Abstract
High-frequency reversible changes in colony morphology were observed in three strains of Cryptococcus neoformans. For one strain (SB4, serotype A), this process produced three colony types: smooth (S), wrinkled (W), and serrated (C). The frequency of switching between colony types varied for the individual colony transitions and was as high as 10−3. Mice infected with colony type W died faster than those infected with other colony types. The rat inflammatory response to infection with colony types S, W, and C was C > S > W and ranged from intense granulomatous inflammation with caseous necrosis for infection with type C to minimal inflammation for infection with type W. Infection with the various colony types was associated with different antibody responses to cryptococcal proteins in rats. Analysis of cellular characteristics revealed differences between the three colony types. High-frequency changes in colony morphology were also observed in two additional strains of C. neoformans. For one strain (24067A, serotype D) the switching occurred between smooth and wrinkled colonies. For the other strain (J32A, serotype A), the switching occurred between mucoid and nonmucoid colonies. The findings indicate that C. neoformans undergoes phenotypic switching and that this process can affect virulence and host inflammatory and immune responses. Phenotypic switching may play a role in the ability of this fungus to escape host defenses and establish chronic infections.
The fungus Cryptococcus neoformans (CN) causes life-threatening infections in both immunocompromised and immunocompetent hosts (1). Cryptococcosis occurs in 6–8% of patients with advanced HIV infection (2). Patients with AIDS who survive cryptococcosis require lifetime suppressive therapy to prevent recurrence of disease (3). Hence, CN has a marked propensity to cause chronic infection, especially in individuals with impaired immunity. However, even in normal individuals, the infection can be chronic and a state of latency with subsequent reactivation may occur (4). CN infections elicit a wide range of inflammatory responses ranging from granuloma formation to the virtual absence of inflammation. Although some differences in inflammatory response are due to the immunological status of the host, there is great variability in the types of inflammation described in both immunocompetent and immunocompromised individuals (5). The mechanism by which CN evades host inflammatory responses and establishes chronic infections is poorly understood.
CN strains can undergo phenotypic variation after in vitro or in vivo passage. Comparison of the capsular polysaccharide of initial and relapse isolates from patients has shown differences in glucuronoxylomannan (GXM) structure for several strains, including SB4, which was used in this study (6). Mouse passage resulted in stable alterations in cell-membrane sterol content (7). Serial CN isolates from patients exhibited differences in virulence for mice (8). Analysis of isolates of a standard laboratory strain maintained in various laboratories revealed significant differences in capsule size, melanin production, growth rates, and virulence for mice (9).
The mechanisms responsible for these effects are not understood. Phenotypic variation can result from many different processes including phenotypic switching, a mechanism by which some microorganisms undergo reversible changes. Phenotypic switching differs from acquired mutations in that it is a reversible and occurs at a much higher frequency than the expected mutation rate. Phenotypic switching has been described for several ascomycetes including Candida albicans, Blastomyces dermatitidis, and Saccharomyces cerevisiae (10–12). Among the pathogenic fungi, phenotypic switching has been extensively studied for C. albicans, where it produces colony types that differ in biochemical characteristics and susceptibility to antifungal drugs (13–16).
In this study, we report phenotypic switching in three strains of CN. Phenotypic switching in CN may contribute to the remarkable variability of strains and to the protean inflammatory reactions associated with cryptococcosis and assist the organism in evading the host immune response.
METHODS
Organisms.
Strains SB4, J32A (both serotype A), and 24067A (serotype D) have been described (8, 9, 17).
Colony Morphology.
The morphology of each SB4 colony type was evaluated after growth in various conditions including temperatures of 30°C, 37°C, and 18°C; Sabouraud’s dextrose agar (SDA); chemically defined medium (18); malt extract agar (Difco); and yeast nitrogen base agar (Difco).
DNA Typing.
Colony types were analyzed for strain identity by electrophoretic karyotype and restriction fragment length polymorphisms with the probe C. neoformans repetitive element-1 (CNRE-1) as described (19, 20).
Switching Frequencies.
Switching frequencies for strains SB4, 24067A, and J32A were determined by counting colonies with altered morphology in agar. Briefly, 21 colonies of each phenotype were collected and suspended in PBS (0.02 M phosphate). After dilution in PBS, each suspension was spread on SDA and incubated at 30°C. Two investigators evaluated the colonies at 72 and 96 h. For strains 24067A and J32A, colony morphology was evaluated at 72 and 120 h. UV irradiation can increase switching frequencies in C. albicans (21). For UV irradiation, single colonies of each SB4 colony type were lifted from SDA plates, grown overnight at 30°C in Sabouraud’s dextrose broth, suspended in PBS (5 × 104 cells per ml), irradiated at 254 nm and 20,000 μJ/cm2 in a Stratolinker (Stratagene) and spread on SDA. Colony morphology was examined after 3 and 4 days of incubation at 30°C. The survival of cells after irradiation was >90% relative to nonirradiated controls.
Capsule Size.
The distance from the cell wall to the outer margin of the capsule and the cell diameter (not including the capsule) were measured in a suspension of india ink, by microscopic examination at ×100 using an eyepiece grid with a resolution of 0.5 μm.
Scanning Electron Microscopy (SEM).
SEM was performed on cells grown to stationary phase in Sabouraud’s dextrose broth at 30°C with a Jeol-6400 microscope (Jeol, Peabody, MA) as described (22).
Growth Rates.
Doubling times were determined by both increases in turbidity and colony-forming units (CFU) as described (9). The doubling time during logarithmic-phase growth was calculated with the equation n = noeat, where n is the number of cells, no is the number of cells initially, a is the slope, and t is the time.
Antifungal Susceptibility.
Amphotericin B (Boehringer Mannheim), fluconazole (Roering-Pfizer, New York), and 5-flurocytosine (Sigma) minimal inhibitory concentrations for SB4 colony types were determined by the macrodilution method (23).
Resistance to Heat and Cold.
Cells from each SB4 colony type were grown for 2 days at 30°C, suspended in PBS at 5 × 103 cells per ml, and either frozen overnight at −20°C or heated to 50°C for 5 min. The suspension was then plated on SDA (n = 4) and incubated for 72 h at 30°C to calculate the survival fraction relative to a control cell suspension.
Murine Experimentation.
A/J mice, (National Cancer Institute, Bethesda, MD), were infected with 1 × 106 cells from each SB4 colony type by tail-vein injection (eight mice per group). Mortality was recorded daily.
Rat Experimentation.
Male Fischer rats from the National Cancer Institute (six rats per group) were infected with 4 × 106 cells from each SB4 colony type by endotracheal inoculation (24). For some rats, dexamethasone-21 phosphate (Sigma) (1.5 mg/liter) was added to the drinking water 7 days later. Rats were killed at day 29–30 of infection by injection of pentobarbital (Anpro Pharmaceutical: Arcadia, CA.). The right lung was fixed in formalin and processed for histopathological studies. Immunohistochemistry for GXM, the major component of the CN capsule, was performed as described (24). The left lung was homogenized in PBS and used for CFU determinations (24).
Protein Extracts and Western Blot Analysis.
CN protein extracts were prepared by a modification of the method described of Hazen and Cutler (25). To enhance cell lysis, specimens were sonicated for two 30-sec intervals with a Sonicator Ultrasonic Processor XL (Misonix, Farmingdale, NY). Approximately 150 μg of protein extract from each colony type was separated in 7.5% SDS/polyacrylamide denaturing gels and transferred to nitrocellulose membranes. Membranes were blocked with 5% dry milk in 0.1% Tween-20 in Tris-buffered saline and incubated with rat serum (1:750 dilution). Goat anti-rat IgG-alkaline phosphatase was used as a secondary antibody, and color was developed by using 5-bromo-4-chloro-3-indolyl phosphate (Sigma).
Statistical Analysis.
The Student t test was used to compare capsule size, average survival fractions after heat and cold exposure, and log-transformed fungal burdens. Mouse survival times were compared by using two-tailed log rank analysis. P values less than 0.05 were considered significant.
RESULTS
A C. neoformans Strain Produces Three Colony Types.
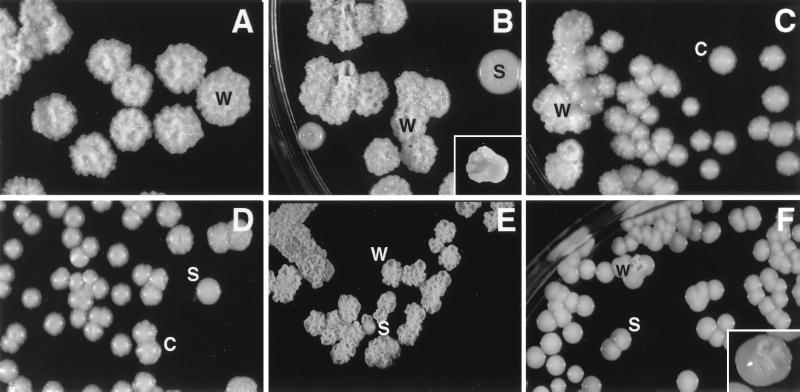
Strain SB4 expressed three colony types, smooth (S), wrinkled (W), and serrated (C) when cultured on SDA at 30°C (Fig. 1). S colonies were round with a smooth dome surface and smooth edges. W colonies had an irregular dome surface with serrated margins. C colonies had a smooth dome and serrated margins. The phenotypes were stable at 30°C after passage in vitro. Sectored colonies composed of wrinkled and smooth regions were noted (Fig. 1). Expression of colony type W was dependent on temperature and nutritional conditions. Colony type W was observed when cells from colony type W were grown at 18°C or 30°C on SDA. At 37°C, cells from colony type W produced smooth and round colonies that were consistently larger and more mucoid (i.e., shiny and moist) than colony types S and C. Colony type W was not observed on other rich medium (malt extract or yeast nitrogen base agar) or on defined agar at 30°C.
Figure 1.
Colony types of strain SB4. (A) Colony types W. (B) Colony type S revertants from colony type W. (Inset) W/S-sectored colony. (C) Colony type W revertants from colony type C. (D) Colony type C with colony type S revertant. Colony types of strain 24067A illustrating colony type S revertant from colony type W (E) and colony type W revertant from colony type S (F). (Inset) W/S-sectored colony. (A and C–F, about ×4; B, about ×2.5.)
DNA typing confirmed that all SB4 colony types were the same strain. CNRE-1 restriction fragment length polymorphisms and electrophoretic karyotype are highly discriminatory DNA typing methods for CN (23, 24). Karyotype analysis of colony types W, S, and C were done on three occasions with different sets of W, S, and C colonies. In two experiments, cells from colony types W, S, and C had indistinguishable karyotypes (data not shown). In the third experiment, a karyotype difference was observed for cells from colony type W that involved two changes in high molecular weight chromosomes (data not shown). Cells from an S revertant colony on a W background did not show any karyotype changes. CNRE-1 restriction fragment length polymorphisms for cells of colony types W, S, and C were the same (data not shown).
Switching Frequencies Between Colony Types.
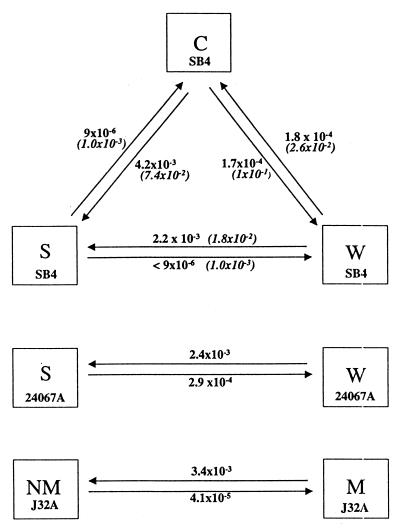
Switching between the various colony types occurred at definable frequencies (Fig. 2). The highest frequency of switching between colony types, occurred for W → S, C → S, and W → C, transitions, each on the order of 10−3 to 10−4. Colony type S was least likely to produce colonies of the other types. No W colony type and only one C colony type was observed when 1.2 × 105 S organisms were examined in 419 plates with 200–300 colonies per plate. Switching frequency determinations were done twice with similar results. Despite the low frequency of S → W conversions, rare S → W colony type revertants were observed during the course of experimentation. UV irradiation increased the frequency of switching between colony types (Fig. 2).
Figure 2.
Switching frequencies for SB4, 24067A, and J32A for cells grown at 30°C on SDA. Frequencies for each strain were determined twice with similar results. Italicized numbers are frequencies after UV irradiation.
Colony Type Characteristics.
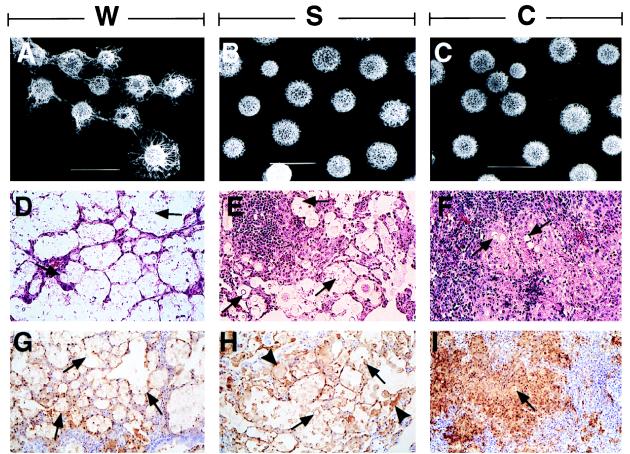
Cells from colony type W had larger capsules than cells from colony type C or S. The average cell diameter (not including the capsule) of cells from colony type W was smaller than cells from colony type S (Table 1). Scanning electron microscopic examination revealed differences in the polysaccharide capsule between the various phenotypes (Fig. 3). Capsular polysaccharide connections between budding organisms were more prominent for cells from colony type W compared with cells from colony type S or C. In addition, cells from colony type W clumped more than cells from colony types S and C. Cells from colony types C and W were less susceptible to killing after exposure to temperatures of −20°C overnight and 50°C for 5 min (Table 1). Other colony type characteristics are listed in Table 1.
Table 1.
Characteristics of SB4: Colony types W, S, and C
| Characteristic | Colony type
|
||
|---|---|---|---|
| W | S | C | |
| Capsule size, μm | 3.9 ± 0.8 | 1.3 ± 0.6 | 1.4 ± 0.6 |
| Cell size, μm | 5.9 ± 0.8 | 7.0 ± 0.9 | 5.6 ± 0.7 |
| Doubling times, h | 1.6 | 1.7 | 2.0 |
| AMPHOTERICIN B MIC, μg/ml | 0.25 | 0.25 | 0.125 |
| FLUCONAZOLE MIC, μg/ml | 2 | 2 | 0.5 |
| 5-FC MIC, μg/ml | 4 | 4 | 2 |
| CN survival at −20°C | 0.22 ± 0.10 | 0.07 ± 0.04 | 0.22 ± 0.11 |
| CN survival at 50°C | 0.19 ± 0.11 | 0.04 ± 0.04 | 0.25 ± 0.04 |
| Mean survival of mice, days | 3.2 ± 1.0 | 6.8 ± 1.3 | 11.9 ± 2.2 |
| Inflamatory response | Minimal | Intermediate | Intense caseous necrosis |
| Polysaccharide localization in vivo | E ≫ 1 | E > 1 | 1 > E |
| Antibody response against W extract | N | 76, 70 kDa | 87, 76, 70, 67, 64, 61, 58, 36 kDa |
| Antibody response aganint S extract | N | 76, 70 kDa | 87, 76, 70, 64, 60, 54, 50 kDa |
| Antibody response against C extract | N | 76, 70, 64 kDa | 76, 70, 67, 64, 62, 61, 54 kDa |
Capsule size is expressed as average capsule radius mean ± 1 SD: (n = 40–50) was determined by light microscopy (P value for S versus W is <0.001: P value for C versus S is 0.28). Cell size is average cell diameter (mean ± 1 SD; n = 40–50, not including the capsule) was determined by light microscopy (P value for S versus W is <0.001: P value for C versus W is 0.05). Susceptibility to amphotericin, fluconazole, or 5-fluorocytosine (5-FL) was determined by National Committee for Clinical Laboratory Standards macrodilution method. Survival fraction after overnight exposure to −20° relative to untreated controls represents the average of four experiments P value for S versus W is 0.025; P value for S versus C is 0.049). Survival fraction after exposure to 50°C for 5 min relative to untreated controls represents the average of three experiments (P value for S versus W is 0.09: P value for S versus C is 0.01). Mouse survival experiments were done twice with similar results (P value for C versus S is 0.002; P value for S versus W is 0.003). Inflammatory responses in rats 4 weeks after endotracheal inoculation are shown. Localization of cryptococcal polysaccharide in rat pulmonary model was determined by immunohistochemistry (E, extracellular; I, intracellular). For antibody responses, the molecular weights of major bands recognized by serum IgG in Western blot analysis are shown. Doubling times were determined by CFU experimentation. N, not detected; MIC, minimal inhibitory concentrations.
Figure 3.
Scanning electron microscopy of SB4 cells from colony types W (A), S (B); and C (C). (×3,000. Bar = 10 μm.) This experiment was done twice using different S, W, and C colonies to ensure the reproducibility of our findings. Hematoxylin and eosin staining of lung tissue form rats infected with SB4 cells from colony types W (D), S (E), and C (F). (×100.) Arrows point to CN. Staining for cryptococcal polysaccharide demonstrates localization of polysaccharide in rats infected with cells from colony types W (G), S (H), and C (I). (×50.) Arrows point to CN and arrowheads point to polysaccharide material inside mononuclear cells.
Animal Studies.
Mice infected i.v. with cells from colony type C lived significantly longer than mice infected with cells from colony types W and S (Table 1).
In rats given intratracheal infection with cells from colony types W, S, and C, the lung fungal burden average (CFU/g; mean ± 1 SD) at day 30 was 7.29 ± 0.06, 6.29 ± 0.63, and 4.42 ± 0.92, respectively (P values for S versus C and W versus C were 0.044 and 0.006, respectively). Dexamethasone treatment was used to reduce the rat immune response and establish whether all colony types had similar virulence in the setting of immunosuppression. The average lung fungal burdens (CFU/g; mean ± 1 SD) for dexamethasone treated rats infected with cells from W, S, and C colonies were 7.24 ± 0.21, 6.94 ± 0.13 and 7.36 ± 0.01, respectively (P values for S versus C and W versus C were 0.39 and 0.1, respectively). The morphology of most colonies isolated from lung homogenates resembled the morphology of the colony type used to infect the animals. For dexamethasone-treated rats infected with cells from colony type C, occasional colony types W and S were isolated from lung homogenates.
There were major differences in the type and intensity of inflammatory responses of immunocompetent rats infected with cells from the various colony types (Fig. 3). Infection with colony type W elicited minimal inflammatory response with little granuloma formation or perivascular cuffing. Most cryptococci were found in large extracellular collections in peribronchiolar areas and inside alveoli. Infection with colony type S elicited occasional granulomas and larger amounts of perivascular cuffing. These granulomas consisted of mature epithelioid cells surrounded by lymphocytes without necrosis. Cryptococci were found in both extracellular collections and inside epithelioid cells. Infection with colony type C resulted in extensive granuloma formation with caseating necrosis, polymorphonuclear leukocyte infiltration, and fibrosis. Most cryptococci were inside epithelioid cells or in areas of caseation.
The distribution of GXM in the rat lung varied significantly depending on the colony type (Fig. 3 and Table 1). In rats infected with colony type C, GXM immunoreactivity was localized to epithelioid cells within granulomas, whereas in rats infected with colony type W, GXM immunoreactivity was localized primarily to extracellular clusters of organisms. In rats infected with colony type S, GXM was localized to epithelioid cells, macrophages, and extracellular clusters of organisms.
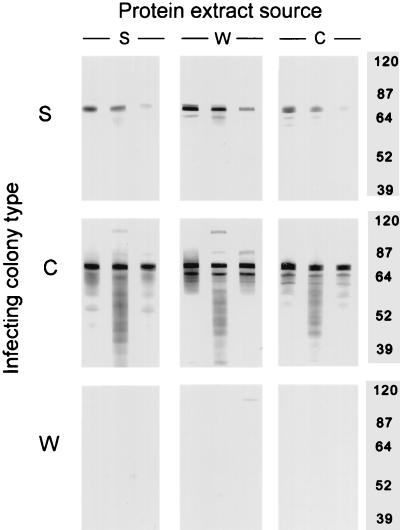
The antibody responses to cryptococcal proteins differed for rats infected with cells from the different colony types. Infection with colony type C elicited the most intense antibody response to cryptococcal proteins, whereas infection with colony type S elicited an intermediate response and infection with colony type W elicited little or no response (Fig. 4, Table 1). Serum from rats infected with cells from colony type W was not reactive even at low dilutions (1:100 dilution; data not shown). The pattern of IgG-reactive bands for rats infected with cells of a particular colony type were similar regardless of which cell protein extract was used. However, there were some differences. Rats infected with cells from colony type C recognized a protein of ≈87 kDa in the protein extract of cells from colony types S and W that was not present in the protein extract of cells from colony type C.
Figure 4.
Western blots of rats infected with cells from the various colony types. The serum of rats (n = 3) infected with each colony type from day 30 of infection was reacted against the protein extracts obtained from colony types S, W, or C.
Experiments with Strains 24067A and J32A.
Strain 24067A exhibited colony types S and W (Fig. 1). Expression of colony type W was dependent on temperature and nutritional conditions. Colony type W was observed when cells from colony type W were grown on SDA at 30°C but not when cells were grown on defined medium plates or when cells were grown on SDA at 37°C. Sectored colonies composed of wrinkled and smooth regions were noted (Fig. 1). Cells from colony type W grown at 30°C tended to clump when grown in Sabouraud’s dextrose broth at 30°C, whereas cells from colony type S grew in suspension. Strain J32A exhibited two different colony types, mucoid and nonmucoid that differed slightly in color (data not shown). Switching between colony types of 24067A and J32A occurred at defined frequencies similar to those found for the SB4 strain (Fig. 2). For both 24067A and J32A, rare additional colony types were also noted, but switching to those colony types was not systematically investigated.
DISCUSSION
Variation in the morphology of CN colonies has been anecdotally described in the literature. In 1950, Benham (26) described variation within individual colonies that resulted in colony sectoring. Drouhet and Couteau (27) in 1951 described three forms of sectored colonies, including mucoid rough- and smooth-sectored colonies. In 1959, Littman (28) suggested that colony sectoring in CN is related to a subpopulation of cells within a colony that produce large capsules or pseudmoycelium. More recently, Granger et al. (29) isolated an avirulent CN clone from a sectored colony. This clone is thinly encapsulated and does not increase polysaccharide production in response to elevated CO2 conditions.
In the course of virulence studies, we noted that SB4 produced unusual colony types in SDA when a liquid culture was left inadvertently at room temperature for several weeks. We confirmed that all colonies belonged to one strain and demonstrated that the various colony types could undergo switching. A similar phenomenon was noted in strains 24067A and J32A. Each of these strains has previously been reported to undergo changes in phenotypic characteristics. Serial clinical isolates of SB4 and J32A had differences in polysaccharide structure and virulence in mice, respectively (6, 8). Strain 24067 was reported to undergo microevolution in vitro resulting in subtypes that differed in: capsule size, growth, and virulence (9). The observation of phenotypic switching in three genetically distinct CN strains that include two serotypes suggests that this phenomenon is a property of the species and extends this process to the Basidiomycota.
Wrinkled colonies were a prominent feature in the phenotypic switching of SB4 and 24067A. Colony wrinkling is probably unrelated to capsule size or cell size alone because these changes are unlikely to promote an irregular colony contour. Furthermore, it is well known and commonly accepted that strains that vary in capsule size produce smooth colony types. Cells from colony types W of both SB4 and 24067A were more adherent to adjacent cells and formed clumps, that could potentially affect cell packing and influence colony morphology. Although colony type W was not observed at 37°C, type W organisms passed in vivo (i.e., at 37°C) retained their ability to produce W colonies, suggesting that phenotypic changes produced by switching are present in the absence of the W colony morphology. Our results suggest that different strains of CN undergo different switching phenomena. In some strains, switching may be associated with readily apparent alterations in colony morphology such as the wrinkled colony type, whereas in others strains the signs of switching may be more subtle. In this regard, phenotypic switching in J32A produced colony types that differed only in mucoid appearance and subtle variation in colony color.
Phenotypic switching resulted in altered susceptibility of the various SB4 colony types to extreme temperatures. As a pathogen with an environmental reservoir, CN is likely to encounter many different conditions (30). Phenotypic switching could provide a mechanism by which CN adapts to various environmental niches. This adaptability could be beneficial in facilitating transitions between environments, including those changes associated with the acquisition of human infection from nature.
The molecular basis for phenotypic switching in CN is not known. In other pathogens, phenotypic switching is mediated by a variety of mechanisms (31). Rearrangement of repetitive DNA elements mediates phenotypic switching for some pathogens including, Haemophilus influenzae (32). For SB4, no DNA rearrangements involving CNRE-1 were detected, however our experiments do not exclude the possibility of rearrangement of other repetitive sequences or small DNA elements as the basis of phenotypic switching in CN. Alternatively, phenotypic switching in CN could be mediated by an epigenetic process, involving silencing and altered chromatin structure. These systems can be very leaky and result in colony sectoring like we observed for SB4 and 24067A (33).
Phenotypic switching has been associated with changes in virulence characteristics for certain pathogens. For C. albicans phenotypic switching produces colony types that differ in several important virulence traits, such as adherence to epithelial cells, production of proteinase, and susceptibility to neutrophil-mediated killing (34–36). Invasive strains of C. albicans are more likely to undergo phenotypic switching than noninvasive isolates (37). In our study, the three colony types of SB4 differed in virulence, as reflected by differences in lung fungal burden in rats and lethality for mice. The colony types that were the most virulent in mice elicited the least inflammation in rats. The mechanism responsible for the differences in inflammatory responses and virulence observed for the different colony types is not known. Administration of dexamethasone to rats infected with the various colony types abolished the differences in inflammatory response and tissue CFU without altering the colony types recovered from the lung. This implies that in immunocompetent rats the differences in lung CFU are the result of the relative efficacy of the tissue responses to the various colony types. The CN capsular polysaccharide has been shown to exert inhibitory effects on the immune system, including inhibition of phagocytosis (38–40). Poorly encapsulated strains of CN evoke more intense inflammatory responses than well-encapsulated strains (41). W type had the largest capsule size in vitro and elicited the least inflammation. The location of polysaccharide antigen in tissue differed for the various colony types and this could influence the inflammatory response. Doubling time may also contribute to differences in virulence. W type cells produced the least inflammation and had the fastest doubling time, whereas C type cells produced intense granulomatous inflammation and had the slowest doubling time. Hence, the lack of inflammation elicited by W type cells may reflect a combination of more polysaccharide antigen during initial infection, extracellular location for polysaccharide antigen, and faster doubling times.
The intensity of tissue inflammatory responses was paralleled by the intensity of the antibody response to cryptococcal protein antigens. W type cells elicited minimal antibody and inflammatory responses. In contrast, C type cells elicited antibodies against multiple CN protein antigens and extensive granuloma formation. These phenomena are probably closely linked. Strong inflammatory responses attract many phagocytic cells that can kill CN, process protein antigens, and present peptides to B and T cells to elicit strong antibody responses. Strong antibody responses may also aid the inflammatory response by producing antibodies that neutralize CN antigens that interfere with the host response. Rats infected with cells from colony type C recognize a CN antigen of ∼87 kDa in the extract of cells from colony types S and W but not in the extract of cells from colony type C. This observation and the isolation of colony types W and S from rats infected with the C type is consistent with the hypothesis that phenotypic switching occurs in vivo.
In summary, we describe phenotypic switching in CN and relate this phenomenon to changes in virulence. Phenotypic switching could contribute to the differences in inflammatory responses described in clinical specimens. For CN infections, granulomatous inflammation has been consistently associated with control and clearance on infection. Our findings indicate that colony type is a variable in the type of inflammatory response and suggest that switching to colony types that elicit minimal inflammation may be a mechanism for persistence of infection.
Acknowledgments
D.L.G. is supported by National Institutes of Health Grant AIO1300. B.F. is supported by a fellowship from the Howard Hughes Medical Institute. A.C. is supported by National Institutes of Health Grants AI22774 and AI33142 and a Burroughs Wellcome Developmental Therapeutics Award.
ABBREVIATIONS
- SDA
Sabouraud’s dextrose agar
- S
smooth
- W
wrinkled
- C
serrated
- CN
C. neoformans
- GXM
glucuronoxylomannan
- CFU
colony-forming unit(s)
References
- 1.Mitchell T G, Perfect J R. Clin Microbiol Rev. 1995;8:515–548. doi: 10.1128/cmr.8.4.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Currie B P, Casadevall A. Clin Infect Dis. 1994;19:1029–1033. doi: 10.1093/clinids/19.6.1029. [DOI] [PubMed] [Google Scholar]
- 3.Sugar A M, Saunders C. Am J Med. 1988;85:481–489. doi: 10.1016/s0002-9343(88)80082-2. [DOI] [PubMed] [Google Scholar]
- 4.Salyer W R, Salyer D C, Baker R D. J Infect Dis. 1974;130:74–77. doi: 10.1093/infdis/130.1.74. [DOI] [PubMed] [Google Scholar]
- 5.Lee S C, Dickson D W, Casadevall A. Hum Pathol. 1996;27:839–847. doi: 10.1016/s0046-8177(96)90459-1. [DOI] [PubMed] [Google Scholar]
- 6.Cherniak R, Morris L C, Belay T, Spitzer E D, Casadevall A. Infect Immun. 1995;63:1899–1905. doi: 10.1128/iai.63.5.1899-1905.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Currie B, Sanati H, Ibrahim A S, Edwards J E, Casadevall A, Ghannoum M A. Antimicrob Agents Chemother. 1995;39:1934–1937. doi: 10.1128/aac.39.9.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fries, B. C. & Casadevall, A. (1998) J. Infect. Dis., in press. [DOI] [PubMed]
- 9.Franzot S P, Mukherjee J, Cherniak R, Chen L, Hamdan J S, Casadevall A. Infect Immun. 1998;66:89–97. doi: 10.1128/iai.66.1.89-97.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Slutsky B, Buffo J, Soll D R. Science. 1985;230:666–669. doi: 10.1126/science.3901258. [DOI] [PubMed] [Google Scholar]
- 11.Clemons K V, Hurley S M, Treat-Clemons L G, Stevens D A. J Med Vet Mycol. 1991;29:165–178. doi: 10.1080/02681219180000271. [DOI] [PubMed] [Google Scholar]
- 12.Clemons K V, Hanson L C, Stevens D A. J Med Vet Mycol. 1996;34:259–264. doi: 10.1080/02681219680000441. [DOI] [PubMed] [Google Scholar]
- 13.Slutsky B, Staebell M, Anderson J, Risen L, Pfaller M, Soll D R. J Bacteriol. 1987;169:189–197. doi: 10.1128/jb.169.1.189-197.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anderson J M, Mihalik R, Soll D R. J Bacteriol. 1990;172:224–235. doi: 10.1128/jb.172.1.224-235.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soll D R, Staebell M, Langtimm C, Pfaller M, Hicks J, Rao T V G. J Clin Microbiol. 1988;26:1448–1459. doi: 10.1128/jcm.26.8.1448-1459.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kennedy M J, Rogers A L, Hanselman L R, Soll D R, Yancey R J. Mycopathology. 1988;109:149–156. doi: 10.1007/BF00437397. [DOI] [PubMed] [Google Scholar]
- 17.Spitzer E D, Spitzer S G, Freundlich L F, Casadevall A. Lancet. 1993;341:595–596. doi: 10.1016/0140-6736(93)90354-j. [DOI] [PubMed] [Google Scholar]
- 18.Wang Y, Casadevall A. Antimicrob Agents Chemother. 1996;40:541–545. doi: 10.1128/aac.40.3.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fries B C, Chen F, Currie B P, Casadevall A. J Clin Microbiol. 1996;34:1531–1534. doi: 10.1128/jcm.34.6.1531-1534.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Franzot S P, Hamdan J S, Currie B P, Casadevall A. J Clin Microbiol. 1997;35:2243–2251. doi: 10.1128/jcm.35.9.2243-2251.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morrow B, Anderson J, Wilson J, Soll D R. J Gen Microbiol. 1989;135:1201–1208. doi: 10.1099/00221287-135-5-1201. [DOI] [PubMed] [Google Scholar]
- 22.Mukherjee J, Cleare W, Casadevall A. J Immunol Meth. 1995;184:139–143. doi: 10.1016/0022-1759(95)00097-t. [DOI] [PubMed] [Google Scholar]
- 23.National Committee for Clinical Laboratory Standards. Reference method for broth dilution antifungal susceptibility testing for yeasts: Proposed standard M 27-P. Villanova, PA: National Committee for Laboratory Standards; 1992. [Google Scholar]
- 24.Goldman D, Lee S C, Casadevall A. Infect Immun. 1994;62:4755–4761. doi: 10.1128/iai.62.11.4755-4761.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hazen K C, Cutler J E. Mycopathologia. 1982;80:113–116. [PubMed] [Google Scholar]
- 26.Benham R W. Ann NY Acad Sci. 1956;50:1299–1314. doi: 10.1111/j.1749-6632.1950.tb39828.x. [DOI] [PubMed] [Google Scholar]
- 27.Drouhet E, Couteau M. Ann Inst Pasteur. 1951;80:456–457. [PubMed] [Google Scholar]
- 28.Littman M L. Am J Med. 1959;27:976–998. doi: 10.1016/0002-9343(59)90181-0. [DOI] [PubMed] [Google Scholar]
- 29.Granger D L, Perfect J R, Durack D T. J Clin Invest. 1985;76:508–516. doi: 10.1172/JCI112000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levitz S M. Rev Infect Dis. 1991;13:1163–1169. doi: 10.1093/clinids/13.6.1163. [DOI] [PubMed] [Google Scholar]
- 31.Deitsch K W, Moxon E R, Wellems T E. Microbiol Mol Biol Rev. 1997;61:281–293. doi: 10.1128/mmbr.61.3.281-293.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hood D W, Deedman M E, Jennings M P, Biseric M, Fleischmann J C, Moxon E R. Proc Natl Acad Sci USA. 1996;93:11121–11125. doi: 10.1073/pnas.93.20.11121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gottschlling D E, Aparicio O M, Billington B L, Zakian V A. Cell. 1990;63:751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- 34.Vargas K, Wertz P W, Drake D, Morrow B, Soll D R. Infect Immun. 1994;62:1328–1335. doi: 10.1128/iai.62.4.1328-1335.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.White T C, Agabian N. J Bacteriol. 1995;177:5215–5221. doi: 10.1128/jb.177.18.5215-5221.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kolotila M P, Diamond R D. Infect Immun. 1990;58:1174–1179. doi: 10.1128/iai.58.5.1174-1179.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones S, White G, Hunter P R. J Clin Microbiol. 1994;32:2869–2870. doi: 10.1128/jcm.32.11.2869-2870.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kozel T R, Gotschlich E. J Immunol. 1982;129:1675–1680. [PubMed] [Google Scholar]
- 39.Murphy J W, Cox R A. Clin Exp Immunol. 1988;73:174–180. [PMC free article] [PubMed] [Google Scholar]
- 40.Dong Z M, Murphy J W. Infect Immun. 1995;63:770–778. doi: 10.1128/iai.63.3.770-778.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Farmer S G, Komorowski R A. Arch Pathol. 1973;96:383–387. [PubMed] [Google Scholar]