Abstract
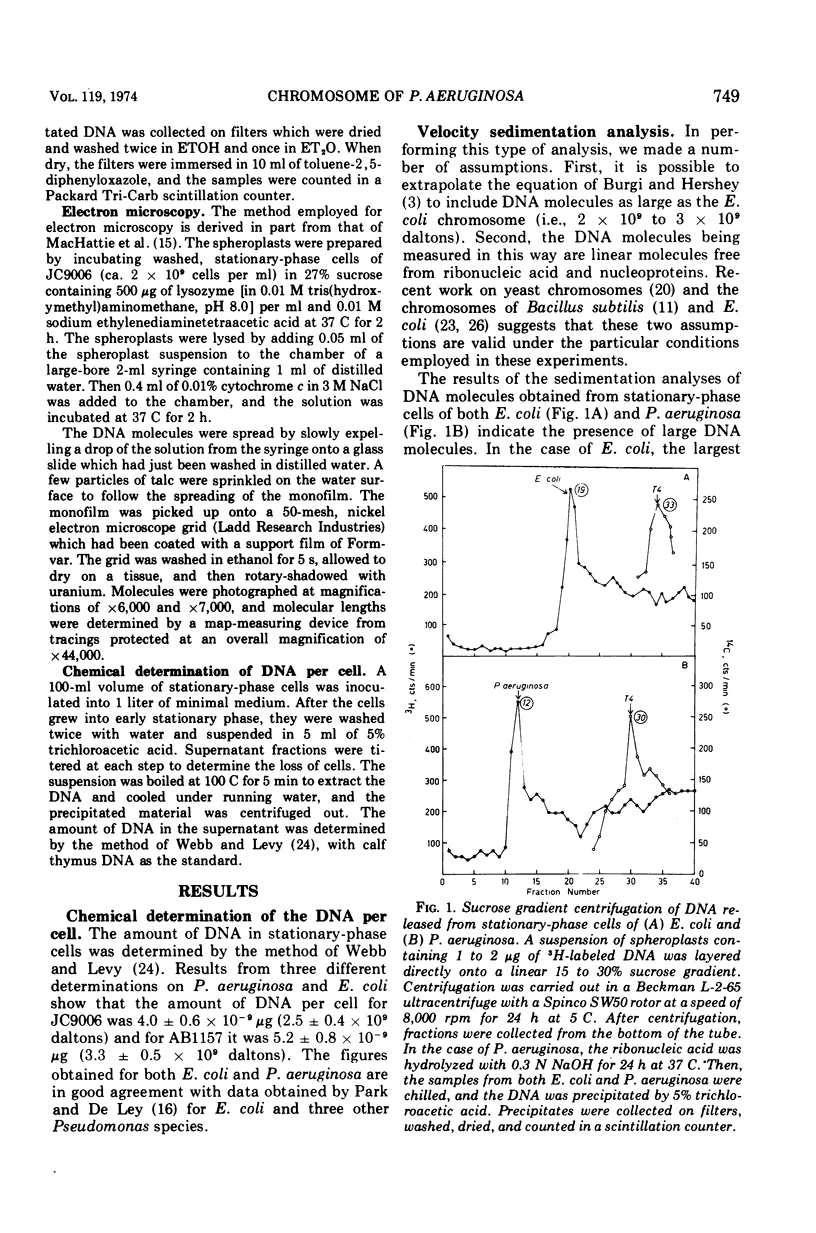
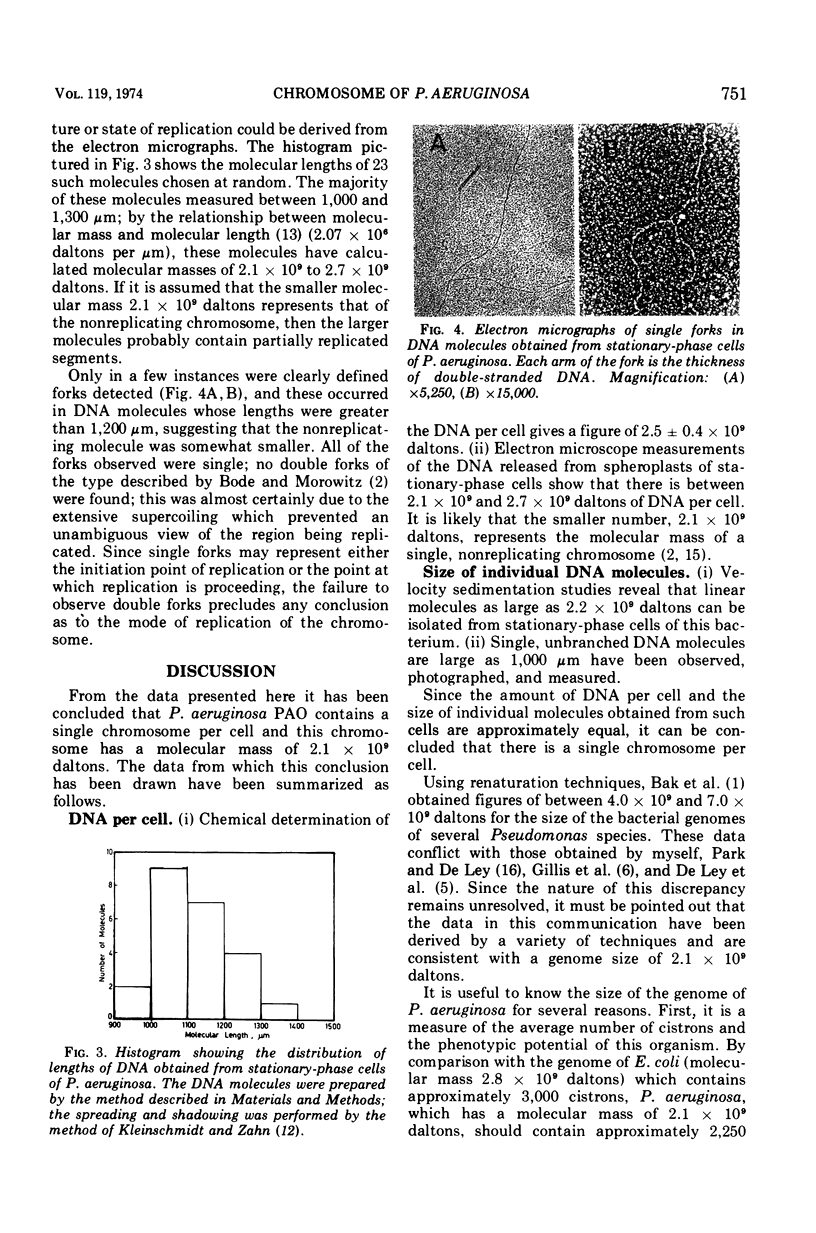
Electron microscope examination and velocity sedimentation analysis of the deoxyribonucleic acid released from Pseudomonas aeruginosa spheroplasts indicate that this organism carries the bulk of its genetic determinants in a single duplex deoxyribonucleic acid molecule having a molecular mass of 2.1 × 109 daltons.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BURGI E., HERSHEY A. D. Sedimentation rate as a measure of molecular weight of DNA. Biophys J. 1963 Jul;3:309–321. doi: 10.1016/s0006-3495(63)86823-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bak A. L., Christiansen C., Stenderup A. Bacterial genome sizes determined by DNA renaturation studies. J Gen Microbiol. 1970 Dec;64(3):377–380. doi: 10.1099/00221287-64-3-377. [DOI] [PubMed] [Google Scholar]
- Bode H. R., Morowitz H. J. Size and structure of the Mycoplasma hominis H39 chromosome. J Mol Biol. 1967 Jan 28;23(2):191–199. doi: 10.1016/s0022-2836(67)80026-3. [DOI] [PubMed] [Google Scholar]
- CAIRNS J. The bacterial chromosome and its manner of replication as seen by autoradiography. J Mol Biol. 1963 Mar;6:208–213. doi: 10.1016/s0022-2836(63)80070-4. [DOI] [PubMed] [Google Scholar]
- De Ley J., Cattoir H., Reynaerts A. The quantitative measurement of DNA hybridization from renaturation rates. Eur J Biochem. 1970 Jan;12(1):133–142. doi: 10.1111/j.1432-1033.1970.tb00830.x. [DOI] [PubMed] [Google Scholar]
- Gillis M., De Ley J., De Cleene M. The determination of molecular weight of bacterial genome DNA from renaturation rates. Eur J Biochem. 1970 Jan;12(1):143–153. doi: 10.1111/j.1432-1033.1970.tb00831.x. [DOI] [PubMed] [Google Scholar]
- Gray H. B., Jr, Hearst J. E. Flexibility of native DNA from the sedimentation behavior as a function of molecular weight and temperature. J Mol Biol. 1968 Jul 14;35(1):111–129. doi: 10.1016/s0022-2836(68)80041-5. [DOI] [PubMed] [Google Scholar]
- HOPWOOD D. A. A CIRCULAR LINKAGE MAP IN THE ACTINOMYCETE STREPTOMYCES COELICOLOR. J Mol Biol. 1965 Jun;12:514–516. doi: 10.1016/s0022-2836(65)80274-1. [DOI] [PubMed] [Google Scholar]
- Holloway B. W., Krishnapillai V., Stanisich V. Pseudomonas genetics. Annu Rev Genet. 1971;5:425–446. doi: 10.1146/annurev.ge.05.120171.002233. [DOI] [PubMed] [Google Scholar]
- Kavenoff R. Characterization of the Bacillus subtilis W23 genome by sedimentation. J Mol Biol. 1972 Dec 30;72(3):801–806. doi: 10.1016/0022-2836(72)90192-1. [DOI] [PubMed] [Google Scholar]
- Lang D. Molecular weights of coliphages and coliphage DNA. 3. Contour length and molecular weight of DNA from bacteriophages T4, T5 and T7, and from bovine papilloma virus. J Mol Biol. 1970 Dec 28;54(3):557–565. doi: 10.1016/0022-2836(70)90126-9. [DOI] [PubMed] [Google Scholar]
- Leighton S. B., Rubenstein I. Calibration of molecular weight scales for DNA. J Mol Biol. 1969 Dec 14;46(2):313–328. doi: 10.1016/0022-2836(69)90424-0. [DOI] [PubMed] [Google Scholar]
- MACHATTIE L. A., BERNE K. I., THOMAS C. A., Jr ELECTRON MICROSCOPY OF DNA FROM HEMOPHILUS INFLUENZAE. J Mol Biol. 1965 Mar;11:648–649. doi: 10.1016/s0022-2836(65)80019-5. [DOI] [PubMed] [Google Scholar]
- Park I. W., De Ley J. Ancestral remnants in deoxyribonucleic acid from Pseudomonas and Xanthomonas. Antonie Van Leeuwenhoek. 1967;33(1):1–16. doi: 10.1007/BF02045528. [DOI] [PubMed] [Google Scholar]
- Pemberton J. M., Clark A. J. Detection and characterization of plasmids in Pseudomonas aeruginosa strain PAO. J Bacteriol. 1973 Apr;114(1):424–433. doi: 10.1128/jb.114.1.424-433.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemberton J. M., Holloway B. W. A new sex factor of Pseudomonas aeruginosa. Genet Res. 1973 Jun;21(3):263–272. doi: 10.1017/s0016672300013458. [DOI] [PubMed] [Google Scholar]
- Pemberton J. M., Holloway B. W. Chromosome mapping in Pseudomonas aeruginosa. Genet Res. 1972 Jun;19(3):251–260. doi: 10.1017/s0016672300014518. [DOI] [PubMed] [Google Scholar]
- Petes T. D., Fangman W. L. Sedimentation properties of yeast chromosomal DNA. Proc Natl Acad Sci U S A. 1972 May;69(5):1188–1191. doi: 10.1073/pnas.69.5.1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SANDERSON K. E., DEMEREC M. THE LINKAGE MAP OF SALMONELLA TYPHIMURIUM. Genetics. 1965 Jun;51:897–913. doi: 10.1093/genetics/51.6.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanisich V. A., Holloway B. W. Chromosome transfer in Pseudomonas aeruginosa mediated by R factors. Genet Res. 1971 Apr;17(2):169–172. doi: 10.1017/s0016672300012179. [DOI] [PubMed] [Google Scholar]
- Stonington O. G., Pettijohn D. E. The folded genome of Escherichia coli isolated in a protein-DNA-RNA complex. Proc Natl Acad Sci U S A. 1971 Jan;68(1):6–9. doi: 10.1073/pnas.68.1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WEBB J. M., LEVY H. B. A sensitive method for the determination of deoxyribonucleic acid in tissues and microorganisms. J Biol Chem. 1955 Mar;213(1):107–117. [PubMed] [Google Scholar]
- Willetts N. S., Clark A. J. Characteristics of some multiply recombination-deficient strains of Escherichia coli. J Bacteriol. 1969 Oct;100(1):231–239. doi: 10.1128/jb.100.1.231-239.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worcel A., Burgi E. On the structure of the folded chromosome of Escherichia coli. J Mol Biol. 1972 Nov 14;71(2):127–147. doi: 10.1016/0022-2836(72)90342-7. [DOI] [PubMed] [Google Scholar]





