi B. Sequence comparison of mecA genes isolated from methicillin-resistant Staphylococcus aureus and Staphylococcus epidermidis. Gene. 1990 Sep 28;94(1):137–138. doi: 10.1016/0378-1119(90)90481-6. [DOI] [PubMed] [Google Scholar]
- Suzuki E., Hiramatsu K., Yokota T. Survey of methicillin-resistant clinical strains of coagulase-negative staphylococci for mecA gene distribution. Antimicrob Agents Chemother. 1992 Feb;36(2):429–434. doi: 10.1128/aac.36.2.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesch W., Ryffel C., Strässle A., Kayser F. H., Berger-Bächi B. Evidence of a novel staphylococcal mec-encoded element (mecR) controlling expression of penicillin-binding protein 2'. Antimicrob Agents Chemother. 1990 Sep;34(9):1703–1706. doi: 10.1128/aac.34.9.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesch W., Strässle A., Berger-Bächi B., O'Hara D., Reynolds P., Kayser F. H. Cloning and expression of methicillin resistance from Staphylococcus epidermidis in Staphylococcus carnosus. Antimicrob Agents Chemother. 1988 Oct;32(10):1494–1499. doi: 10.1128/aac.32.10.1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasz A., Drugeon H. B., de Lencastre H. M., Jabes D., McDougall L., Bille J. New mechanism for methicillin resistance in Staphylococcus aureus: clinical isolates that lack the PBP 2a gene and contain normal penicillin-binding proteins with modified penicillin-binding capacity. Antimicrob Agents Chemother. 1989 Nov;33(11):1869–1874. doi: 10.1128/aac.33.11.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubukata K., Nonoguchi R., Matsuhashi M., Konno M. Expression and inducibility in Staphylococcus aureus of the mecA gene, which encodes a methicillin-resistant S. aureus-specific penicillin-binding protein. J Bacteriol. 1989 May;171(5):2882–2885. doi: 10.1128/jb.171.5.2882-2885.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]



