Abstract
It is well known that the functional activity of the diphtheria toxin repressor DtxR is controlled by iron, which serves as an essential cofactor necessary for activation of target DNA binding by this regulatory element. In this communication, we describe the isolation and characterization of a unique series of DtxR mutants that are constitutively active and repress the expression of β-galactosidase from a diphtheria tox promoter/operator–lacZ transcriptional fusion, even in the absence of iron. These self-activating mutants of DtxR (SAD) were isolated through the use of a positive selection system for the cloning of functional dtxR alleles and target DNA operator sites. Of the four independently isolated SAD mutants that were characterized, two (SAD2 and SAD11) were found to carry a single missense mutation (E175K) in their respective C-terminal SH3-like domains. In contrast, the mutant allele encoding SAD3 was found to carry a total of six missense mutations distributed throughout the N- and C-terminal domains of the repressor. Partial diploid analysis of strains carrying both native dtxR and alleles encoding either SAD2 or SAD3 demonstrate that these iron-independent mutants possess a positive dominant phenotype in the regulation of β-galactosidase expression from a diphtheria tox promoter/operator–lacZ transcriptional fusion.
Keywords: diphtheria tox repressor/PCR mutagenesis/positive dominant phenotype
The diphtheria toxin repressor DtxR from Corynebacterium diphtheriae has been shown to be an iron-activated regulatory element that controls the expression of a series of iron-sensitive genes, including the diphtheria toxin structural gene, genes involved in siderophore production, and heme oxygenase (1–4). Partial x-ray structures of apo-DtxR, transition metal ion-bound DtxR, and the ternary Ni(II)-DtxR-tox operator (toxO) complex have been solved (5–9). These studies have shown that the N-terminal 136-aa region of DtxR carries at least three functional domains: a helix-turn-helix motif involved in target DNA recognition, a protein–protein interaction domain that stabilizes the metal ion-activated dimeric form of the repressor, and the primary activation and ancillary metal-ion binding sites (5–7). The activation of apo-DtxR involves the coordination of a transition metal ion in the primary binding site, which is composed of Met10, Cys102 [Asp102 in DtxR(C102D)], Glu105, His106, and a water molecule (7). In contrast, relatively little is known of the function of the C-terminal 90 amino acids of DtxR. The x-ray crystal structure of this region of DtxR was shown by Qui et al. (8) to be structurally homologous to SH3 domains. Recently, Logan and co-workers (P.D. Twigg, G.P. Wylie, G. Wang, D.C.D. Caspar, J.R.M., and T.M. Logan, unpublished work) have shown by multidimensional heteronuclear NMR that the structure of the C-terminal peptide DtxR(130–226) in solution adopts an SH3-like conformation. Of importance, this study also demonstrated that the isolated C-terminal domain of DtxR is capable of binding a 15-aa proline-rich sequence derived from the unstructured region between the N- and C-terminal domains of DtxR.
Monomeric apo-DtxR appears to be in weak equilibrium with a dimeric form of the repressor (10). The binding of Ni(II) to the apo-repressor is cooperative and appears to cause a conformational change(s) that results in stabilizing the dimeric form of DtxR (10). A caliper-like movement of the helix-turn-helix motif was postulated to allow binding of each monomer in the dimeric structure to a major groove in the target DNA sequence (6). White et al. (9) recently have shown that the binding of DtxR to the toxO involves not only this caliper-like movement but also a helix-to-coil transition of amino acids 1–6 and the interaction of the guanidium moiety of Arg60 with the minor groove.
We describe here the development of a positive genetic selection system for the cloning of dtxR alleles and DtxR DNA target sites [the positive selection of DtxR alleles and targets (PSDT)]. This system is based on expression of the chloramphenicol acetyltransferase gene (cat), which is carried by the indicator Escherichia coli strain and under the control of the tetracycline resistance determinant repressor TetR, which in turn is controlled by DtxR. Therefore, in the presence of a functional DtxR/toxO genetic circuit, the cat gene is expressed, and the indicator strain of E. coli becomes chloramphenicol-resistant (CmR). In the absence of either dtxR or the toxO, or in the presence of the iron chelator 2,2′-dipyridyl (DP), the indicator strain becomes Cm-sensitive (CmS). In the present study, we have exploited the sensitivity of the PSDT system to DP and have selected iron-independent, functionally active mutants of DtxR on Luria–Bertani (LB) agar medium supplemented with chloramphenicol and DP. We demonstrate that these iron-independent mutants of DtxR are constitutively active and repress the expression of β-galactosidase from a diphtheria tox promoter/operator (toxPO)-lacZ transcriptional fusion. Moreover, partial diploid analysis of two of the mutants, self-activating DtxR 2 (SAD2) and SAD3, demonstrates that they each exhibit a positive dominant phenotype in the regulation of lacZ expression. Because SAD2 and SAD11 each carry a single missense mutation (E175K) in their respective C-terminal SH3-like domains, future structural studies of these mutants are likely to shed additional light on the mechanism and requirements for transition metal ion-activation of DtxR and its subsequent binding to target DNA sequences.
MATERIALS AND METHODS
Bacterial Strains, Plasmids, Phages, and Medium.
The bacterial strains, plasmids and phages used in this study are described in Results and Table 1. E. coli strains were grown in LB (10 g of tryptone, 10 g of NaCl, and 5 g of yeast extract per liter). LB broth and LB agar were supplemented with ampicillin (Ap; 100 μg/ml), kanamycin (Kn; 25 μg/ml), Cm (12.5 μg/ml), and 5-bromo-4-chloro-3-indolyl β-d-galactopyranoside (40 μg/ml) as indicated. The iron chelator DP was added to a final concentration of 200 μM to LB agar and as indicated to LB broth.
Table 1.
Bacterial strains, bacteriophages, and plasmids used in this study
| Strain/phage/plasmid | Genotype/phenotype | Source or reference |
|---|---|---|
| Strains | ||
| E. coli TOP10 | mcrAΔ(mrr-hsdRMS-mcrBC) Φ80Δlac | Invitrogen |
| ΔM15ΔlacX74 deoR recA1 endA1 | ||
| E. coli DH5a | F−(Φ80d lacZ ΔM15) Δ(lacZYA-argF) | BRL |
| recA1 endA gyrA thi1 hsdR17 (rk−, mk−) | ||
| supE44 relA1 U196 | ||
| Bacteriophages | ||
| λRS45toxPO | toxPO-LacZ KnR | Ref. 1 |
| λRS65 | tetAPO-cat-lacZYA CmR | Ref. 10 |
| λRS65T | tetAPO-cat-lacZ′YA CmR | Ref. 10 |
| Plasmids | ||
| pBR322 | ColE1 ori APR | New England Biolabs |
| pACYC184 | p15A ori CmR | New England Biolabs |
| pRDA | ColE1 ori dtxR ApR | This study |
| pLS-2 | ColE1 ori ΔdtxR ApR | This study |
| pSC6 | pSC101 ori toxPO-tetR KnR | Ref. 10 |
| pSC6M1 | pSC101 ori toxPO-tetR KnR | Ref. 10 |
| pSDM2 | pSC101 ori SAD2 KnR | This study |
| pSDM3 | pSC101 ori SAD3 KnR | This study |
| pSDM11 | pSC101 ori SAD11 ApR | This study |
| pSDM5 | ColE1 ori SAD5 ApR | This study |
| pDM2 | ColE1 ori SAD2 ApR | This study |
| pDM3 | ColE1 ori SAD3 ApR | This study |
| pDM11 | ColE1 ori SAD11 ApR | This study |
| pDM5 | ColE1 ori SAD5 ApR | This study |
| pDM2A | P15A ori SAD2 CmR | This study |
| pDM3A | p15A ori SAD3 CmR | This study |
Nucleic Acids.
DNA cloning, plasmid preparation, and DNA sequence analysis were performed according to standard methods as described (11, 12). Restriction endonucleases, T4 polynucleotide kinase, and Klenow fragment of DNA polymerase (New England Biolabs) were used according to the manufacturer’s specifications. PCR mutagenesis of dtxR was based on the method of Vartanian et al. (13). In brief, BglII-tagged primers 1515 (5′-ACCAGATCTGCCGAAAAACTTCGA-3′) and 1516 (5′-ACCAGATCTCCGCCTTTAGTATTTA-3′) were used to PCR amplify dtxR from plasmid pRDA, which carries the wild-type dtxR operon. The products of the amplification then were digested with BglII and either were ligated into BglII-linearized pSC6M1 and were transformed into E. coli TOP10/λRS65T or were ligated into BamHI digested pBR322 and were transformed into E. coli TOP10/λRS65T/pSC6. Iron-independent mutants of DtxR then were selected on LB agar plates supplemented with Cm and DP.
β-Galactosidase Assay.
β-galactosidase activity was measured essentially as described by Miller (14). In brief, 0.5 ml of an overnight culture at A600 of ≈1.0 was lysed by the addition of lysis mix (chloroform:10% SDS, 2:1), vortexing the mixture for 10 sec and transferring 200 μl to 800 μl Z buffer (60 mM Na2HPO4/40 mM NaH2PO4/10 mM KCl/1 mM MgSO4/50 mM β-mercaptoethanol). The reaction was initiated by adding 200 μl of o-nitrophenyl β-d-galactopyranoside (Sigma) (4 mg/ml). After incubation at room temperature (5 min-1 hr), the reaction was stopped by the addition of 0.5 ml of 1M sodium carbonate. Absorbance was measured at 420 and 550 nm, and β-galactosidase units were calculated according to Miller (14).
RESULTS
Development of the PSDT System.
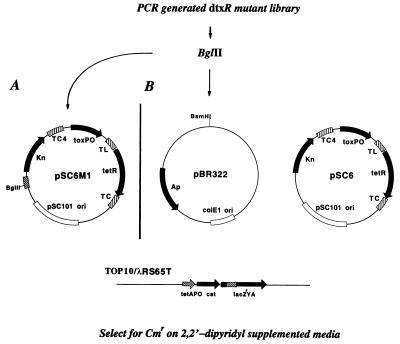
The PSDT system consists of a lysogenic E. coli TOP10 host strain, which carries the reporter gene cat on an integrated λ phage, λRS65T, and a set of detector plasmids (Fig. 1). Expression of cat on λRS65T is controlled by the tetA promoter/operator (tetAPO), and, in the absence of the tetracycline repressor (TetR), E. coli TOP10/λRS65T is resistant to chloramphenicol (CmR). We next constructed the detector plasmid pSC6, which carries the tetR gene under the control of the diphtheria toxPO. When pSC6 is transformed into E. coli TOP10/λRS65T, the bacterial host strain becomes Cm-sensitive (CmS) by virtue of the constitutive expression of tetR, which recognizes and binds to the tetAO and represses cat gene expression. However, if a functional dtxR allele is introduced into the bacterial host on a second compatible plasmid, pRDA, the interaction between DtxR and the toxO will repress the expression of tetR, and the bacterial host, E. coli TOP10/λRS65T/pSC6/pRDA, then will regain its CmR phenotype. Furthermore, because the iron chelator DP is known to inactivate DtxR, the addition of DP to the growth medium should result in a phenotypic conversion from CmR to CmS. As shown in Table 2, the addition of DP to the growth medium did result in the conversion to a CmS phenotype. Finally, to demonstrate the requirement for a functional DtxR in the PSDT system, we digested pRDA with EcoRV to delete a 713-bp fragment and thereby knock out the dtxR gene (Fig. 3). The resulting plasmid, pLS-2, then was transformed into E. coli TOP10/λRS65T/pSC6. As anticipated, in the absence of a functional DtxR, the indicator strain becomes CmS. Table 2 summarizes the antibiotic resistance phenotypes of derivatives of E. coli TOP10/λRS65T that carry one or more of the PSDT detector plasmids.
Figure 1.
Strategy used for the selection of SAD mutants. The indicator strain, E. coli TOP10/λRS65T, carries the cat gene under the control of the tetA promoter/operator (tetAPO), which is regulated by TetR encoded on either pSC6M1 (A) or pSC6 (B). PCR-generated mutant libraries of dtxR either were ligated into the BglII site of pSC6M1 and were transformed into TOP10/λRS65T or were ligated into the BamHI site of pBR322 and were transformed into TOP10/λRS65T/pSC6. In these indicator strains, tetR is under the control of a functional dtxR:toxO genetic circuit. SAD mutants of DtxR were selected on LB agar supplemented with Cm and the iron chelator (DP). TC, transcriptional terminators; TL, translational terminators.
Table 2.
Antibiotic resistance phenotype of E. coli TOP10/λRS65T and derivative strains carrying various plasmids used in the PSDT system
| Strains and plasmids | Growth on medium supplemented with antibiotics and DP
|
||||||
|---|---|---|---|---|---|---|---|
| Ap | Kn | Cm | Ap/Kn | Ap/Kn/Cm | Ap/Kn/DP | Ap/Kn/Cm/DP | |
| TOP10/λRS65T | − | − | +++ | − | − | − | − |
| pSC6 | − | +++ | − | − | − | − | − |
| pRDA | +++ | − | +++ | − | − | − | − |
| pSC6/pRDA | +++ | +++ | +++ | +++ | +++ | − | − |
| pSC6/pLS-2 | +++ | +++ | +++ | +++ | − | − | − |
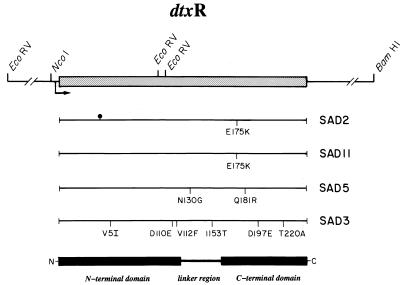
Figure 3.
Schematic representation of the dtxR structural gene, the relative position and identification of individual point mutations in the iron-independent repressors SAD2, SAD11, SAD5, and SAD3, and the relative positions of the N- and C-terminal domains of DtxR. Arrow, dtxR promoter and direction of transcription; (•|), the T to C transition at nucleotide 123 in this dtxR allele.
Isolation of Iron-Independent SAD Mutants.
Although the PSDT system initially was developed for the positive selection of dtxR alleles, homologs, and DtxR DNA target sites from genomic libraries, we have exploited the DP sensitivity of this system to isolate a unique class of iron-independent, constitutively active mutants of DtxR. For this purpose, we constructed the detector plasmid pSC6M1, in which the introduction of a unique BglII restriction endonuclease site allows the cloning of dtxR alleles in the same vector that carries the toxPO-tetR transcriptional fusion. The overall scheme for the selection of SAD mutants is shown in Fig. 1. The wild-type dtxR allele carried by pRDA was mutagenized by PCR amplification according to the method of Vartanian et al.(13) by using BglII tagged primers 1515 and 1516. After PCR mutagenesis, the amplified DNA was digested with BglII and either was ligated into the BglII site of pSC6M1 and was transformed into TOP10/λRS65T or was ligated into the BamHI site of pBR322 and was transformed into TOP10/λRS65T/pSC6. Because wild-type DtxR is inactivated in the presence of the iron chelator DP, potential SAD mutants were selected on LB agar supplemented with both Cm and 200 μM DP. In a typical experiment, ≈105 colony forming units were plated on LB/Cm/DP agar plates, and after 24-hr incubation at 37°C, the colonies that developed were picked and colony purified, and their respective plasmids were analyzed.
In each instance, restriction endonuclease digestion analysis demonstrated that the plasmids had an insertion of the size anticipated for dtxR. Of the nine colonies that were isolated using the PSDT system, four clones subsequently were characterized fully. The plasmids named pSDM2, pSDM3, pSDM11 are derivatives of the detector plasmid pSCM61, and pSMD5 was derived from pBR322 (Fig. 1). The mutant DtxRs that are encoded by these plasmids were designated SAD2, SAD3, SAD11, and SAD5, respectively.
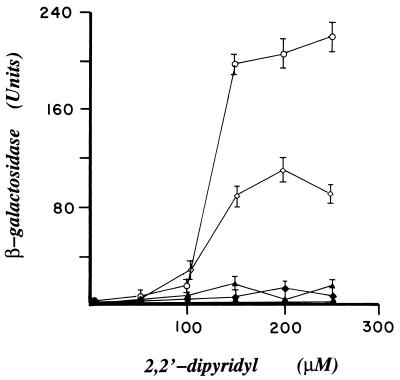
To demonstrate that each isolate carried an active iron-independent mutant of DtxR, their respective dtxR alleles were recloned into the NcoI/BamHI sites of pRDA to replace the wild-type dtxR. The resulting plasmids were designated pDM2, pDM3, pDM11, and pDM5, respectively. The pDM series of plasmids then were transformed individually into E. coli DH5α/λRS45toxPO. This reporter strain carries a transcriptional fusion in which lacZ gene is under the control of the diphtheria toxPO, toxPO-lacZ, and, as a result, the synthesis of β-galactosidase is regulated by their respective dtxR gene products. Individual strains were grown in LB broth supplemented with appropriate antibiotics in either the absence or presence of increasing concentrations of DP. As shown in Fig. 2, β-galactosidase assays indicated that, in marked contrast to transformants that expressed wild-type DtxR, those strains that expressed either SAD2, SAD11, or SAD3 maintained complete repression of lacZ, even in the presence of 250 μM DP. In contrast, the transformant that expressed SAD5 displayed an intermediate iron-independent phenotype. These results are consistent with the observation that the growth of E. coli TOP10/λRS65T/pSC6/pSDM5 is inhibited partially by Cm in the presence of 200 μM DP. In comparison, E. coli TOP10/λRS65T strains that carry either pSDM2, pSDM3, or pDM11 are completely CmR.
Figure 2.
β-galactosidase activity of E. coli DH5α/λRS45toxPO strains that carry plasmid pRDA (○), pDM2 (♦), pDM3 (▴), pDM11 (•), or pDM5 (⋄). These plasmids encode either DtxR or the iron-independent mutants SAD2, SAD3, SAD11, or SAD5, respectively. E. coli strains were grown in the absence or presence of increasing concentrations of DP. The error bars represent the SD of three parallel experiments.
To further demonstrate that SAD2 and SAD3 were iron-independent mutants of DtxR, each allele was knocked out by the deletion of a 713-bp EcoRV restriction endonuclease fragment that encompasses the promoter and N-terminal encoding portion of their structural genes (Fig. 3). As anticipated, transformation of E. coli DH5α/λRS45toxPO with plasmids encoding defective SAD2 and SAD3 genes failed to confer detectable repression of lacZ (data not shown).
DNA Sequence Analysis of SAD Mutants.
Because each of the SAD mutants of DtxR were iron-independent and constitutively active, we determined the DNA sequence of their respective dtxR alleles. The structural gene encoding SAD2 was found to carry two point mutations: a T to C transition at position 123 that results in a silent mutation (V41V), and a G to A transition at position 523 that introduces the missense mutation E175K. It is of interest to note that the independently isolated SAD11 mutant also was found to carry an identical single missense mutation, E175K. In contrast, the structural gene encoding SAD3 carries a total of eight point mutations that result in six missense mutations (V5I, D110E, V112F, I153T, D197E, and T220A) (Fig. 3) whereas the dtxR allele encoding partial iron-independent SAD5 mutant carries two missense mutations (N130G and Q181R). In an attempt to identify a mutation in SAD3 that imparts the iron-independent active phenotype, we introduced separately each of the six point mutations into wild-type dtxR by site-directed mutagenesis. The mutants DtxR(D110E) and DtxR(V112F) were found to have lost all repressor activity whereas the remaining four mutations (V5I, I153T, D197E, and T220A) were phenotypically silent (data not shown).
Determination of the Apparent Relative toxO Binding Affinities of DtxR, SAD2, and SAD3.
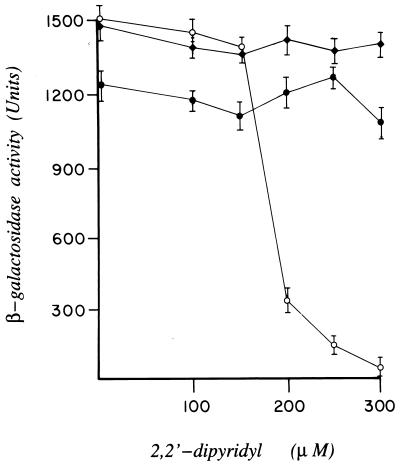
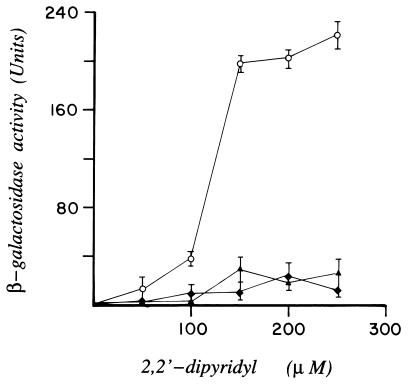
To determine the apparent relative binding affinities of DtxR, SAD2, and SAD3 for the toxO, we constructed the reporter strain of E. coli TOP10/λRS65 in which lacZ was placed under the control of tetAPO. In this instance, the expression of lacZ is negatively controlled by TetR. E. coli TOP10/λRS65/pSC6 then was transformed independently with plasmids pRDA, pDM2 or pDM3, which carry the structural genes encoding DtxR, SAD2, or SAD3, respectively. As anticipated in this indicator strain, transformation with pRDA results in the expression of maximal levels of β-galactosidase in the presence of excess iron in the growth medium (i.e., DtxR-mediated repression of tetR). The addition of increasing concentrations of the chelator DP to the growth medium inactivates wild-type DtxR, resulting in the expression of tetR and subsequent repression of lacZ (Fig. 4). In marked contrast, maximal levels of β-galactosidase were expressed in strains of E. coli TOP10/λRS65/pSC6 that carried either pDM2 or pDM3 regardless of the concentration of iron in the growth medium. It is of interest to note that the maximal level of β-galactosidase expressed in E. coli TOP10/λRS65/pSC6/pDM3 was consistently 20% lower than that found in the isogenic strain that carried pDM2. Because immunoblot analysis with polyclonal anti-DtxR antibodies of serial dilutions of total cellular extracts suggest that the relative expression of SAD2 and SAD3 are equivalent (data not shown), SAD3 may have a lower apparent affinity for the toxO than either wild-type DtxR or SAD2. If this is the case, the slight decrease in maximal β-galactosidase levels expressed in E. coli TOP10/λRS65/pSC6/pDM3 may be caused by leaky expression of tetR.
Figure 4.
Assessment of the apparent relative toxO binding affinity and DP sensitivity of DtxR, SAD2, and SAD3 as measured by β-galactosidase activity. E. coli TOP10/λRS65 was cotransformed with pSC6 and either pRDA (encoding DtxR; ○), pDM2 (encoding SAD2; ♦), or pDM3 (encoding SAD3; •). β-galactosidase activity was assayed after the growth of E. coli strains in the absence or presence of increasing concentrations of DP. The error bars represent the SD of three parallel experiments.
SAD2 and SAD3 Are Positive Dominant Mutants of DtxR.
Because both SAD2 and SAD3 displayed an iron-independent mutant phenotype, we next determined by partial diploid analysis whether the mutant phenotype would dominate over wild-type DtxR. Accordingly, we constructed a reporter strain of E. coli that carried both the wild-type dtxR and either SAD2 or SAD3 alleles. For this purpose, SAD2 and SAD3 were recloned individually into the low copy number plasmid pACYC184 to form pDM2A and pDM3A, respectively. E. coli DH5α/λRS45toxPO then was transformed with both pRDA and either pDM2A or pDM3A, and levels of β-galactosidase in the transformants were determined in the presence of increasing concentrations of DP. As shown in Fig. 5, lacZ remained repressed in E. coli DH5α/λRS45toxPO that were diploid for dtxR and either SAD2 or SAD3. These results clearly demonstrate that iron-independent SAD2 and SAD3 mutant repressors are dominant over DtxR and continue to bind to the diphtheria toxO and regulate lacZ expression in the absence of iron.
Figure 5.
β-galactosidase activity of E. coli DH5α/λRS45toxPO strains that carried the wild-type dtxR (pRDA, ○) and partial diploid strains of E. coli DH5α/λRS45toxPO that carry both the wild-type dtxR and mutant alleles encoding either SAD2 or SAD3, pRDA/pDM2A [♦], or pRDA/pDM3A [▴]. β-galactosidase activity was assayed after growth of E. coli strains in the absence or presence of increasing concentrations of DP. The error bars represent the SD of three parallel experiments.
DISCUSSION
We have described the PSDT system, which, in this instance, has been used for the positive selection of iron-independent SAD mutants. This selection is based on two sequential levels of gene regulation that lead to cat gene expression and the emergence of a CmR phenotype in the E. coli indicator strain. In this system, cat gene expression is controlled by TetR, which in turn is regulated by a functional DtxR:toxO circuit. It is well known that wild-type DtxR must be activated by Fe(II), or other transition metal ions in vitro, to bind to the toxO. Therefore, the addition of the iron chelator DP to the growth medium results in the inactivation of the repressor. In the PSDT system, addition of DP to the growth medium results in the derepression of tetR and leads to a CmS phenotype in the indicator strain. Because the power of the PSDT selection system is on the order of 10−8 (data not shown), we have used this system in the isolation of a class of iron-independent mutants of DtxR that remain constitutively active even in the absence of iron.
In the present work, we randomly have mutagenized the dtxR gene by PCR amplification. After digestion of the amplified DNA with BglII and ligation into the BglII site of pSC6M1, E. coli TOP10/λRS65T was transformed and plated on LB agar supplemented with Cm and 200 μM DP. Alternatively, BglII digested amplified DNA was ligated into the BamHI site of pBR322, E. coli TOP10/λRS65T/pSC6 was transformed, and transformants were selected on LB agar supplemented with Cm and 200 μM DP. By using this system, a total of nine iron-independent SAD mutants were isolated, of which four were characterized extensively.
The independently isolated SAD2, SAD3, and SAD11 mutants were isolated from derivatives of pSC6M1 in the TOP10/λRS65T strain of E. coli. In each instance, recombinant E. coli that carried either pSDM2, pSDM3, or pSDM11 were completely CmR in the presence of DP. As anticipated, SAD2, SAD3, and SAD11 were found to be completely iron-independent and resistant to the inhibitory effect of DP at concentrations as high as 250 μM.
In addition, we isolated several mutants of DtxR that displayed a partial iron-independent phenotype. One of these mutants, SAD5, was fully characterized. In this instance, the SAD5-encoding allele of dtxR initially was cloned in the BamHI site of pBR322, a moderate copy number plasmid with a ColE1 ori. Because SAD5 regulates the expression of tetR on pSC6 (a low copy number plasmid with a pSC101 ori) in trans, it is likely that its initial isolation on LB/Cm/DP agar medium, and its ability to confer an intermediate iron-independent phenotype to E. coli DH5α/λRS45toxPO, is related to its relative affinity for the toxO. Although this mutant clearly is activated by iron and capable of mediating complete repression of lacZ, the partial derepression of lacZ in the presence of DP suggests that SAD5 is still able to bind partially to the toxO in the absence of iron.
Compared with wild-type DtxR, DNA sequence analysis has revealed that both SAD2 and SAD11 carry a single missense mutation, E175K, which gives rise to their respective iron-independent phenotype. Because we initially had anticipated that this class of DtxR mutants might involve an amino acid substitution(s) in close proximity to the primary metal ion-activation site (7), the results of the DNA sequence analysis of SAD2 and SAD11 were surprising. Furthermore, the independent isolation of two dtxR alleles that encode SAD mutants with identical mutations in their respective C-terminal domains underscores the importance of this mutation in giving rise to an iron-independent active phenotype.
The x-ray crystallographic analysis of apo-DtxR and DtxR transiton metal ion complexes and the recent solution of the ternary complex of DtxR-Ni(II)-toxO all have revealed partial structures in which only the N-terminal 136-aa portion of the repressor was solved (5–9). Furthermore, this region was found to carry the helix-turn-helix motif involved in binding to DNA, a protein–protein interaction domain involved in the stabilization of a dimeric form of the repressor, and to both the primary activating and ancillary metal ion binding sites. In contrast, the C-terminal 90 amino acids of DtxR exhibited large thermal factors that prevented complete chain tracing. Recently, Qiu et al. (8) described higher resolution structures of Co(II) and Mn(II) bound forms of DtxR in which the C-terminal 90-aa region was solved largely to between 2.8- and 2.0-Å resolution. Amino acids 160–188 of DtxR were shown to fold into an SH3-like domain; however, the thermal B factors of this domain were still 50 Å, which indicates a high degree of fluctuation in atomic positions. It is most interesting that the E175K mutation, which gives rise to the iron-independent repressor activity in SAD2 and SAD11, is positioned in this apparently flexible C-terminal domain of DtxR. Although there are several possible models for SAD2 and SAD11 self-activation, the apparent flexibility of the SH3-like domain may allow it to fold over the face of the repressor and may result in the insertion of the ɛ-amino moiety of Lys175 into the primary metal ion binding site, where it may act as a surrogate for iron. It is of additional interest to note that the partial iron-independent phenotype of SAD5 is caused by the mutations N130G and Q181R. The proximity of Q181R to E175K in the C-terminal domain of DtxR, and the similarity between the ɛ-amino group of lysine and the δ-guanidium group of arginine, suggests that the terminal amino moiety of each amino acid may be involved in the activation of DtxR. There is precedence for the ɛ-amino moiety of lysine occupying a metal ion-binding site of other proteins without the induction of a fundamental change in the native structure. Allen et al.(18) have characterized the E108K mutation in d-xylose isomerase. Though this mutant was capable of opening the sugar ring, it was not able to catalyze the isomerization of glucose to fructose. X-ray crystallographic analysis of both the wild-type and E108K mutant forms of d-xylose isomerase revealed that the ɛ-amino group of K108 was able to structurally replace the Mg(II) normally found in this binding site. Alternatively, in the absence of metal ion binding, the SH3-like C-terminal domain of DtxR may bind to the proline-rich linker between the N- and C-terminal domains of the repressor and may stabilize the inactive monomeric form (P. D. Twigg, G. P. Wylie, G. Wang, D. L. D. Caspar, J.R.M., and T. M. Logan, unpublished work). In this instance, mutations in the C-domain that disrupt this association may give rise to active dimeric forms of the repressor even in the absence of divalent transition metal ions. Finally, in the case of SAD2 and SAD11 and the absence of structural studies, we cannot rule out the possibility that the E175K mutation may result in a more global conformational change in the repressor that leads to their iron-independent binding to the toxO. In the case of Drosophila melanogaster calmodulin, the E to K (BK) mutations in the four loop regions were found to trigger profound conformational changes in and surrounding the Ca(II) binding sites (19).
It is noteworthy that partial diploid analysis of E. coli strains that carry dtxR and the allele that encodes either SAD2 or SAD3 clearly demonstrate that these iron-independent mutants are positive dominant over the wild-type DtxR. Because DtxR in C. diphtheriae and its homolog IdeR in Mycobacterium tuberculosis (20) are likely to be global regulatory elements that control the expression of iron-sensitive genes involved in iron acquisition and virulence, it will be of interest to examine whether partial diploids that express DtxR/SAD2 and/or IdeR/SAD2 in these prokaryotic pathogens will result in an attenuation of their respective virulence.
Acknowledgments
This work was supported by Public Health Service Grant AI-21628 from the National Institute of Allergy and Infectious Diseases.
ABBREVIATIONS
- Ap
ampicillin
- Kn
kanamycin
- Cm
chloramphenicol
- CmR
Cm-resistant
- CmS
Cm-sensitive
- DP
2,2′-dipyridyl
- PSDT
positive selection of DtxR alleles and targets
- SAD
self-activating DtxR
- toxO
tox operator
- toxPO
tox promoter/operator
- LB
Luria–Bertani
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. M34239).
References
- 1.Boyd J, Oza M N, Murphy J R. Proc Natl Acad Sci USA. 1990;87:5986–5972. [Google Scholar]
- 2.Tao X, Boyd J, Murphy J R. Proc Natl Acad Sci USA. 1992;89:5897–5901. doi: 10.1073/pnas.89.13.5897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schmitt M P, Holmes R K. Infect Immun. 1991;59:1899–1904. doi: 10.1128/iai.59.6.1899-1904.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmitt M P. Infect Immun. 1997;65:4634–4641. doi: 10.1128/iai.65.11.4634-4641.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qui X, Verlinde C L M J, Zhang S, Schmitt M P, Holmes R K, Hol W G J. Structure. 1995;3:87–100. doi: 10.1016/s0969-2126(01)00137-x. [DOI] [PubMed] [Google Scholar]
- 6.Schiering N, Tao X, Zeng H, Murphy J R, Petsko G A, Ringe D. Proc Natl Acad Sci USA. 1995;92:9843–9850. doi: 10.1073/pnas.92.21.9843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ding X, Zeng H, Schiering N, Ringe D, Murphy J R. Nat Struct Biol. 1996;3:382–387. doi: 10.1038/nsb0496-382. [DOI] [PubMed] [Google Scholar]
- 8.Qui X, Pohl E, Holmes R K, Hol W G L. Biochemistry. 1996;35:12292–12302. doi: 10.1021/bi960861d. [DOI] [PubMed] [Google Scholar]
- 9.White, A., Ding, X., vanderSpek, J., Murphy, J. R. & Ringe, D. Nature (London), 394, 502–506. [DOI] [PubMed]
- 10.Tao X, Zeng H, Murphy J R. Proc Natl Acad Sci USA. 1995;92:6803–6807. doi: 10.1073/pnas.92.15.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1988. [Google Scholar]
- 12.Sanger F, Nicklen S, Coulsen A R. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vartanian J-P, Henry S, Wain-Hobson S. Nucleic Acids Res. 1996;24:2627–2631. doi: 10.1093/nar/24.14.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller J H. Experiments in Molecular Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1972. [Google Scholar]
- 15.Tao X, Murphy J R. J Biol Chem. 1992;30:21761–21764. [PubMed] [Google Scholar]
- 16.Schmitt M P, Holmes R K. J Bacteriol. 1994;176:1141–1149. doi: 10.1128/jb.176.4.1141-1149.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tao X, Murphy J R. Proc Natl Acad Sci USA. 1994;91:9646–9650. doi: 10.1073/pnas.91.20.9646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allen K N, Lavie A, Glasfield A, Tanada T N, Gerrity D P, Carlson S C, Farber G K, Petsko G A, Ringe D. Biochemistry. 1994;33:1488–1494. doi: 10.1021/bi00172a027. [DOI] [PubMed] [Google Scholar]
- 19.Maune J F, Beckingham K, Martin S R, Bayley P M. Biochemistry. 1992;31:7779–7786. doi: 10.1021/bi00149a006. [DOI] [PubMed] [Google Scholar]
- 20.Schmitt M P, Predich M, Doukhan L, Smith I, Holmes R K. Infect Imun. 1995;63:4284–4289. doi: 10.1128/iai.63.11.4284-4289.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simons R W, Houman F, Kleckner N. Gene. 1987;53:69–85. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- 22.Tao X, Murphy J R. Proc Natl Acad Sci USA. 1993;90:8524–8528. doi: 10.1073/pnas.90.18.8524. [DOI] [PMC free article] [PubMed] [Google Scholar]