Abstract
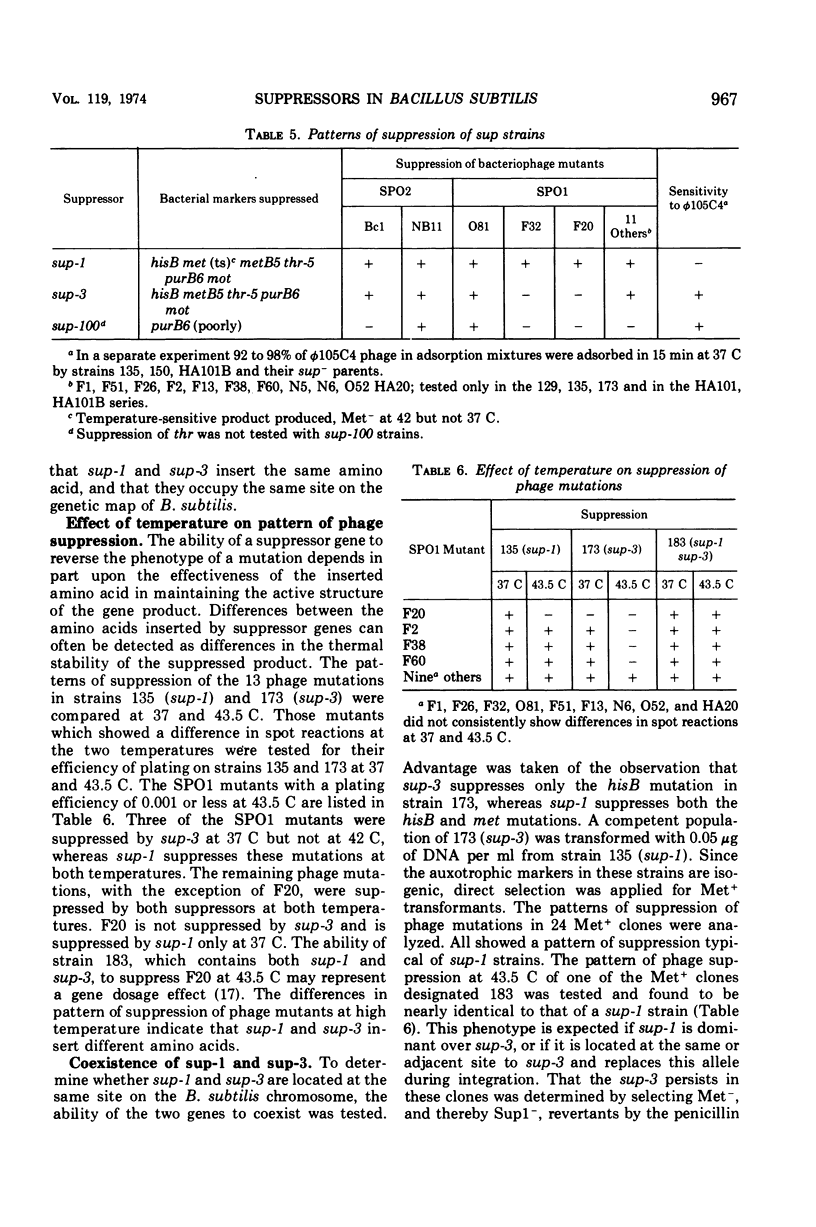
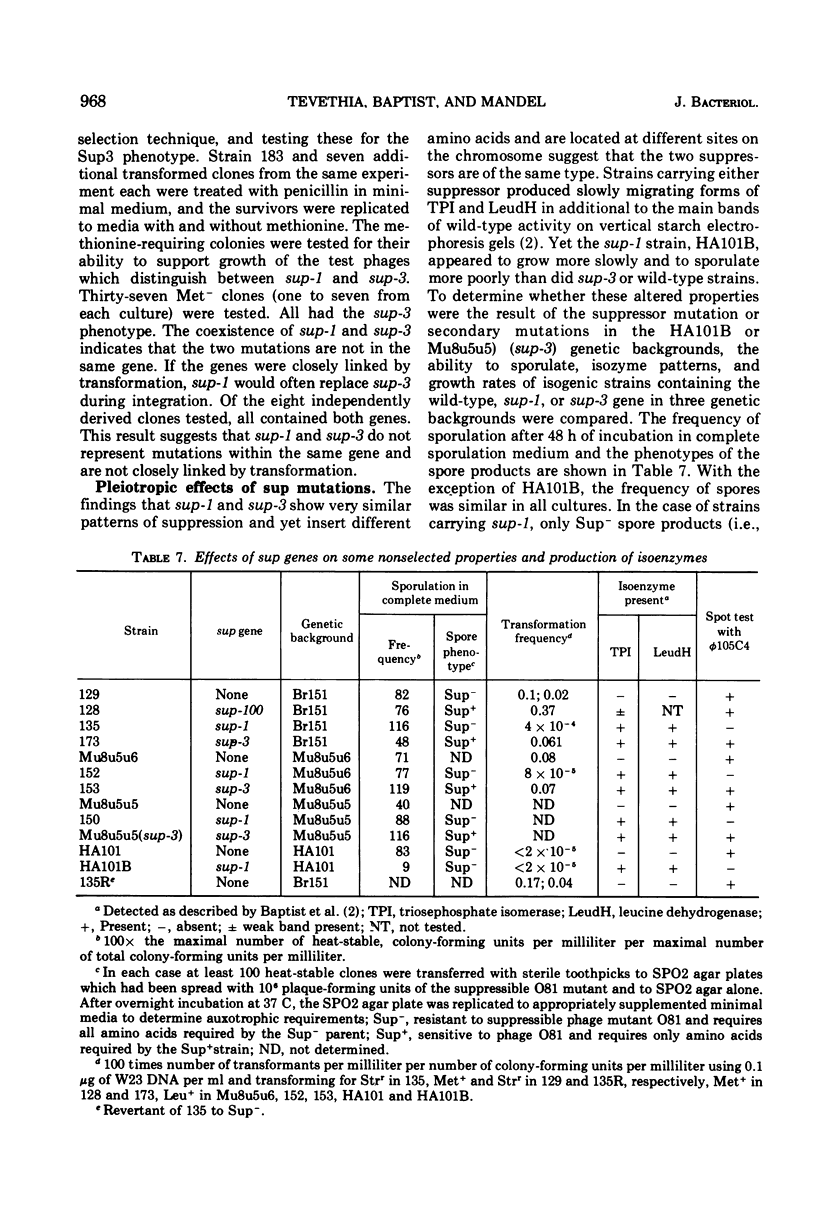
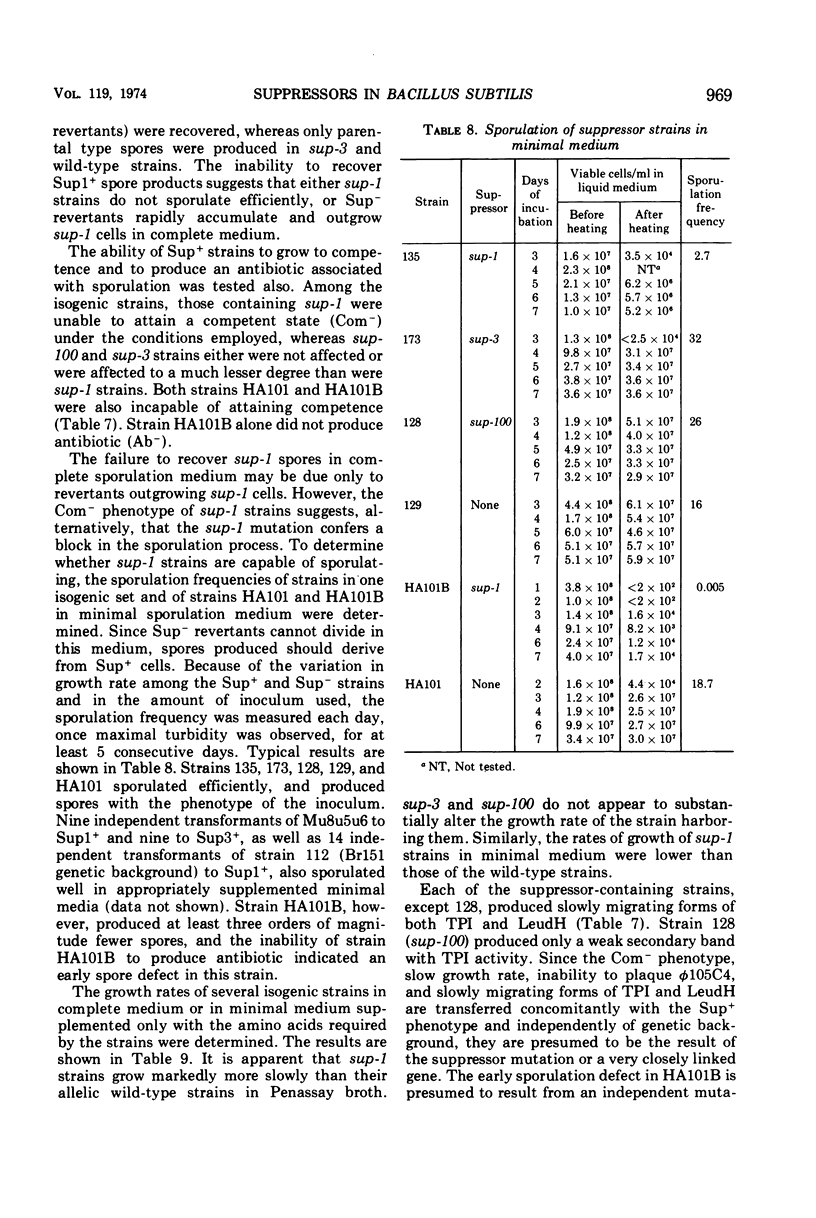
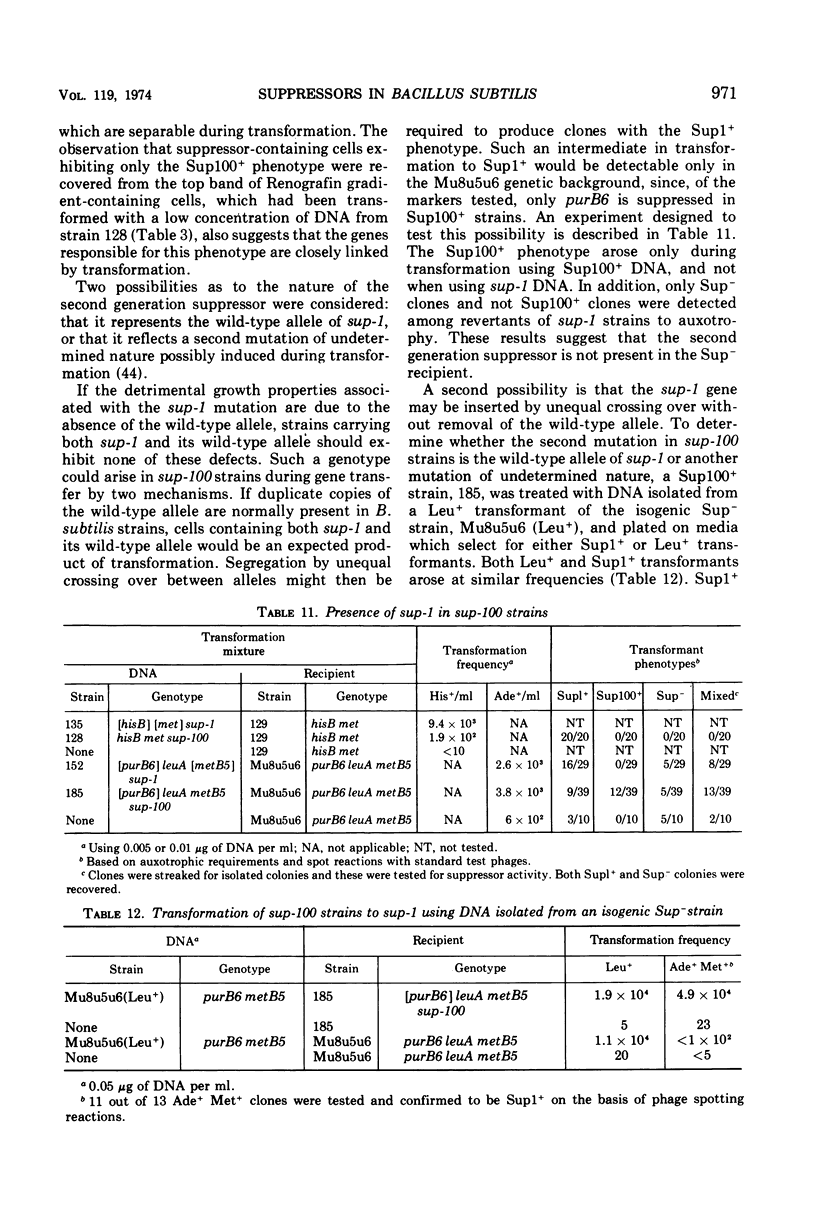
Isogenic strains of Bacillus subtilis carrying sup-1 (26), sup-3 (10), or their wild-type alleles were constructed in three genetic backgrounds. The patterns of suppression at 37 and 43.5 C, identity of mapping site, effects of the suppressor genes on growth rate, sporulation, and production of altered enzymes were examined. The similarity of the suppression pattern by sup-1 and sup-3 suggests that the suppressors are of the same type. They do not, however, represent mutations in the same gene, since, based on differences in temperature sensitivity of phage mutants in suppressor-containing hosts, sup-1 and sup-3 insert different amino acids and can coexist within the same cell. The ability to produce slow-migrating forms of enzymes of the type described in the accompanying paper was co-transferred with either of the suppressor genes during transformation, was lost on reversion of the suppressor mutations, and was independent of the genetic background. Similarly, transformation and reversion studies indicate that the additional pleiotropic properties such as slow growth rate and inability to attain competence or to yield plaques with φ105C4, which are characteristic of the Okubo sup-1 strain (HA101B) but not its early sporulation defect, result from the presence of the suppressor mutation. The possible mechanisms by which altered enzyme forms and the additional pleiotropic effects are produced in suppressor strains are discussed. In addition, a newly recognized suppressor phenotype is described and partially characterized.
Full text
PDF










Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alberts B. M., Doty P. Characterization of a naturally occurring, cross-linked fraction of DNA. 1. Nature of the cross-linkage. J Mol Biol. 1968 Mar 14;32(2):379–403. doi: 10.1016/0022-2836(68)90017-x. [DOI] [PubMed] [Google Scholar]
- Baptist J. N., Tevethia M. J., Mandel M., Shaw C. R. Altered proteins with triosephosphate isomerase activity in suppressor-containing strains of Bacillus subtilis. J Bacteriol. 1974 Sep;119(3):976–985. doi: 10.1128/jb.119.3.976-985.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbon J., Squires C., Hill C. W. Glycine transfer RNA of Escherichia coli. II. Impaired GGA-recognition in strains containing a genetically altered transfer RNA; reversal by a secondary suppressor mutation. J Mol Biol. 1970 Sep 28;52(3):571–584. doi: 10.1016/0022-2836(70)90420-1. [DOI] [PubMed] [Google Scholar]
- Contreras R., Ysebaert M., Jou W. M., Fiers W. Bacteriophage Ms2 RNA: nucleotide sequence of the end of the a protein gene and the intercistronic region. Nat New Biol. 1973 Jan 24;241(108):99–101. doi: 10.1038/newbio241099a0. [DOI] [PubMed] [Google Scholar]
- DONNELLAN J. E., Jr, NAGS E. H., LEVINSON H. S. CHEMICALLY DEFINED, SYNTHETIC MEDIA FOR SPORULATION AND FOR GERMINATION AND GROWTH OF BACILLUS SUBTILIS. J Bacteriol. 1964 Feb;87:332–336. doi: 10.1128/jb.87.2.332-336.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demerec M., Adelberg E. A., Clark A. J., Hartman P. E. A proposal for a uniform nomenclature in bacterial genetics. Genetics. 1966 Jul;54(1):61–76. doi: 10.1093/genetics/54.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau D., Goldthwaite C., Smith I., Marmur J. Genetic mapping in Bacillus subtilis. J Mol Biol. 1967 Jul 14;27(1):163–185. doi: 10.1016/0022-2836(67)90358-0. [DOI] [PubMed] [Google Scholar]
- Erickson R. J., Braun W. Apparent dependence of transformation on the stage of deoxyribonucleic acid replication of recipient cells. Bacteriol Rev. 1968 Dec;32(4 Pt 1):291–296. [PMC free article] [PubMed] [Google Scholar]
- Fujita D. J., Ohlsson-Wilhelm B. M., Geiduschek E. P. Transcription during bacteriophage SPO1 development: mutations affecting the program of viral transcription. J Mol Biol. 1971 Apr 28;57(2):301–317. doi: 10.1016/0022-2836(71)90348-2. [DOI] [PubMed] [Google Scholar]
- Georgopoulos C. P. Suppressor system in Bacillus subtilis 168. J Bacteriol. 1969 Mar;97(3):1397–1402. doi: 10.1128/jb.97.3.1397-1402.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh H. P., Ghosh K., Ganoza M. C. Mechanism of polypeptide chain termination. Translation of tandem amber termination codons by an amber suppressor transfer ribonucleic acid. J Biol Chem. 1972 Sep 10;247(17):5322–5326. [PubMed] [Google Scholar]
- Goldschmidt R. In vivo degradation of nonsense fragments in E. coli. Nature. 1970 Dec 19;228(5277):1151–1154. doi: 10.1038/2281151a0. [DOI] [PubMed] [Google Scholar]
- Goldthwaite C., Dubnau D., Smith I. Genetic mapping of antibiotic resistance in markers Bacillus subtilis. Proc Natl Acad Sci U S A. 1970 Jan;65(1):96–103. doi: 10.1073/pnas.65.1.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorini L. Informational suppression. Annu Rev Genet. 1970;4:107–134. doi: 10.1146/annurev.ge.04.120170.000543. [DOI] [PubMed] [Google Scholar]
- Hayashi H., Söll D. Purification of an Escherichia coli leucine suppressor transfer ribonucleic acid and its aminoacylation by the homologous leucyl-transfer ribonucleic acid synthetase. J Biol Chem. 1971 Aug 25;246(16):4951–4954. [PubMed] [Google Scholar]
- Hill C. W., Foulds J., Soll L., Berg P. Instability of a missense suppressor resulting from a duplication of genetic material. J Mol Biol. 1969 Feb 14;39(3):563–581. doi: 10.1016/0022-2836(69)90146-6. [DOI] [PubMed] [Google Scholar]
- Hoffman E. P., Wilhelm R. C. Effect of Sul gene dosage on the production of suppressed alkaline phosphatase in Escherichia coli K12. J Mol Biol. 1970 Apr 14;49(1):241–244. doi: 10.1016/0022-2836(70)90390-6. [DOI] [PubMed] [Google Scholar]
- Kano-Sueoka T., Sueoka N. Characterization of a modified leucyl-tRNA of Escherichia coli after bacteriophage T2 infection. J Mol Biol. 1968 Nov 14;37(3):475–491. doi: 10.1016/0022-2836(68)90116-2. [DOI] [PubMed] [Google Scholar]
- Kano-Sueoka T., Sueoka N. Leucine tRNA and cessation of Escherichia coli protein synthesis upon phage T2 infection. Proc Natl Acad Sci U S A. 1969 Apr;62(4):1229–1236. doi: 10.1073/pnas.62.4.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu P., Rich A. The nature of the polypeptide chain termination signal. J Mol Biol. 1971 Jun 14;58(2):513–531. doi: 10.1016/0022-2836(71)90368-8. [DOI] [PubMed] [Google Scholar]
- Miller C. G., Roth J. R. Recessive-lethal nonsense suppressors in Salmonella typhimurium. J Mol Biol. 1971 Jul 14;59(1):63–75. doi: 10.1016/0022-2836(71)90413-x. [DOI] [PubMed] [Google Scholar]
- Natori S., Garen A. Molecular heterogeneity in the amino-terminal region of alkaline phosphatase. J Mol Biol. 1970 May 14;49(3):577–588. doi: 10.1016/0022-2836(70)90282-2. [DOI] [PubMed] [Google Scholar]
- Nichols J. L. Nucleotide sequence from the polypeptide chain termination region of the coat protein cistron in bacteriophage R17 RNA. Nature. 1970 Jan 10;225(5228):147–151. doi: 10.1038/225147a0. [DOI] [PubMed] [Google Scholar]
- Ohlsson B. M., Strigini P. F., Beckwith J. R. Allelic amber and ochre suppressors. J Mol Biol. 1968 Sep 14;36(2):209–218. doi: 10.1016/0022-2836(68)90376-8. [DOI] [PubMed] [Google Scholar]
- Okubo S., Romig W. R. Comparison of ultraviolet sensitivity of Bacillus subtilis bacteriophage SPO2 and its infectious DNA. J Mol Biol. 1965 Nov;14(1):130–142. doi: 10.1016/s0022-2836(65)80235-2. [DOI] [PubMed] [Google Scholar]
- Okubo S., Yanagida T. Isolation of a suppressor mutant in Bacillus subtilis. J Bacteriol. 1968 Mar;95(3):1187–1188. doi: 10.1128/jb.95.3.1187-1188.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Person S., Osborn M. The conversion of amber suppressors to ochre suppressors. Proc Natl Acad Sci U S A. 1968 Jul;60(3):1030–1037. doi: 10.1073/pnas.60.3.1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt T., Miller J. H., Weber K. In vivo degradation of mutant lac repressor. Nature. 1970 Dec 19;228(5277):1154–1156. doi: 10.1038/2281154a0. [DOI] [PubMed] [Google Scholar]
- Russell R. L., Abelson J. N., Landy A., Gefter M. L., Brenner S., Smith J. D. Duplicate genes for tyrosine transfer RNA in Escherichia coli. J Mol Biol. 1970 Jan 14;47(1):1–13. doi: 10.1016/0022-2836(70)90397-9. [DOI] [PubMed] [Google Scholar]
- Rutberg L. Mapping of a temperate bacteriophage active on Bacillus subtilis. J Virol. 1969 Jan;3(1):38–44. doi: 10.1128/jvi.3.1.38-44.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spizizen J. TRANSFORMATION OF BIOCHEMICALLY DEFICIENT STRAINS OF BACILLUS SUBTILIS BY DEOXYRIBONUCLEATE. Proc Natl Acad Sci U S A. 1958 Oct 15;44(10):1072–1078. doi: 10.1073/pnas.44.10.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strigini P., Gorini L. Ribosomal mutations affecting efficiency of amber suppression. J Mol Biol. 1970 Feb 14;47(3):517–530. doi: 10.1016/0022-2836(70)90319-0. [DOI] [PubMed] [Google Scholar]
- Taylor A. L. Current linkage map of Escherichia coli. Bacteriol Rev. 1970 Jun;34(2):155–175. doi: 10.1128/br.34.2.155-175.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tevethia M. J., Caudill C. P. Relationship between competence for transformation of Bacillus subtilis with native and single-stranded deoxyribonucleic acid. J Bacteriol. 1971 Jun;106(3):808–811. doi: 10.1128/jb.106.3.808-811.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tevethia M. J., Mandel M. Nature of the ethylenediaminetetraacetic acid requirement for transformation of Bacillus subtilis with single-stranded deoxyribonucleic acid. J Bacteriol. 1970 Mar;101(3):844–850. doi: 10.1128/jb.101.3.844-850.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters L. C., Novelli G. D. A new change in leucine transfer RNA observed in Escherichia coli infected with bacteriophage T2. Proc Natl Acad Sci U S A. 1967 Apr;57(4):979–985. doi: 10.1073/pnas.57.4.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigert M. G., Lanka E., Garen A. Amino acid substitutions resulting from suppression of nonsense mutations. 3. Tyrosine insertion by the Su-4 gene. J Mol Biol. 1967 Feb 14;23(3):401–404. doi: 10.1016/s0022-2836(67)80114-1. [DOI] [PubMed] [Google Scholar]
- Weiner A. M., Weber K. Natural read-through at the UGA termination signal of Q-beta coat protein cistron. Nat New Biol. 1971 Sep 15;234(50):206–209. doi: 10.1038/newbio234206a0. [DOI] [PubMed] [Google Scholar]
- Yasunaka K., Tsukamoto H., Okubo S., Horiuchi T. Isolation and properties of suppressor-sensitive mutants of Bacillus subtilis bacteriophage SP02. J Virol. 1970 Jun;5(6):819–821. doi: 10.1128/jvi.5.6.819-821.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa H. DNA synthesis during germination of Bacillus subtilis spores. Proc Natl Acad Sci U S A. 1965 Jun;53(6):1476–1483. doi: 10.1073/pnas.53.6.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa H. Mutations Resulting from the Transformation of BACILLUS SUBTILIS. Genetics. 1966 Nov;54(5):1201–1214. doi: 10.1093/genetics/54.5.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]



