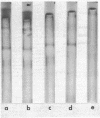
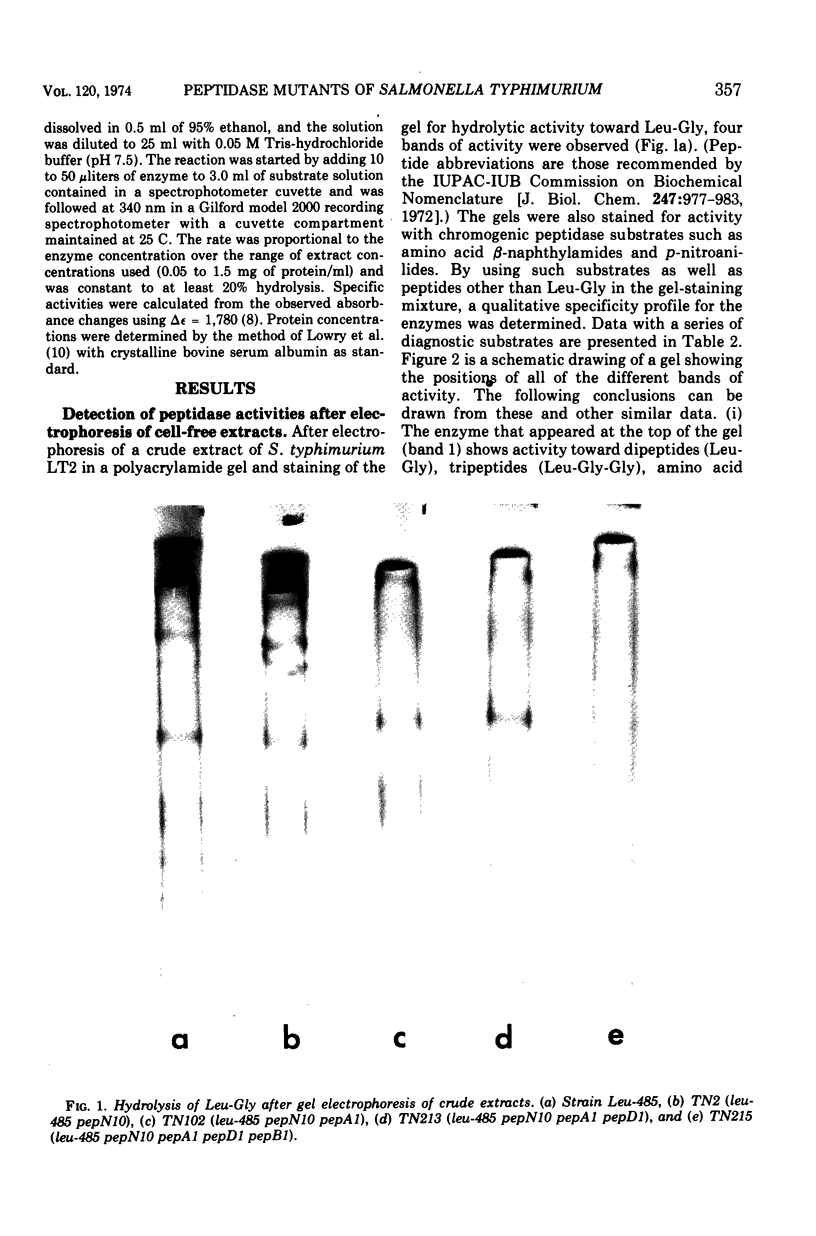
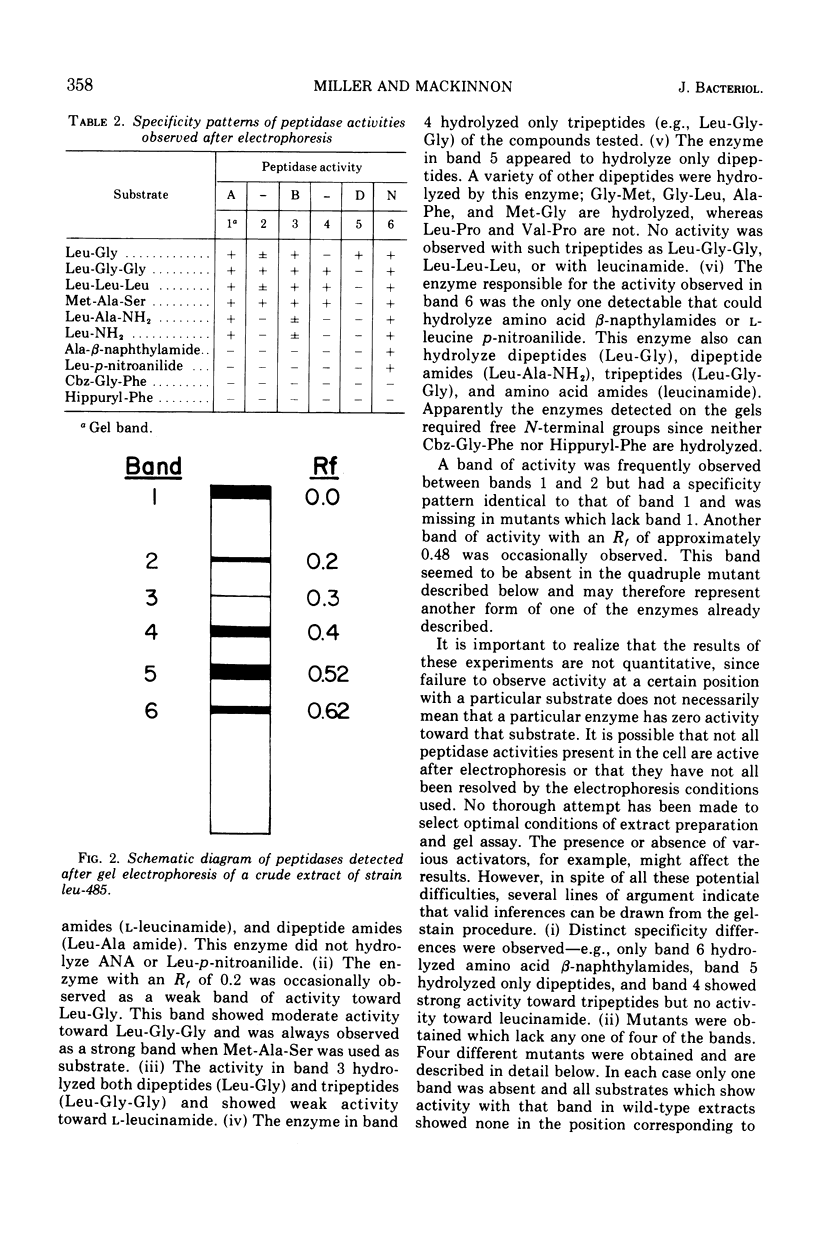
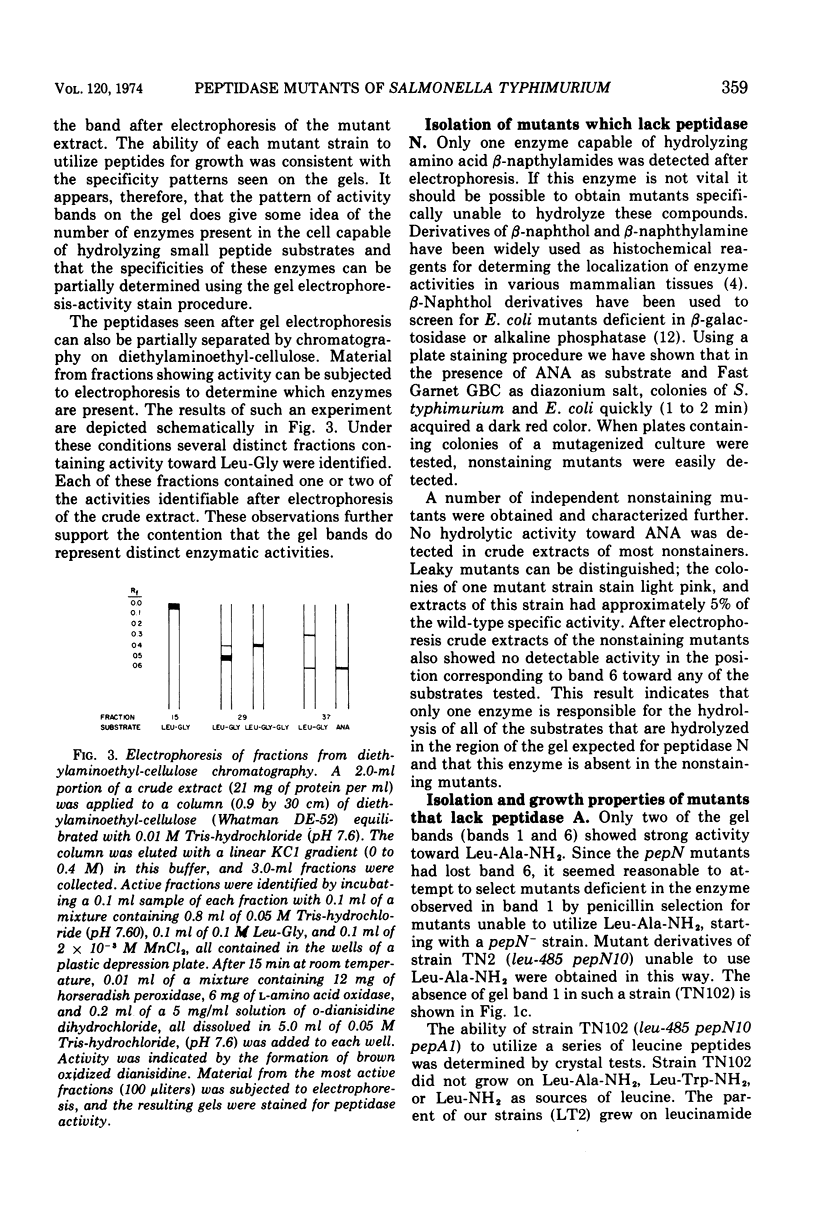
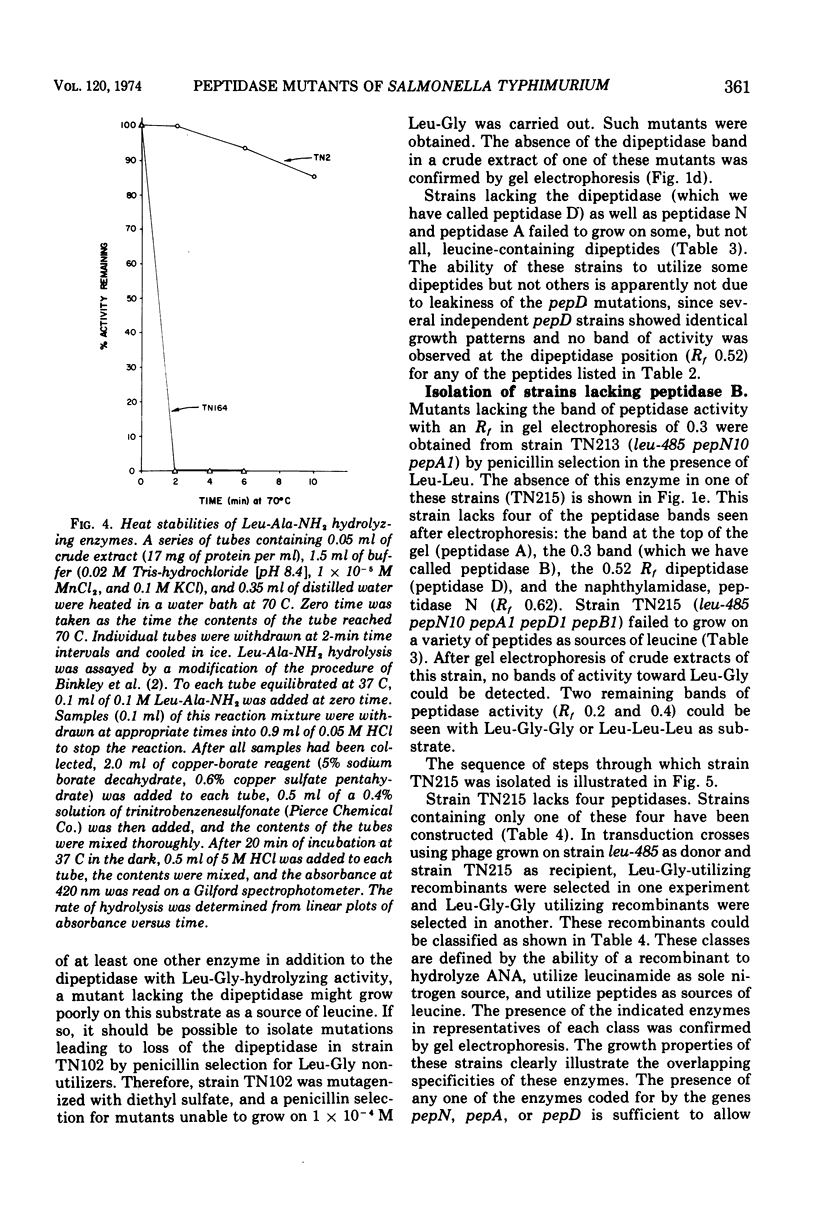
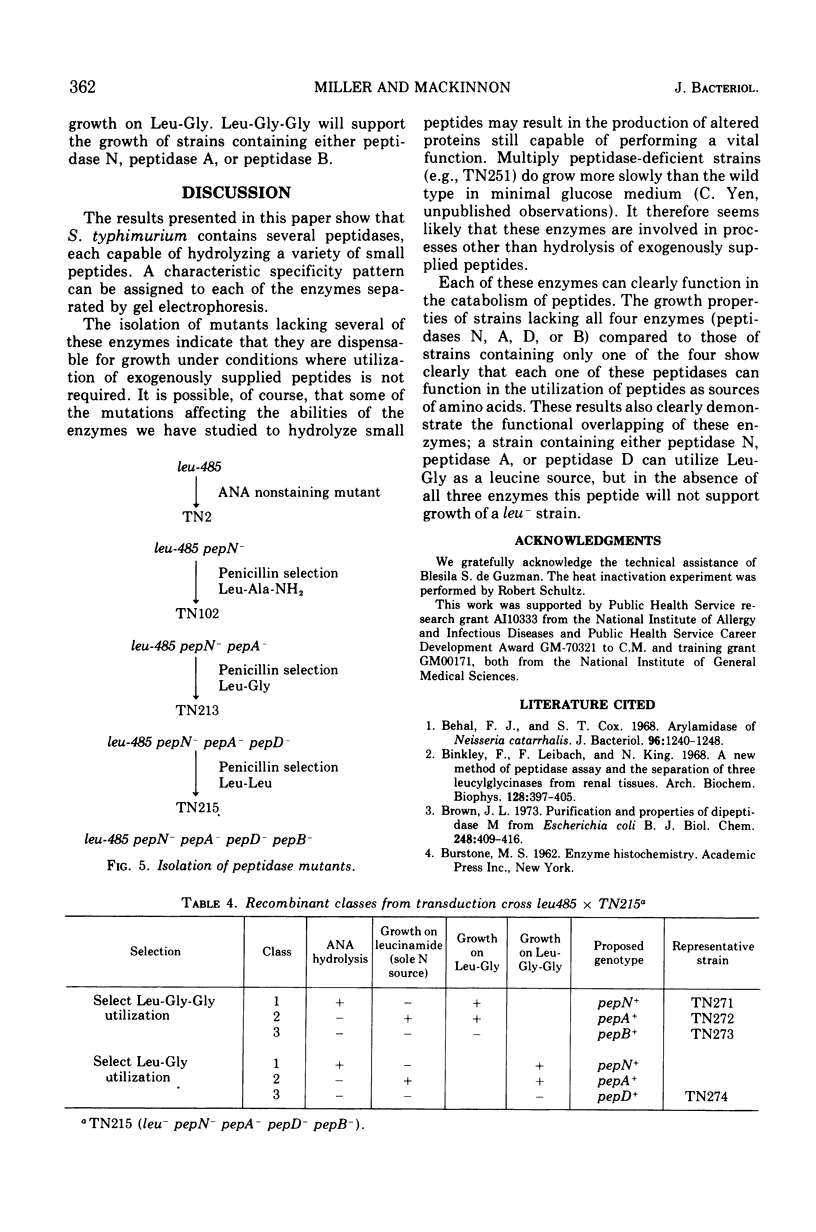
Abstract
Six peptidase activities have been distinguished electrophoretically in cell extracts of Salmonella typhimurium with the aid of a histochemical stain. The activities can also be partially separated by chromatography on diethylaminoethyl-cellulose. These peptidases show overlapping substrate specificities. Mutants (pepN) of the parent strain leu-485 lacking one of these enzymes (peptidase N) were obtained by screening for colonies that do not hydrolyze the chromogenic substrate l-alanyl-β-naphthylamide. The absence of this broad-specificity peptidase in leu-485 pepN− mutants allowed the selection of mutants unable to use l-leucyl-l-alaninamide as a leucine source. These mutants (leu-485 pepN−pepA−) lack a broad-specificity peptidase (peptidase A) similar to aminopeptidase I previously described in Escherichia coli. Mutants (pepD) lacking a dipeptidase (peptidase D) have been isolated from a leu-485 pepN−pepA− parent by penicillin selection for mutants unable to use l-leucyl-l-glycine as a leucine source. Mutants (pepB) lacking a fourth peptidase (peptidase B) have been isolated from a leu-485 pepN−pepA−pepD− strain by penicillin selection for failure to utilize l-leucyl-l-leucine as a source of leucine. Single recombinants were obtained by transduction for each of the peptidases missing in a leu-485 pepN−pepA−pepD−pepB− strain. The growth response of these recombinants to leucine peptides shows that all of these peptidases can function in the catabolism of peptides and that they display overlapping substrate specificities in vivo.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Behal F. J., Cox S. T. Arylamidase of Neisseria catarrhalis. J Bacteriol. 1968 Oct;96(4):1240–1248. doi: 10.1128/jb.96.4.1240-1248.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binkley F., Leibach F., King N. A new method of peptidase assay and the separation of three leucylglycinases of renal tissues. Arch Biochem Biophys. 1968 Nov;128(2):397–405. doi: 10.1016/0003-9861(68)90046-5. [DOI] [PubMed] [Google Scholar]
- Brown J. L. Purification and properties of dipeptidase M from Escherichia coli B. J Biol Chem. 1973 Jan 25;248(2):409–416. [PubMed] [Google Scholar]
- DAVIS B. J. DISC ELECTROPHORESIS. II. METHOD AND APPLICATION TO HUMAN SERUM PROTEINS. Ann N Y Acad Sci. 1964 Dec 28;121:404–427. doi: 10.1111/j.1749-6632.1964.tb14213.x. [DOI] [PubMed] [Google Scholar]
- Gutnick D., Calvo J. M., Klopotowski T., Ames B. N. Compounds which serve as the sole source of carbon or nitrogen for Salmonella typhimurium LT-2. J Bacteriol. 1969 Oct;100(1):215–219. doi: 10.1128/jb.100.1.215-219.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KESSEL D., LUBIN M. On the distinction between peptidase activity and peptide transport. Biochim Biophys Acta. 1963 Jun 4;71:656–663. doi: 10.1016/0006-3002(63)91139-9. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Lee H. J., LaRue J. N., Wilson I. B. A simple spectrophotometric assay for amino acyl arylamidases (naphthylamidases, aminopeptidases). Anal Biochem. 1971 Jun;41(2):397–401. doi: 10.1016/0003-2697(71)90157-6. [DOI] [PubMed] [Google Scholar]
- Lewis W. H., Harris H. Human red cell peptidases. Nature. 1967 Jul 22;215(5099):351–355. doi: 10.1038/215351a0. [DOI] [PubMed] [Google Scholar]
- McHugh G. L., Miller C. G. Isolation and characterization of proline peptidase mutants of Salmonella typhimurium. J Bacteriol. 1974 Oct;120(1):364–371. doi: 10.1128/jb.120.1.364-371.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messer W., Vielmetter W. High resolution colony staining for the detection of bacterial growth requirement mutants using naphthol azo-dye techniques. Biochem Biophys Res Commun. 1965 Oct 26;21(2):182–186. doi: 10.1016/0006-291x(65)90106-3. [DOI] [PubMed] [Google Scholar]
- Neu H. C., Heppel L. A. The release of enzymes from Escherichia coli by osmotic shock and during the formation of spheroplasts. J Biol Chem. 1965 Sep;240(9):3685–3692. [PubMed] [Google Scholar]
- Pine M. J. Turnover of intracellular proteins. Annu Rev Microbiol. 1972;26:103–126. doi: 10.1146/annurev.mi.26.100172.000535. [DOI] [PubMed] [Google Scholar]
- Sussman A. J., Gilvarg C. Peptidases in Escherichia coli K-12 capable of cleaving lysine homopeptides. J Biol Chem. 1970 Dec 25;245(24):6518–6524. [PubMed] [Google Scholar]
- Sussman A. J., Gilvarg C. Peptide transport and metabolism in bacteria. Annu Rev Biochem. 1971;40:397–408. doi: 10.1146/annurev.bi.40.070171.002145. [DOI] [PubMed] [Google Scholar]
- VOGEL H. J., BONNER D. M. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956 Jan;218(1):97–106. [PubMed] [Google Scholar]
- Vogt V. M. Purification and properties of an aminopeptidase from Escherichia coli. J Biol Chem. 1970 Sep 25;245(18):4760–4769. [PubMed] [Google Scholar]