Abstract
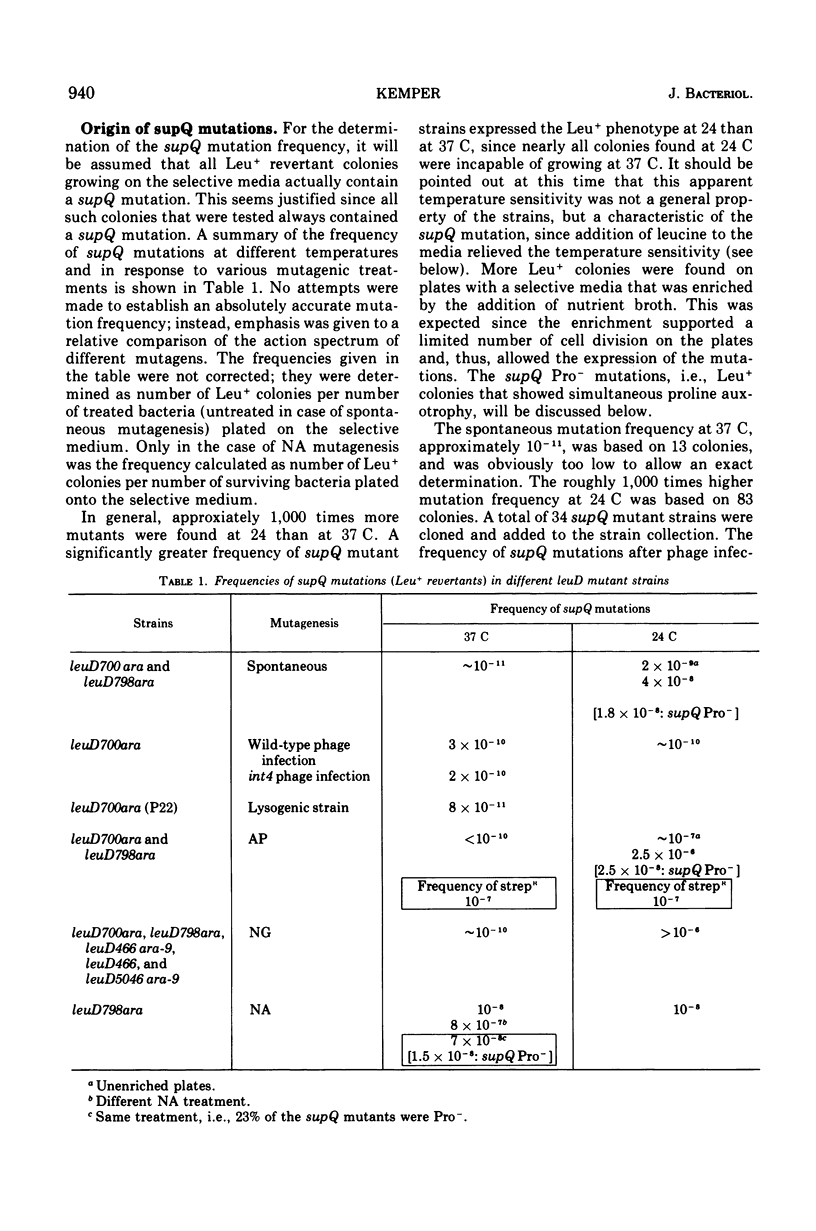
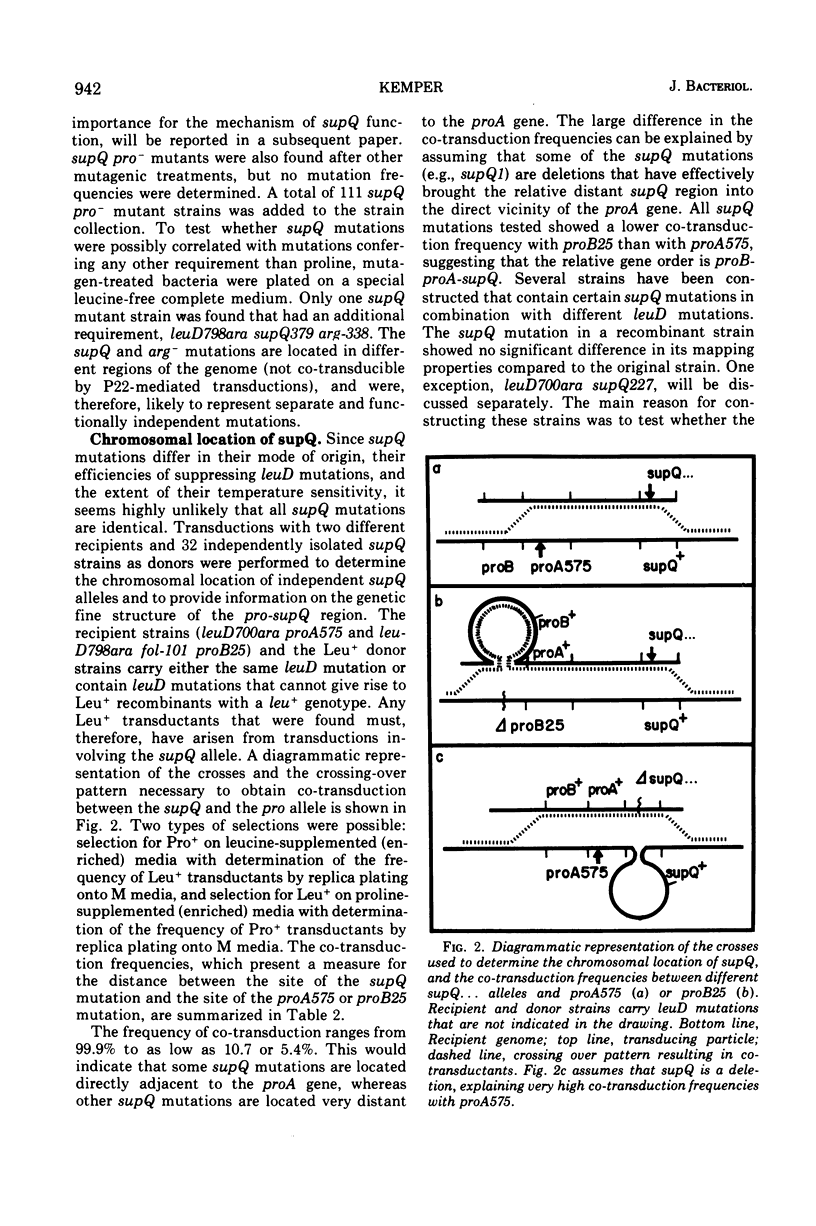
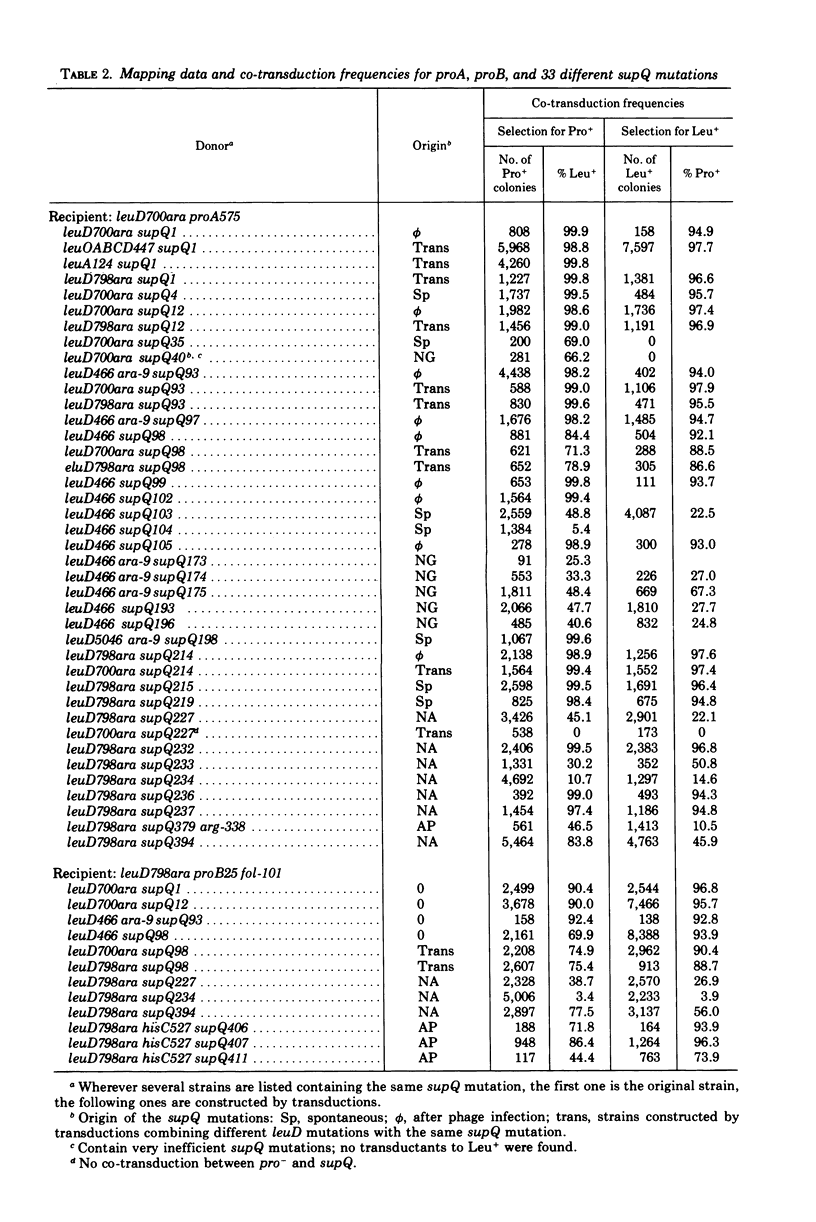
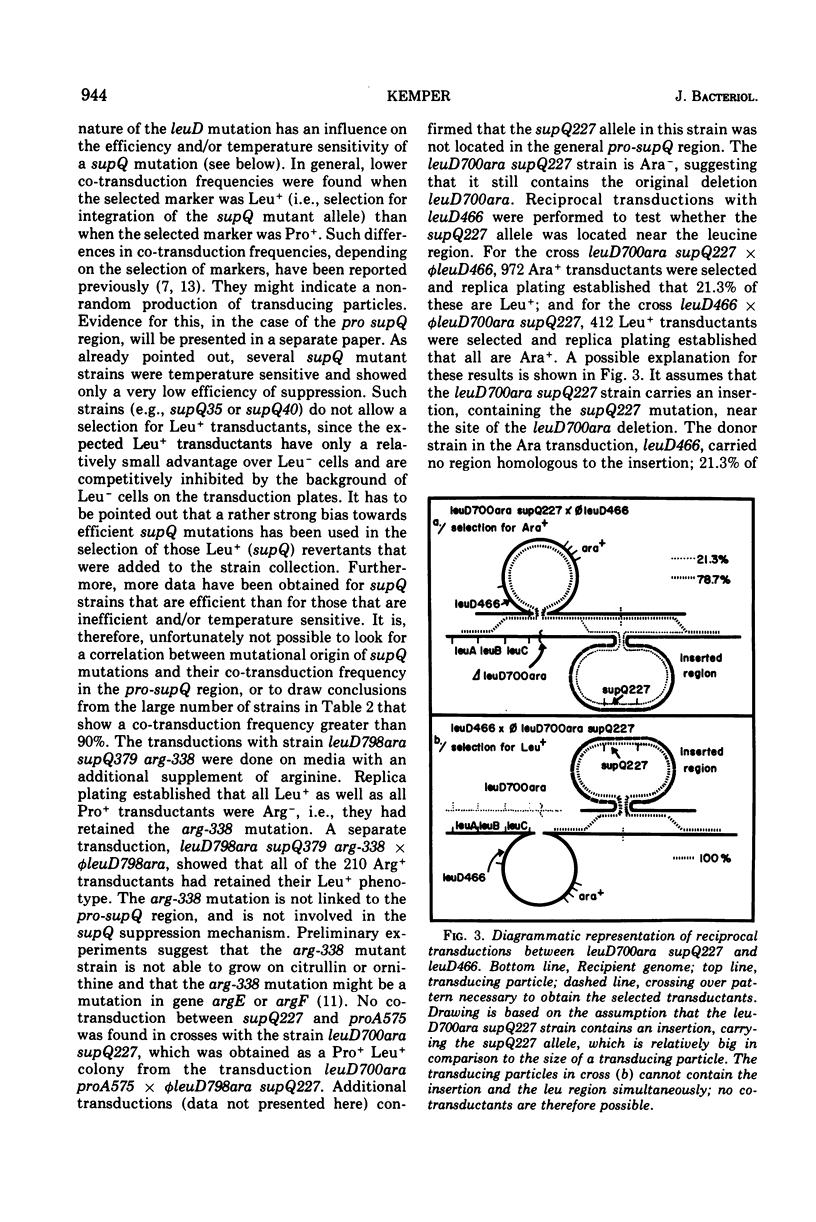
Alpha-isopropylmalate isomerase, the second specific enzyme in the biosynthesis of leucine, is coded for by two genes, leuC and leuD. Leucine auxotrophs, harboring leuD mutations including a deletion of the entire leuD gene, revert to leucine prototrophy owing to mutations at a locus, supQ, substantially distant to the leucine operon. A large number of independently isolated supQ mutations were characterized. A significant increase in the spontaneous frequency of supQ mutations was found after mutagenesis with 2-aminopurine, N-methyl-N′-nitro-N-nitrosoguanidine, diethyl sulfate, and nitrous acid. The supQ function in most of these strains is temperature sensitive, resulting in more efficient suppression with decreasing temperature. At higher temperatures, the supQ limits the growth rate of leuD supQ mutant strains. All supQ mutations are co-transducible with proA and proB, with co-transduction frequencies ranging from 5.4 to 99.9% for different supQ mutations. Many supQ mutations were isolated, especially after nitrous acid mutagenesis, that had acquired a simultaneous proline requirement. The data support the idea of two genes, supQ and newD, whose protein products form a complex. The newD gene product, without any genetic alteration, is capable of substituting for the missing leuD protein. However, mutations in the supQ gene (point mutations or deletions) are necessary to make the newD protein available, which is normally tied up in a complex with the supQ protein.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Burns R. O., Calvo J., Margolin P., Umbarger H. E. Expression of the leucine operon. J Bacteriol. 1966 Apr;91(4):1570–1576. doi: 10.1128/jb.91.4.1570-1576.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo J. M., Worden H. E. A multisite-mutation map of the leucine operon of Salmonella typhimurium. Genetics. 1970 Feb;64(2):199–214. doi: 10.1093/genetics/64.2.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell J. H., Lengyel J. A., Langridge J. Evolution of a second gene for beta-galactosidase in Escherichia coli. Proc Natl Acad Sci U S A. 1973 Jun;70(6):1841–1845. doi: 10.1073/pnas.70.6.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GROSS S. R., BURNS R. O., UMBARGER H. E. THE BIOSYNTHESIS OF LEUCINE. II. THE ENZYMIC ISOMERIZATION OF BETA-CARBOXY-BETA-HYDROXYISOCAPROATE AND ALPHA-HYDROXY-BETA-CARBOXYISOCAPROATE. Biochemistry. 1963 Sep-Oct;2:1046–1052. doi: 10.1021/bi00905a023. [DOI] [PubMed] [Google Scholar]
- Guerola N., Ingraham J. L., Cerdá-Olmedo E. Induction of closely linked multiple mutations by nitrosoguanidine. Nat New Biol. 1971 Mar 24;230(12):122–125. doi: 10.1038/newbio230122a0. [DOI] [PubMed] [Google Scholar]
- Hartman P. E., Roth J. R. Mechanisms of suppression. Adv Genet. 1973;17:1–105. doi: 10.1016/s0065-2660(08)60170-4. [DOI] [PubMed] [Google Scholar]
- Kemper J. Gene order and co-transduction in the leu-ara-fol-pyrA region of the Salmonella typhimurium linkage map. J Bacteriol. 1974 Jan;117(1):94–99. doi: 10.1128/jb.117.1.94-99.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemper J., Margolin P. Suppression by gene substitution for the leuD gene of Salmonella typhimurium. Genetics. 1969 Oct;63(2):263–279. doi: 10.1093/genetics/63.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARGOLIN P. Genetic fine structure of the leucine operon in Salmonella. Genetics. 1963 Mar;48:441–457. doi: 10.1093/genetics/48.3.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake T, Demerec M. Proline Mutants of Salmonella Typhimurium. Genetics. 1960 Jun;45(6):755–762. doi: 10.1093/genetics/45.6.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NOVICK R. P., MAAS W. K. Control by endogenously synthesized arginine of the formation of ornithine transcarbamylase in Escherichia coli. J Bacteriol. 1961 Feb;81:236–240. doi: 10.1128/jb.81.2.236-240.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson K. E. Linkage map of Salmonella typhimurium, edition IV. Bacteriol Rev. 1972 Dec;36(4):558–586. doi: 10.1128/br.36.4.558-586.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz D. O., Beckwith J. R. Mutagens which cause deletions in Escherichia coli. Genetics. 1969 Feb;61(2):371–376. doi: 10.1093/genetics/61.2.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. O., Levine M. A phage P22 gene controlling integration of prophage. Virology. 1967 Feb;31(2):207–216. doi: 10.1016/0042-6822(67)90164-x. [DOI] [PubMed] [Google Scholar]
- TESSMAN I. The induction of large deletions by nitrous acid. J Mol Biol. 1962 Oct;5:442–445. doi: 10.1016/s0022-2836(62)80033-3. [DOI] [PubMed] [Google Scholar]
- ZINDER N. D., LEDERBERG J. Genetic exchange in Salmonella. J Bacteriol. 1952 Nov;64(5):679–699. doi: 10.1128/jb.64.5.679-699.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]



