Abstract
Type IV pili of Neisseria gonorrhoeae, the Gram-negative etiologic agent of gonorrhea, facilitate colonization of the human host. Gonococcal PilT, a protein belonging to a large family of molecules sharing a highly conserved nucleotide binding domain motif, has been shown to be dispensable for organelle biogenesis but essential for twitching motility and competence for genetic transformation. Here, we show that the defect in pilus biogenesis resulting from mutations in the pilC gene, encoding a putative pilus-associated adhesin for human tissue, can be suppressed by the absence of functional PilT. These data conclusively demonstrate that PilT influences the Type IV pilus biogenesis pathway and strongly suggest that organelle expression is a dynamic process. In addition, these findings imply that PilT antagonizes the process of organelle biogenesis and provide the basis for a model for how the counteractive roles of PilT and PilC might relate mechanistically to the phenomenon of twitching motility.
The adherence of pathogenic bacteria to mammalian tissue represents a crucial step in the pathogenesis of infections with the specificity of adhesin–receptor interactions defining host range and tissue tropism. In virtually all Gram-negative pathogens that colonize mucosal surfaces, attachment is mediated by supramolecular polymeric fibers termed pili or fimbriae (1).
Many bacterial pathogens, including Neisseria gonorrhoeae, Neisseria meningitidis, enteropathogenic E. coli, Vibrio cholerae, Moraxella bovis, Pseudomonas aeruginosa, and Dichelobacter nodosus, express the Type IV pilus colonization factor. These pili share structural, biochemical, antigenic, and functional features (2). In addition, the prepilin subunits of Type IV pili (Tfp) display a high degree of identity to each other within the N terminus, the domain that functions in inner membrane insertion and proteolytic processing and is predicted to form the central helical core of the pilus filament (3). Further evidence for the relatedness of these organelles can be found in the conservation of genes and gene products required for their biogenesis. These include prepilin peptidases, soluble proteins with essential nucleotide-binding motifs, polytopic inner membrane proteins, and a family of outer membrane proteins that appear to function as gated channels or pores through which pili are extruded (4). Homologues of some Tfp biogenesis machinery components have been shown to be required for secretion of toxins and hydrolases by Gram-negative bacteria (5), competence for transformation in Bacillus subtilis (6), and filamentous phage morphogenesis (7). Therefore, it appears that these elements are part of a highly conserved pathway for membrane translocation of macromolecules. However, with the exception of the prepilin peptidases (8), the precise functions of these conserved components remain unknown.
Gonococcal Tfp appear to play an important role in promoting adherence to mucosal epithelial cells, a crucial step in colonization of the human host (9). This function requires the simultaneous expression of pili and the outer membrane protein PilC, which originally was identified by virtue of its presence in pilus preparations (10, 11). Although nonpiliated, PilC+ mutants fail to adhere, PilC appears to be the adhesive moiety of gonococcal pili because purified PilC binds specifically to cells of human origin and can competitively block the adherence of piliated gonococci (12). Attempts at isolating piliated, PilC− mutants to confirm this have been complicated because PilC is required for organelle biogenesis (10).
In addition to their role in adherence, gonococcal Tfp are required for a novel mode of flagella-independent surface translocation known as twitching motility (13, 14). Tfp-associated motility also is found in P. aeruginosa as well as in Myxococcus xanthus, where it is manifest as social gliding motility (15, 16). The exact role of Tfp in this phenomenon has yet to be elucidated. Mutations in the highly conserved pilT gene lead to a piliated but nonmotile phenotype in each of these three species (17–19). PilT proteins are members of a large family of putative nucleotide binding proteins (TrbB-like proteins) that are involved in membrane translocation of macromolecules in prokaryotes (20). Phylogenetic analysis has shown that PilT proteins are most similar to the other TrbB-like family members required for Tfp biogenesis (M.K., unpublished data). In addition, the M. xanthus pilT gene is clustered with other genes whose products are required for Tfp biogenesis (17). It seemed plausible then that PilT proteins influence the Tfp biogenesis pathway. However, no alterations in Tfp structure, morphology, or expression have been noted in any of the paralyzed pilT mutants except in P. aeruginosa, in which mutants have been found to be hyperpiliated (17–19). These findings prompted us to examine whether gonococcal mutants lacking both PilT and PilC, a known Tfp biogenesis component, might display unique phenotypes.
Here we show that PilT influences the Tfp biogenesis pathway. We discovered that loss of function mutations in pilT alleviate the requirement for PilC in pilus biogenesis. This result implies that PilT antagonizes the biogenesis process, consistent with its potential role in organelle retraction or destabilization. This finding also made it possible to characterize genetically defined mutants that express Tfp in the absence of PilC. We demonstrate that these mutants are incapable of adhering to human epithelial tissue.
MATERIALS AND METHODS
Bacterial Strains and Plasmids.
The bacterial strains used in this study are described in Table 1. E. coli and gonococcal strains were grown as described (21). Gonococcal strain N401 was created by replacing the isopropyl β-d-thiogalactopyranoside (IPTG)-inducible recA allele (recA6) linked marker (tetM) in strain N400 (21) with a kanamycin resistance gene. This was accomplished by replacing the SacI fragment of pVD300recA6 (22) with a HincII fragment from pUCKan (accession no. X06404) containing the kanamycin resistance gene to yield pVD300recA6(Kan). This plasmid then was used to transform strain N400. Transformants were selected for resistance to kanamycin (50 μg/ml) and sensitivity to tetracycline (4 μg/ml). The pilT-inducible strain MW4 was created by transforming N401 with placP-PilT as described (18).
Table 1.
Genotype and phenotype of N. gonorrhoeae strains used in this study
| Strain | Parent strain | Relevant genotype | Piliation* | Adherence† | Reference |
|---|---|---|---|---|---|
| N400 | VD300 | recA6(tetM)‡ | ++ | +++ | 24 |
| N401 | N400 | recA6(kan) | ++ | +++ | This study |
| MW1 | N400 | pilC1∷erm | ++ | +++ | This study |
| MW2 | N400 | pilC2∷cat | + | + | This study |
| MW3 | MW1 | pilC1∷erm, pilC2∷cat | − | − | This study |
| MW4 | N401 | pilTind | ++ | +++ | This study |
| MW5 | MW4 | pilTind, pilC1∷erm | ++ | +++ | This study |
| MW6 | MW4 | pilTind, pilC2∷cat | ++ | + | This study |
| MW7 | MW5 | pilTind, pilC1∷erm, pilC2∷cat | ++ | − | This study |
| GT101 | N400 | pilTdud1 | ++ | +++ | 18 |
| MW8 | GT101 | pilTdud1, pilC2off | ++ | + | This study |
| GT102 | N400 | pilTΔQSL | ++ | +++ | 18 |
| MW9 | GT102 | pilTΔQSL, pilC2? | ++ | + | This study |
| MW10 | GT102 | pilTΔQSL, pilC2off | ++ | + | This study |
Piliation was accessed by transmission electron microscopy: −, <1 pilus/diplococci; +, 1–5 pili/bacterial cell; ++, >10 pili/bacterial cell.
Adherence was determined by the average number of bacterial cells associated with each primary human corneal epithelial cell: −, <0.1 bacterium/epithelial cell; +, 1–10 bacteria/epithelial cell; +++, >100 bacteria/epithelial cell.
recA6 is an IPTG-inducible allele of recA.
pilC null alleles were generated by PCR amplification of internal fragments of each gene. pilC1 was amplified by using primers AJM.C1.589F (5′-CGAATTCAGACGGCGACCTCATCCTTGCTTCCATTCAG) and AJM.C1.1672B (5′-CGAATTCAGACGGCTATACGCACAGAACAGGAGCAGGCTC). Primers contained gonococcal DNA uptake sequences (bold) and flanking EcoRI restriction sites (italicized). The product spans nucleotides 589-1672 of pilC1 (accession no. Z50180). A fragment of pilC2 was amplified by using primers AJM.C2.262F (5′-GAATTCAGACGGCATACGAGCCAGAGAAACTGGAAC) and AJM.C2.1581B (5′-GAATTCAGACGGCTTCTTTTGTGCGGATGCGG). The pilC2 product spans nucleotides 262-1581 (accession no. Z49120). The PCR fragments were cloned into the EcoRI site of pUC19. An internal EcoRV fragment in pilC1 (nucleotides 1034–1188) was replaced with a fragment carrying an erythromycin resistance gene to yield pSPAM2. The same approach was used to replace an internal EcoRV fragment of pilC2 (nucleotides 722-1266) with a chloramphenicol acetyl-transferase gene cassette to produce pSPAM3. The wild-type gonococcal strain N400 and the pilT-inducible strain MW4 were transformed with pSPAM2 and/or pSPAM3 and were selected on the appropriate antibiotics to generate the pilC null strains described in Table 1.
SDS/PAGE, Immunoblotting, and Staining.
Procedures for SDS/PAGE and immunoblotting have been described (23). PilE and PilC were detected by immunoblotting pilus preps and whole cell lysates by using rabbit polyclonal antibodies and alkaline phosphatase coupled goat anti-rabbit antibodies (Tago). PilC specific sera was a gift of A.-B. Jonsson (Karolinska Institute, Stockholm). PilE specific sera (lot 2–66) was described (24). The pilin subunit from purified pili was detected by Coomassie staining of SDS/PAGE gels. Pilus purification was carried out as described (18).
Protein Quantitation.
Total cellular protein was extracted by resuspending gonococcal cell pellets in lysis buffer (0.2 M potassium phosphate buffer, pH 7.5/20% acetone/40 mM EDTA/0.1% Triton X-100) and incubating on ice for 15 min. Protein concentration was determined by using the Bio-Rad Protein Assay and BSA standards according to the manufacturer’s instructions.
DNA Sequencing.
Fragments containing the poly G stretch of pilC1 and pilC2 were PCR amplified by using Vent DNA polymerase and primers PilC1–5′ (5′-ACTTTGAAACGGCAGGTTTTCCGC), PilC1–3′ (5′-ATTCTGCTTTACCTCGGGCTGGTT), PilC2–5′ (5′-AACTTTAAAAAGGCGGGTTTTCCGC), and PilC2–3′ (5′-TTTCTCTGGCTCGTATGGCTAAATG). PCR products were sequenced by using the Thermo Sequenase radiolabeled terminator cycle sequencing kit (Amersham) according to the manufacturer’s protocol.
Epithelial Cell Adherence Assay.
Primary cultures of human corneal epithelial cells were established as described (25). For use in adherence assays, epithelial cells were grown on 12-mm circular glass or thermanox coverslips in 1 ml of medium. Before the start of the infection, the medium was replaced with 1 ml DMEM supplemented with 5% fetal calf serum. Gonococci were suspended in tissue culture medium and were added to the cells (2 × 107 per well). When appropriate, IPTG was present during the assay. After 1 h incubation (37°C, 10% CO2), the infection was stopped by rinsing the cells three times with 1 ml of Dulbecco’s PBS to remove unbound bacteria, followed by fixation (at least 30 min at room temperature) in 0.1% glutaraldehyde/1% paraformaldehyde in Dulbecco’s PBS. Specimens were stained with crystal violet (0.007% in distilled water), and bacterial adherence was scored with an Olympus (New Hyde Park, NY) BH-2 microscope.
Electron Microscopy.
Colonies of bacteria grown on GC agar plates for 12 h were touched gently with pioloform-coated grids and were air-dried. Grids subsequently were stained with 1% ammonium molybdate in water for 2 min, were rinsed once with water, were air-dried, and were viewed in a Hitachi (Tokyo) HU-11E-1 electron microscope.
RESULTS
An Assay To Examine the Potential Influence of PilT on Tfp Biogenesis.
Because PilT molecules are closely related to other proteins within the TrbB-like family that are essential for Tfp biogenesis, we theorized that new insights into PilT function might be gained from studying mutants that carry both pilT mutations and lesions in genes whose products are essential for Tfp expression. Both nonpiliated and PilT mutants are nontransformable owing to defects at the level of DNA uptake (18), a situation that made it problematic to construct double mutants by standard allelic replacement methods. As a means to circumventing this obstacle, we used the strain MW4, which carries the pilT gene under control of an inducible promoter. In the absence of induction, no detectable PilT is produced whereas addition of the inducer (IPTG) leads to gene expression and wild-type Tfp-associated phenotypes (18). In this background, mutations in biogenesis genes could be introduced into the genome by transformation after transient pilT expression.
Lack of Transcription of pilT Suppresses the Biogenesis Defect in pilC Mutants.
Mutants that fail to express intact PilC have been reported to have dramatically reduced levels of Tfp (10). The gonococcal genome contains two complete, unlinked alleles of pilC whose expression is phase-variable owing to frameshifting within a poly G stretch early in their ORFs. To assess the influence of PilC on Tfp biogenesis in the parental strain N400, inactivated alleles of pilC1 and pilC2 (containing deletions in the pilC ORFs in conjunction with the insertion of selectable gene markers) were introduced by transformation. Strain MW1, which carries a disruption of pilC1, was phenotypically wild-type (Fig. 1B) whereas inactivation of pilC2 (strain MW2) yielded cells that were phenotypically nonpiliated (Fig. 1C). Sequencing of the poly G stretches within both genes confirmed that the pilC1 allele is out-of-frame (12 G residues) whereas that of pilC2 is in-frame (13 G residues). These findings demonstrate that the strains used here express PilC from the pilC2 allele whereas the pilC1 allele is in a phase-off configuration, confirming similar findings made in previous studies of related strains (10, 11).
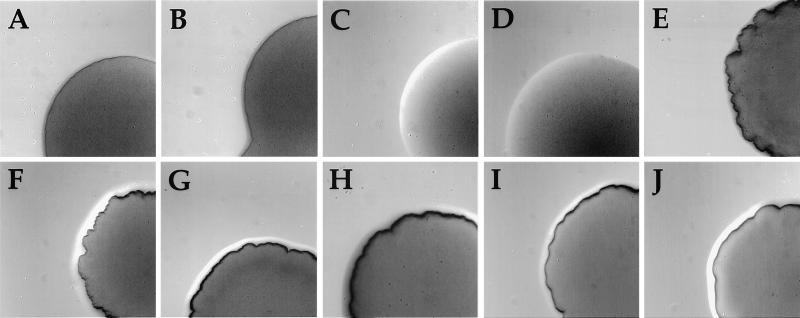
Figure 1.
Contributions of pilC and pilT to Tfp expression as assessed by colony morphologies, a readout of pilus-dependent autoagglutination. Colonies were photographed after 24 h of growth on Gc agar plates at a magnification of 70× using a stereomicroscope. (A) N400 (wild-type). (B) MW1 (pilC1∷erm). (C) MW2 (pilC2∷cat). (D) MW3 (pilC1∷erm, pilC2∷cat). (E) MW4 (pilTind). (F) MW5 (pilTind, pilC1∷erm). (G) MW6 (pilTind, pilC2∷cat). (H) MW7 (pilTind, pilC1∷erm, pilC2∷cat). (I) MW8 (pilTdud1, pilC2off). (J) MW9 (pilTΔQSL, pilC2?—a variant in which PilC2 does not associate with pili).
Soluble, truncated forms of pilin (termed S-pilin) lacking the first 40 residues of the mature protein are seen in all Tfp biogenesis mutants characterized by our group (4). This altered processing of pilin appears to be a consequence of a disrupted biogenesis pathway, and although its precise basis is not clear, its presence provides a sensitive marker for reduced or disrupted Tfp expression. Consistent with their defect in Tfp expression, strain MW2 carrying the defective pilC2 allele produced S-pilin (data not shown). However, when examined by electron microscopy, these cells expressed low levels of pili, with ≈10% of cells exhibiting one or more pilus filaments (Fig. 2D). Piliation in this background, albeit reduced, was confirmed by the ability to recover low amounts of pili by a standard purification scheme (Fig. 3, lane 3). To assess the basis for residual pilus expression in this background, strain MW3 was constructed in which both alleles of pilC were disrupted. This strain was phenotypically nonpiliated (Fig. 1D), and no pili were detected by purification (Fig. 3, lane 4) or by electron microscopic examination (data not shown) whereas S-pilin was readily detectable (Fig. 4, lane 2).
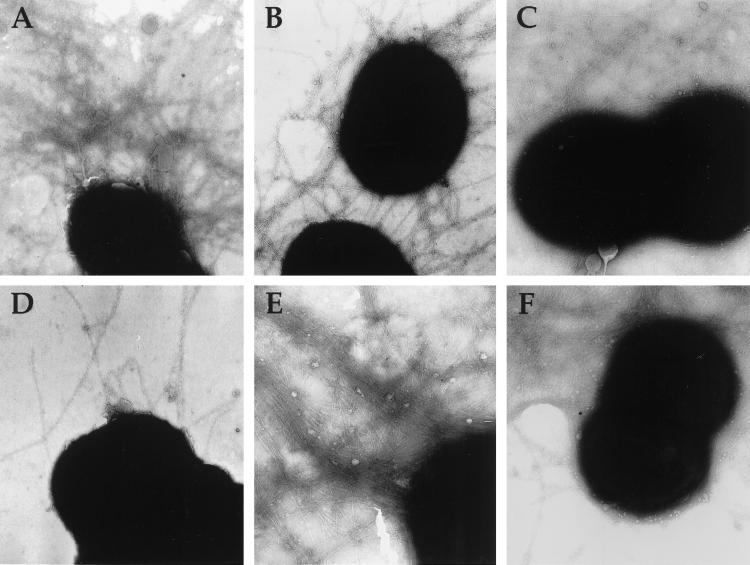
Figure 2.
Piliation of gonococcal strains analyzed by transmission electron microscopy. (A) N400 (wild-type). (B) MW4 (pilTind). (C) GT102 (pilTΔQSL). (D) MW2 (pilC2∷cat). (E) MW7 (pilTind, pilC1∷erm, pilC2∷cat). (F) MW9 (pilTΔQSL, pilC2?). Magnification at 90,000×.
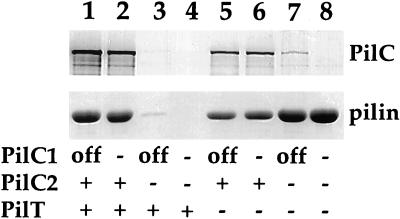
Figure 3.
Quantitative and qualitative analysis of pili purified from gonococcal strains. (Upper) Immunoblotting of purified pili by using rabbit antibodies specific for PilC1 and PilC2. (Lower) Coomassie-stained SDS/PAGE gel showing the relative amounts of pilin subunit in purified pili. The amounts of samples loaded were equalized based on the total protein concentration of whole cells. Lanes: 1, N400 (wild-type); 2, MW1 (pilC1∷erm); 3, MW2 (pilC2∷cat); 4, MW3 (pilC1∷erm, pilC2∷cat); 5, MW4 (pilTind); 6, MW5 (pilTind, pilC1∷erm); 7, MW6 (pilTind, pilC2∷cat); 8, MW7 (pilTind, pilC1∷erm, pilC2∷cat). With regard to PilC expression, “+” denotes expression, “off” denotes a phase-off allele, and “−” denotes a knockout mutant. For PilT, “+” denotes wild-type expression while “−” denotes the noninduced pilTind allele.
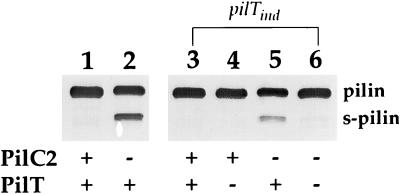
Figure 4.
Detection of S-pilin, a correlate of a biogenesis defect, in a PilC− background requires pilT expression. Shown is immunoblotting of whole cell lysates by using the pilin-specific mAb MC02. Lanes: 1, N400 (wild-type); 2, MW3 (pilC1∷erm, pilC2∷cat); 3, MW4 (pilTind); 4, MW4 (pilTind); 5, MW7 (pilTind, pilC1∷erm, pilC2∷cat); 6, MW7 (pilTind, pilC1∷erm, pilC2∷cat). With regard to PilT expression, in lanes 1 and 2, “+” denotes the wild-type pilT allele; in lanes 3–6, “+” denotes addition of IPTG and “−” denotes no IPTG.
We then examined the influence of these same pilC mutations in strain MW4, the background in which pilT transcription was repressed. Strain MW5, created by introduction of the pilC1 knockout mutation, was phenotypically indistinguishable from its parent (Fig. 1 E and F and Table 1). In contrast, introduction of the inactivated pilC2 allele into MW4 (creating MW6) yielded transformants with colony morphologies normally representative of highly piliated variants but slightly different from that found for the parental strain (Fig. 1G). These pilC mutants expressed levels of Tfp as high, if not higher, than that found for N400 or MW4. In addition, the gross morphology of the pilus fibers was indistinguishable from those seen in the other parental strains (data not shown). Immunoblotting revealed that the pilin present in these mutants was not processed detectably into S-pilin (data not shown). Given the possibility that a product derived from the out-of-frame pilC1 allele might contribute to Tfp biogenesis, a derivative of MW5 was constructed in which both pilC alleles were disrupted, giving rise to strain MW7. As found for the pilC2 knockout mutant in this background, this mutant had a piliated colony morphology (Fig. 1H), showed high levels of piliation by electron microscopy (Fig. 2E), and did not express S-pilin (Fig. 4, lane 6).
Next, we assessed the relative levels of Tfp and the status of putative Tfp-associated proteins in these strains. For this purpose, Tfp were purified from the isogenic derivatives and were examined after SDS/PAGE by protein staining and immunoblotting (Fig. 3). In all cases, the migrations of pilins in these backgrounds were identical to one another, although the relative abundance of pilin was reduced in strains MW4 and MW5 (Fig. 3, lanes 5 and 6). Reduction in pilin found for these strains did not reflect a diminution of Tfp levels because electron microscopy does not reveal discernible differences between wild-type and pilT mutants. Rather, it appears to represent the propensity of Tfp from pilT mutants to pellet during the centrifugation step used to remove whole cells after shearing off of pili (unpublished data). Coomassie stained protein profiles of pili isolated from MW6 and MW7, the PilC-lacking derivatives of MW4, were identical to that of the other strains save for the absence of a 110-kDa protein (data not shown). Immunoblotting demonstrated that this molecule was PilC and also revealed that the pili from MW6 (pilC2∷cat) contained low levels of reactive material (Fig. 3, lane 7) that was shown to be derived from the pilC1 allele (Fig. 3, lane 8). These findings established that gonococcal Tfp can be expressed at high levels in the absence of PilC, providing that transcription of pilT is abolished.
Given that the suppression of the PilC− biogenesis defect occurred in the absence of pilT transcription, we tested whether restoration of PilT expression would reverse this effect. When grown in the presence of IPTG strains, MW6 and MW7 behaved identically to PilC− strains. The phenotypes manifest in these strains propagated under pilT-inducing conditions included a dramatic reduction in Tfp expression (data not shown) and elaboration of pilin as S-pilin (Fig. 4, lane 5).
Characterization of Piliated, PilC Phase-Off Variants Arising in pilT Null Mutants.
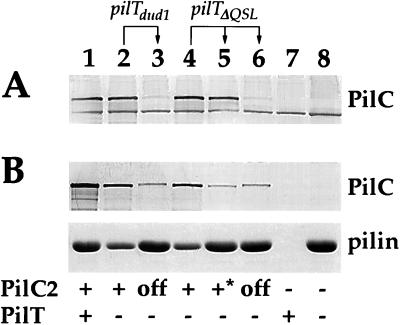
In earlier studies, alleles of pilT that carry loss-of-function mutations were identified and characterized (18). These include a frameshift mutation occurring early in the pilT ORF (the pilTdud1 allele) and an in-frame deletion mutation that results in the loss of three amino acid residues from the C-terminal part of the protein (the pilTΔQSL allele). Strains bearing these defective alleles are nontransformable, because of a block at the level of DNA uptake, making it impossible to introduce defined pilC mutations into these backgrounds. However, earlier studies had demonstrated that PilC expression is subject to high frequency phase variation as a consequence of frameshift mutations within the poly G stretch positioned early in the pilC ORFs (10, 11). We reasoned that spontaneous PilC2 phase-off mutants would arise at a readily detectable frequency in these backgrounds. Colony variants with the same morphology seen for the piliated, PilC− knockout mutants were found at frequencies of ≈10−3 in all of these pilT mutants (Fig. 1 I and J), and, in each case, these derivatives expressed high levels of Tfp as detected by TEM (Fig. 2F). Whole cell lysates of such variants arising in strains GT101 and GT102, bearing the pilTdud1 and pilTΔQSL alleles, respectively, were analyzed in more detail by immunoblotting, and, in all but one case, PilC protein was detected at reduced levels equivalent to the previously characterized pilC phase-off mutants (Fig. 5A, lanes 3 and 6). Direct nucleotide sequencing of the early region of the pilC2 ORF in the variants showed that the reduction of expression resulted from frameshifting in the poly G stretch (data not shown). In line with these observations, variants that displayed the colony morphotype of their original PilT− progenitors appeared at frequencies of 10−3 in these backgrounds, and, in all cases examined, they regained high level expression of PilC (data not shown). In the one exception noted above, a variant isolated in the pilTΔQSL background (MW9), which had a colony morphology characteristic of strains having simultaneous defects in pilT and pilC, retained unaltered levels of immunoblot-detectable PilC2 (Fig. 5A, lane 5).
Figure 5.
Quantitative analysis of pili and PilC in pilC2 phase-off variants arising in pilT null mutants. (A) Immunoblotting of whole cell lysates using rabbit antibodies specific for PilC1 and PilC2. (B) Purified pili. (Upper) Immunoblotting using rabbit antibodies specific for PilC1 and PilC2. (Lower) Coomassie-stained SDS/PAGE gel showing the relative amounts of pilin subunit. Amounts of pilus preparations loaded were standardized based on total protein concentration of extracted cells. Lanes: 1, N400 (wild-type); 2, GT101 (pilTdud1); 3, MW8 (pilTdud1, pilC2off); 4, GT102 (pilTΔQSL); 5, MW9 (pilTΔQSL, pilC2?); 6, MW10 (pilTΔQSL, pilC2off); 7, MW3 (pilC1∷erm, pilC2∷cat); 8, MW7 (pilTind, pilC1∷erm, pilC2∷cat). Note that strain MW9 (lane 5) expresses wild-type levels of PilC2 antigen in whole cell lysates but has reduced amounts in purified pili and has a PilT−, PilC− phenotype. Strains MW3 and MW7 (lanes 7 and 8) are pilC1, pilC2 double-knockout mutants whereas all other strains used carry the pilC1 phase-off allele.
Examination of protein profiles of pili purified from these phase variants by staining demonstrated that, in every case, the only discernible alteration in the variant pili was the absence of the PilC protein (data not shown). When PilC antisera was used in immunoblotting of the same samples, a low but clearly reproducible level of PilC protein was seen in these fractions (Fig. 5B, lanes 3 and 6). Surprisingly, strain MW9, which has a phenotype similar to the phase-off variants but expressed wild-type levels of PilC2 in whole cell lysates, contained reduced levels of this molecule in purified pili (Fig. 5B, lane 5). This finding demonstrates that the piliated, PilC−-related colony morphology can be correlated not only with a quantitative defect in PilC expression but also with the failure of PilC to be associated with purified pili.
Piliated, PilC− Strains Are Deficient in Human Epithelial Cell Adherence.
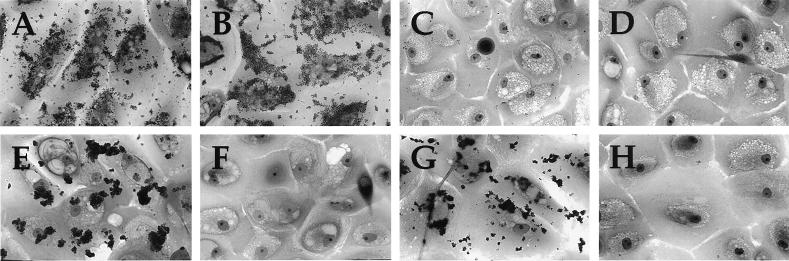
Genetically defined mutants expressing Tfp in the absence of PilC have not been available previously. The isolation of such strains here made it possible to address directly the relative contribution of PilC to Tfp-associated adherence for human epithelial cells. The capacity of derivatives of N400 bearing knockout mutations in pilC2 to adhere to human primary corneal epithelial cells was dramatically diminished (Fig. 6C; ≈5 diplococci/epithelial cell for MW2 vs. >100 diplococci/epithelial cell seen for the wild-type strain). We recovered the adherent cells seen using MW2 in this assay and found that only 10% of the colonies arising corresponded to piliated, pilC1 phase-on variants whereas the rest were identical to the inoculating strain. When retested, those pilC1 phase-on variants adhered as well to these epithelial cells as the piliated, pilC2 phase-on strains (data not shown), confirming that PilC1 and PilC2 are functionally interchangeable, not only with respect to Tfp biogenesis but also in promoting epithelial cell adherence. The pilC1, pilC2 double knockout mutant strain MW3 adhered at levels below that seen with the pilC2 knockout (Fig. 6D; <0.1 diplococcus/epithelial cell vs. ≈5 diplococci/epithelial cell), a finding consistent with the hypothesis that partial pilus biogenesis and adhesive functions can be imparted by the pilC1 phase-off allele.
Figure 6.
Adherence of genetically defined gonococcal mutants and variants for human corneal epithelial cells. Adherent gonococci are stained with crystal violet. (A) N400 (wild-type). (B) MW1 (pilC1∷erm). (C) MW2 (pilC2∷cat). (D) MW3 (pilC1∷erm, pilC2∷cat). (E) MW4 (pilTind). (F) MW7 (pilTind, pilC1∷erm, pilC2∷cat). (G) GT102 (pilTΔQSL). (H) MW9 (pilTΔQSL, pilC2?).
Strains with pilT mutations adhere to the human epithelial cell line ME180 as well as their isogenic wild-type parent, although they bind in a more localized fashion because of their increased autoagglutination (18). Virtually identical results were found here by using the human primary corneal epithelial cells (Fig. 6 E and G and Table 1). A severe defect in epithelial cell adherence was found for all piliated strains failing to express functional PilT in combination with either of the pilC knockout mutations (Fig. 6F) or the phase-off alleles (data not shown and Table 1), regardless of the particular combinations of mutations tested. In the strains using the phase-off pilC alleles, spontaneous variants in which a phase-on switch in pilC had occurred (identified by virtue of their PilT− colony morphology) regained epithelial cell adherence (data not shown). These findings ruled out the possibility that a second mutation may have occurred in these backgrounds to account for the adherence defect.
Given the strong association between PilC and epithelial cell adherence, we examined the adhesive property of strain MW9, which expresses both Tfp (Fig. 2F) and PilC2 but in which PilC2 fails to copurify with pili (Fig. 5, lane 5). Despite the presence of these two components, this strain was as nonadherent for human primary corneal epithelial cells (Fig. 6H) as the other piliated, PilC− mutants (Fig. 6F and Table 1).
DISCUSSION
The ability of most Gram-negative pathogens to colonize mucosal sites requires the expression of pili. The primary role of these proteinaceous appendages as colonization factors appears to be the presentation of adhesive molecules in forms capable of binding to their specific epithelial cell receptor. The strongest previous evidence for PilC being the Tfp-associated epithelial cell adhesin comes from the finding that purified PilC is capable of competitively blocking the human epithelial cell adherence of both piliated gonococci and meningococci (12). Here, we found that genetically defined gonococcal pilT, pilC mutants expressing high levels of Tfp were incapable of binding to human primary corneal epithelial cells. These results concur with results obtained by using a piliated pilC mutant that was reported to carry at least one and perhaps two other uncharacterized mutations that were responsible for suppression of the pilC biogenesis defect (11). All pilT mutants characterized here adhered quantitatively as well, if not better, to human epithelial cells than to wild-type cells. Therefore, PilC appears to function as an adhesin under conditions in which it is not required for biogenesis. Nonetheless, its ability to function as an adhesin appears to correlate with its capacity to associate with Tfp as evidenced by the adherence defect of the mutant that expresses Tfp and wild-type levels of PilC2 protein but in which PilC2 fails to copurify with pili. Future studies should determine whether this mutant carries a mutation in pilC2 or in a second gene whose product is required for PilC to associate with Tfp. This particular class of mutant also demonstrates how strains lacking PilT provide a unique background in which to probe the relationships between the structure of PilC proteins and their capacity to function as Tfp-associated adhesins.
It was surprising that, when expression of PilC2 was blocked by gene disruption in a pilT background, low levels of PilC antigen still could be detected by immunoblotting purified Tfp. Low levels of PilC antigen also were seen in whole cell lysates and purified Tfp from the strains in which both pilC alleles were in their phase-off configuration. The simplest explanation would be that this PilC antigen reflects the presence of phase-on variants within the cell population. However, the frequencies with which phase-on variants arise (10−3) is too low to account for the levels of PilC antigen seen (data not shown). We believe this PilC antigen represents polypeptide translated from the out-of-frame mRNA caused by ribosomal frameshifting. A similar phenomenon in which functional levels of gene product are expressed from an out-of-frame allele has been observed for the N. gonorrhoeae lsi2 gene, which also contains a poly G stretch in its ORF (26). Regardless of the basis for this phenomenon, Tfp purified from pilT mutants with phase-off pilC alleles contains low levels of PilC antigen, and those strains adhere poorly to human epithelial tissue but at levels higher than that found for the pilC double-knockout mutants. This may indicate that a certain threshold level of PilC associated with Tfp needs to be achieved in order for efficient adherence. Alternatively, the PilC found with purified Tfp in these backgrounds may not promote adherence because it is not displayed properly or associated with Tfp in the same manner as in the strains expressing high levels of adherence.
Although it is now clear through studies of P. aeruginosa, M. xanthus, and N. gonorrhoeae that Tfp expression is required for twitching motility, the molecular basis for these associations remains unknown. Studies in all three species have shown that Tfp are necessary, but not sufficient, for this property because, in a remarkable convergence of findings, piliated but nonmotile mutants in all three species have been found to carry loss-of-function mutations in pilT. Here, we provide direct evidence that gonococcal PilT influences the Tfp biogenesis pathway as assessed by the genetic suppression of the defect in pilC mutants when functional PilT is absent. Viewed in a slightly different way, these findings show that pilC mutants are deficient in Tfp expression because they express PilT.
The isolation of extragenic suppresser mutations often implies that the products of the two genes either interact with one another or act on a common pathway. PilC is reported to localize to the outer membrane (27) as well as to Tfp fibers (12) whereas gonococcal PilT has been shown to be localized to the cytoplasm and the inner membrane (28). Although one must interpret protein localization findings with caution, it seems likely that these molecules do not interact directly with one another. The fact that loss of PilT function suppresses the pilC defect appears to indicate that the two molecules have antagonistic effects on Tfp biogenesis. Although the role of PilC in biogenesis is not understood, the following observations may relate to its function. First, gonococcal mutants lacking functional PilQ, the secretin family member that, in its multimeric form, appears to act as the outer membrane channel for Tfp biogenesis, shed PilC into the extracellular milieu (29). Second, immunogold labeling has been interpreted as indicating that PilC colocalizes with Tfp filaments (12). As such, we propose that PilC functions as a biogenesis factor by virtue of its physical association with Tfp. Specifically, we envision that PilC acts either as an initiator of Tfp fiber extrusion or as a chaperone for Tfp as they are formed. Twitching motility has been proposed to be a consequence of pilus retraction (15), but, for technical reasons, direct evidence for this possibility is lacking. Still, the retraction hypothesis suggests a mechanism for the findings presented here as follows: Tfp formation occurs in the absence of PilC, but those pili are not stabilized and are retracted rapidly. Suppression of the pilus biogenesis defect in pilC mutants might then result from a defect in the retraction step because of loss-of-function mutations in pilT. Although speculative, this scenario lends itself to a number of testable hypotheses based on the notion that PilC and PilT have counteractive effects on Tfp biogenesis/extrusion that lead to the phenomenon of twitching motility.
In summary, we have demonstrated that the PilT protein influences the gonococcal Tfp biogenesis pathway and have provided evidence that Tfp biogenesis is a dynamic process. Given that gonococcal PilT is required for DNA uptake during genetic transformation and twitching motility, these results provide a link for how PilT expression might physically influence these processes. In addition to the potential relevance to studies of twitching motility in other species, this gonococcal system may provide insights into the Type II protein secretion pathway of Gram-negatives and the gene transfer systems of both Gram-negative and Gram-positive species, which use a high-related set of components.
Acknowledgments
We thank Alexey Merz in the So laboratory, Oregon Health Sciences University, Portland, OR for providing the MS11 pilC knockout alleles. Human corneas were generously provided by Dr. P. Buck, Montana Eye Bank, Missoula, MT. This work was supported by Public Health Service Grant A127837 (to M.K.) and National Institutes of Health Clinical Research Center grant M01 RR 00042.
ABBREVIATIONS
- Tfp
Type IV pili
- IPTG
isopropyl β-d-thiogalactopyranoside
References
- 1.Hultgren S J, Abraham S, Caparon M, Falk P, St. Geme J W, Normark S. Cell. 1993;73:887–901. doi: 10.1016/0092-8674(93)90269-v. [DOI] [PubMed] [Google Scholar]
- 2.Strom M S, Lory S. Annu Rev Microbiol. 1993;47:565–596. doi: 10.1146/annurev.mi.47.100193.003025. [DOI] [PubMed] [Google Scholar]
- 3.Parge H E, Forest K T, Hickey M J, Christensen D A, Getzoff E D, Tainer J A. Nature (London) 1995;378:32–38. doi: 10.1038/378032a0. [DOI] [PubMed] [Google Scholar]
- 4.Tønjum T, Koomey M. Gene. 1997;192:155–163. doi: 10.1016/s0378-1119(97)00018-8. [DOI] [PubMed] [Google Scholar]
- 5.Pugsley A P. Microbiol Rev. 1993;57:50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dubnau D. Gene. 1997;192:191–198. doi: 10.1016/s0378-1119(96)00804-9. [DOI] [PubMed] [Google Scholar]
- 7.Russel M, Linderoth N A, Sali A. Gene. 1997;192:23–32. doi: 10.1016/s0378-1119(96)00801-3. [DOI] [PubMed] [Google Scholar]
- 8.Lory S, Strom M S. Gene. 1997;192:117–121. doi: 10.1016/s0378-1119(96)00830-x. [DOI] [PubMed] [Google Scholar]
- 9.Swanson J. J Exp Med. 1973;137:571–589. doi: 10.1084/jem.137.3.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jonsson A B, Nyberg G, Normark S. EMBO J. 1991;10:477–488. doi: 10.1002/j.1460-2075.1991.tb07970.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rudel T, van Putten J P M, Gibbs C P, Hass R, Meyer T F. Mol Microbiol. 1992;6:3439–3450. doi: 10.1111/j.1365-2958.1992.tb02211.x. [DOI] [PubMed] [Google Scholar]
- 12.Rudel T, Scheurerpflug I, Meyer T F. Nature (London) 1995;373:357–359. doi: 10.1038/373357a0. [DOI] [PubMed] [Google Scholar]
- 13.Swanson J. Infect Immun. 1978;19:320–331. doi: 10.1128/iai.19.1.320-331.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henrichsen J. Annu Rev Microbiol. 1983;37:81–93. doi: 10.1146/annurev.mi.37.100183.000501. [DOI] [PubMed] [Google Scholar]
- 15.Bradley D E. Can J Microbiol. 1980;26:146–154. doi: 10.1139/m80-022. [DOI] [PubMed] [Google Scholar]
- 16.Wu S, Kaiser D. Mol Microbiol. 1995;18:547–558. doi: 10.1111/j.1365-2958.1995.mmi_18030547.x. [DOI] [PubMed] [Google Scholar]
- 17.Wu S, Wu J, Kaiser D. Mol Microbiol. 1997;23:109–121. doi: 10.1046/j.1365-2958.1997.1791550.x. [DOI] [PubMed] [Google Scholar]
- 18.Wolfgang M, Lauer P, Park H-S, Brossay L, Hebert J, Koomey M. Mol Microbiol. 1998;29:321–330. doi: 10.1046/j.1365-2958.1998.00935.x. [DOI] [PubMed] [Google Scholar]
- 19.Whitchurch C B, Hobbs M, Livingston S P, Krishnapillai V, Mattick J S. Gene. 1991;101:33–44. doi: 10.1016/0378-1119(91)90221-v. [DOI] [PubMed] [Google Scholar]
- 20.Lessl M, Lanka E. Cell. 1994;77:321–324. doi: 10.1016/0092-8674(94)90146-5. [DOI] [PubMed] [Google Scholar]
- 21.Freitag N, Seifert H S, Koomey M. Mol Microbiol. 1995;16:575–586. doi: 10.1111/j.1365-2958.1995.tb02420.x. [DOI] [PubMed] [Google Scholar]
- 22.Seifert H. Gene. 1997;188:215–220. doi: 10.1016/s0378-1119(96)00810-4. [DOI] [PubMed] [Google Scholar]
- 23.Koomey M, Bergstrom S, Blake M, Swanson J. Mol Microbiol. 1991;5:279–287. doi: 10.1111/j.1365-2958.1991.tb02108.x. [DOI] [PubMed] [Google Scholar]
- 24.Tønjum T, Freitag N E, Normark E, Koomey M. Mol Microbiol. 1995;16:451–464. doi: 10.1111/j.1365-2958.1995.tb02410.x. [DOI] [PubMed] [Google Scholar]
- 25.van Putten J P M, Paul S M. EMBO J. 1995;14:2144–2154. doi: 10.1002/j.1460-2075.1995.tb07208.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burch C L, Danaher R J, Stein D C. J Bacteriol. 1997;179:982–986. doi: 10.1128/jb.179.3.982-986.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rahman M, Kallstrom H, Normark S, Jonsson A-B. Mol Microbiol. 1997;25:11–26. doi: 10.1046/j.1365-2958.1997.4601823.x. [DOI] [PubMed] [Google Scholar]
- 28.Brossay L, Paradis G, Fox R, Koomey M, Hebert J. Infect Immun. 1994;62:2302–2308. doi: 10.1128/iai.62.6.2302-2308.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Drake S L, Sandstedt S A, Koomey M. Mol Microbiol. 1996;23:657–668. doi: 10.1046/j.1365-2958.1997.2511618.x. [DOI] [PubMed] [Google Scholar]