Abstract
The endogenous plant hormones salicylic acid (SA) and jasmonic acid (JA), whose levels increase on pathogen infection, activate separate sets of genes encoding antimicrobial proteins in Arabidopsis thaliana. The pathogen-inducible genes PR-1, PR-2, and PR-5 require SA signaling for activation, whereas the plant defensin gene PDF1.2, along with a PR-3 and PR-4 gene, are induced by pathogens via an SA-independent and JA-dependent pathway. An Arabidopsis mutant, coi1, that is affected in the JA-response pathway shows enhanced susceptibility to infection by the fungal pathogens Alternaria brassicicola and Botrytis cinerea but not to Peronospora parasitica, and vice versa for two Arabidopsis genotypes (npr1 and NahG) with a defect in their SA response. Resistance to P. parasitica was boosted by external application of the SA-mimicking compound 2,6-dichloroisonicotinic acid [Delaney, T., et al. (1994) Science 266, 1247–1250] but not by methyl jasmonate (MeJA), whereas treatment with MeJA but not 2,6-dichloroisonicotinic acid elevated resistance to Alternaria brassicicola. The protective effect of MeJA against A. brassicicola was the result of an endogenous defense response activated in planta and not a direct effect of MeJA on the pathogen, as no protection to A. brassicicola was observed in the coi1 mutant treated with MeJA. These data point to the existence of at least two separate hormone-dependent defense pathways in Arabidopsis that contribute to resistance against distinct microbial pathogens.
To defend themselves against microbial invaders, plants use a variety of defense systems; some of these are preformed, and others are inducible. The components of these defense systems, whether preformed or inducible, usually include specific cell-wall polymers and a mixture of antimicrobial secondary metabolites and antimicrobial proteins or peptides. The efficiency of these physical and chemical barriers to a particular microbial invader, as well as the rate and extent at which they are erected in inducible reactions, is believed to determine whether a plant becomes infected by the potential pathogen.
In vegetative plant tissues such as leaves, inducible defense responses are particularly well elaborated, probably because they are more economic in terms of energy and element utilization compared with anticipatory defense components. Inducible defense responses are activated on recognition by plant cell receptors of elicitor molecules derived from the invading microorganisms. This recognition event triggers a signal-transduction cascade leading to, among other things, the production of endogenous signaling compounds. Such signaling compounds are spread systemically to tissues that are distant from the initial infection site, where they activate antimicrobial protein/peptide (AMP) genes. A well-known inducer of AMPs is salicylic acid (SA), whose levels are raised significantly both in infected tissues and in tissues distant from infection sites (1, 2). In leaves of the plant Arabidopsis thaliana, infection by pathogens, as well as exogenous application of SA, results in the induction of a set of AMP genes (3, 4) encoding PR-1, PR-2, (a β-1,3-glucanase), and PR-5, (a thaumatin-like protein). NahG and npr1–1 are Arabidopsis genotypes that are blocked in their response to SA, either because of expression of a chimeric transcription unit encoding an SA hydroxylase (for NahG plants; ref. 5) or to a point mutation in an IκB-like signal-transduction component acting downstream of SA (for npr1–1 mutants; ref. 6). Neither NahG nor npr1–1 is able to induce the PR-1, PR-2, or PR-5 gene (5–8), indicating that SA is essential in this process. However, SA is not the only signaling compound produced by plants in response to infection. We have recently shown that levels of jasmonic acid (JA), an oxylipin-like hormone derived from oxygenated linolenic acid, increased strongly on challenge of Arabidopsis with the fungus Alternaria brassicicola, both in inoculated leaves and in untreated leaves of inoculated plants (9). Challenge-inoculation with A. brassicicola, as well as exogenous application of methyl jasmonate (MeJA), but not SA, resulted in the induction of the PDF1.2 gene. PDF1.2 encodes an antifungal peptide belonging to the family of plant defensins. Local, as well as systemic, induction of PDF1.2 on challenge-inoculation with A. brassicicola was abolished in the Arabidopsis mutant coi1–1 (9), which is affected in a signal-transduction component acting downstream of JA (10). In contrast, induction of PDF1.2 was not affected in the SA-response mutants NahG and npr1–1. Thus, it appears that Arabidopsis contains, in addition to an SA-dependent defense-response pathway, a JA-dependent pathway leading to the activation of the effector gene PDF1.2 (9).
In the present paper we investigated whether other effector genes are coregulated with PDF1.2 along the JA-dependent pathway and whether this pathway is important for resistance of Arabidopsis plants against pathogens.
MATERIALS AND METHODS
Biological Material.
The transgenic Arabidopsis thaliana line containing the NahG gene (5) was obtained from J. Ryals (Novartis, Research Triangle Park, NC). The mutants npr1–1, coi1–1, and pad3 were provided by X. Dong (Duke University, Durham, NC), J. Turner (University of East Anglia, Norwich, U.K.), and J. Glazebrook (University of Maryland, College Park, MD), respectively. All of the mutant and transgenic lines listed above are derived from the Columbia (Col-0) ecotype. Arabidopsis plants were grown as described (9). Growth and spore-harvesting of the fungus Alternaria brassicicola (strain MUCL20297; Mycothèque Université Catholique de Louvain, Louvain-la-Neuve, Belgium) and Botrytis cinerea (strain iMi 169558, International Mycological Institute, Kew, U.K.) was done as described (11). Peronospora parasitica strains Wela and Noco were maintained on living Arabidopsis plants of the ecotypes Weiningen and Col-0, respectively.
Plant Inoculations.
Inoculation of Arabidopsis plants with A. brassicicola was performed on 4-week-old soil-grown plants by placing three 5-μl drops of a suspension of 5 × 105 conidial spores per ml of water on each leaf. For the chemical protection assay, only expanded upper rosette leaves were inoculated.
Inoculation with B. cinerea was performed on 4-week-old soil-grown plants. Three needle-prick wounds were applied to the leaves, and the fresh wounds were covered with 5-μl drops of a suspension of 5 × 105 conidial spores per ml in 12 g/l potato dextrose broth (Difco).
P. parasitica was inoculated by spraying until droplet runoff occurred, using a suspension of 105 conidial spores per ml of water. Inoculated plants were incubated at 18°C at 100% relative humidity in propagator flats covered with a clear polystyrene lid for the time periods indicated.
Determination of Fungal Biomass.
A. brassicicola (strain MUCL20297) was transformed by using biolistics with a plasmid containing a hygromycin-based selection marker gene consisting of the glyceraldehyde-3-phosphate dehydrogenase (GPD1) promoter and the Escherichia coli uidA coding region (12). A transgenic A. brassicicola strain showing β-glucuronidase expression was selected. Arabidopsis plants were inoculated with three 5-μl drops per leaf in water from a suspension of 5 × 105 conidial spores per ml of this strain. Inoculated plants were incubated at 100% relative humidity. For each time point and each genotype, 30 discs (12.6 mm2) were punched out from inoculated leaves around an infection spot, and RNA was extracted (13), quantified, and hybridized with uidA probe (13) after dot-blotting of serial dilutions. Hybridization signals were quantified by using a luminescence charge-coupled device camera system and related to hybridization signals obtained with RNA extracted from fungal mycelia grown in vitro. This allowed us to calculate the percentage of fungal RNA in total RNA extracted from the discs.
RNA Gel Blot Analysis.
RNA was extracted from tissues of Arabidopsis by the phenol-LiCl method according to Eggermont et al. (13). RNA gel-blot analysis was performed as described (9). Riboprobes for PDF1.2 and β-Tubulin 1 were synthesized as described (9). Sequences corresponding to PR-1, PR-3, and PR-4 were amplified by PCR using the primer sets 5′-CTCGCTCGCCCACCACAAGATTATCTAAGGG-3′/5′-CTGGTTCGGCCCACCTGCATATGATGCTCCT-3′, 5′-AATGAATTCTGGATGGGCTACAGCACC-3′/5′-AATAATAAGCTTAATAGCAGCTTCGAGGAGG-3′, 5′-AATGGATCCACAATGCGGTCGTCAAGG-3′/5′-AATGAATTCTTCTGGAATCAGGCTGCC-3′, respectively. The PCR products were subcloned in plasmid pSPT18 (Boehringer Mannheim), and riboprobes were synthesized as described (9).
RESULTS
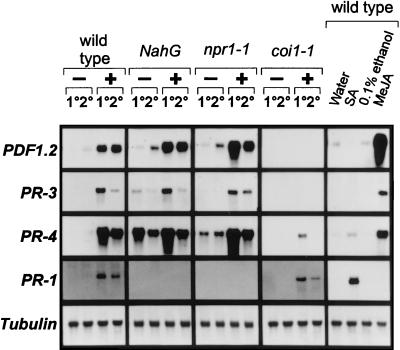
To determine whether other genes are activated coordinately with PDF1.2 in the JA-dependent defense-response pathway, we further assessed pathogen-induced activation of previously identified antimicrobial protein genes in Arabidopsis. The genes investigated were the pathogen-inducible PR-3, which encodes a basic chitinase (14), and PR-4, which encodes a hevein-like protein (15). These two genes are, like PDF1.2, induced on inoculation with the fungus A. brassicicola of either Arabidopsis wild-type (Col-0), npr1–1, or NahG plants, both in inoculated leaves and nontreated leaves of inoculated plants (Fig. 1). However, none of these genes is induced in either inoculated or uninoculated leaves of pathogen-challenged JA-insensitive coi1–1 plants. In contrast, pathogen-induced activation of the SA-dependent PR-1 gene is abolished in NahG-expressing plants and npr1–1 mutants, but not in the coi1–1 mutants. In addition, PDF1.2, PR-3, and PR-4 are induced by exogenous application of MeJA but not by SA applied at a concentration at which it induces PR-1 (Fig. 1). Hence, the pathogen-induced JA-dependent defense-response pathway in Arabidopsis leads to activation of at least three genes encoding proteins with known antimicrobial properties (PDF1.2 and PR-3; refs. 9 and 17, respectively) or proteins homologous to antimicrobial proteins from other plants (PR-4; ref. 16).
Figure 1.
Induction of pathogenesis-related genes in Arabidopsis in response to infection with A. brassicicola or to hormone treatment. Four-week-old soil-grown wild-type (Col-0), NahG, npr1–1, and coi1–1 plants were infected with A. brassicicola and harvested 48 hr after treatment. RNA blots were hybridized with the probes indicated on the left of the figure. Symbols above the lanes are as follows: −, mock-inoculated with water; +, inoculated with A. brassicicola spore suspension; 1°, treated lower rosette leaves; 2°, untreated upper rosette leaves. In addition, four-week-old soil-grown wild-type (Col-0) plants were treated with water, 5 mM SA, 0.1% ethanol, or 50 μM MeJA in 0.1% ethanol (MeJA). Plants were harvested 48 hr after treatment, and RNA blots were hybridized with the probes indicated on the left.
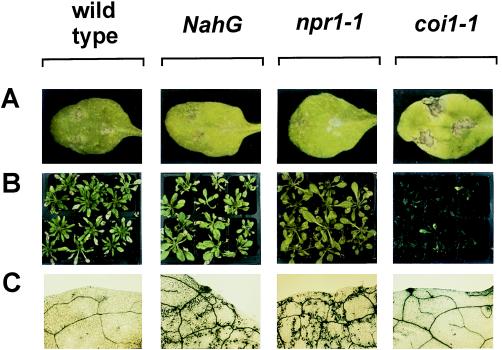
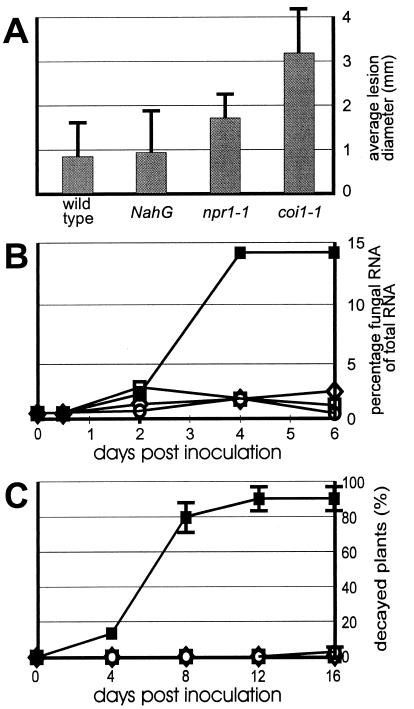
When challenged with the fungus A. brassicicola, 4-week-old wild-type plants (Col-0) produced small brown necrotic lesions (Fig. 2A). Microscopic examination indicated that, although some spore germination and penetration attempts between epidermal cells occurred, hyphae did not grow beyond the epidermal cell layer (results not shown). A similar restriction of the pathogen was observed in inoculated NahG and npr1–1 plants (Fig. 2A). In contrast, coi1–1 challenged with A. brassicicola produced spreading lesions (Fig. 2A) that were heavily colonized by fungal hyphae. The average diameter of lesions caused by A. brassicicola was determined 6 days after inoculation and found to be at least 3-fold greater in coi1–1 plants than in wild-type plants (Fig. 3A). To verify that the more severe disease symptoms observed on coi1–1 were indeed caused by increased fungal colonization, A. brassicicola was transformed with a chimeric heterologous reporter gene to permit estimation of fungal biomass by measuring transcripts from the heterologous reporter gene. As shown in Fig. 3B, the ratio of fungal RNA versus total RNA in the lesions 6 days after inoculation reached at least 7-fold higher levels in coi1–1 plants compared with wild-type, NahG, or npr1–1 plants. It was also verified that homozygous COI1 (nonmutant) plants selected from a population segregating for the coi1–1 mutation behaved essentially as Col-0 wild-type plants with respect to colonization by A. brassicicola (results not shown). Increased susceptibility for fungal infection of the JA-insensitive coi1–1 plants was also observed after challenge-inoculation of 4-week-old plants with the fungus Botrytis cinerea. Of the coi1–1 mutants treated with B. cinerea, 86% succumbed during the 16-day period after inoculation because of complete rotting of all above-ground organs (Figs. 2B and 3C). Decay of coi1–1 plants was followed by abundant sporulation. In marked contrast, inoculation of wild-type, npr1–1, and NahG plants resulted in necrosis on the challenged leaves, but 99%, 100%, and 100% of all plants survived the treatment, respectively (Figs. 2B and 3C). Inoculated wild-type, npr1–1, and NahG plants eventually flowered and set seed the same as untreated plants. Homozygous COI1 (nonmutant) plants selected from a population segregating for the coi1–1 mutation were as resistant as wild-type plants to B. cinerea (results not shown).
Figure 2.
Disease development in Arabidopsis inoculated with different fungal pathogens. (A) Four-week-old Arabidopsis plants inoculated with A. brassicicola. Pictures show lesion formation 6 days after inoculation. A typical example of each genotype is shown. (B) Four-week-old Arabidopsis plants inoculated with B. cinerea. Pictures were made 12 days after inoculation (8 days after inoculation for coi1–1). (C) Four-week-old Arabidopsis plants inoculated with P. parasitica strain Wela. Plants were stained with lactophenol-trypan blue (32). Microscopy pictures show blue-stained fungal structures and damaged plant cells in a leaf from plants 11 days after inoculation. A representative example of each genotype is shown. Genotypes are indicated above the pictures.
Figure 3.
Quantification of disease development in Arabidopsis inoculated with different fungal pathogens. (A) Average diameter of lesions formed 6 days after inoculation with A. brassicicola. Data represent averages with SDs from measurements of 60 lesions. (B) Percentage fungal RNA of total RNA in infection sites at different times after inoculation of leaves with A. brassicicola. Data points represent measurements on 30 leaf discs. ○, wild-type (Col-0) plants; ⋄, NahG plants; □, npr1–1 plants; ■, coi1–1 plants. (C) Decay of Arabidopsis plants inoculated with B. cinerea. The percentage of dead plants is expressed as a function of time after inoculation. Plants were considered dead when their hearts were completely rotten. Data represent averages with sSDs of four independent experiments performed with 20 plants per genotype. Symbols are as for B.
NahG-expressing plants and npr1–1 mutants have previously been shown to be more susceptible than wild-type plants to the biotrophic fungal pathogen P. parasitica strain Wela (5). To test whether the JA-dependent defense-response pathway is important for resistance against this pathogen, 4-week-old coi1–1 plants were inoculated with P. parasitica strain Wela, and the disease rate was assessed by using macroscopic observation and staining of fungal hyphae in leaf tissue 11 days after inoculation. Inoculated wild-type plants as well as inoculated coi1–1 mutants remained healthy, and no hyphae could be detected in any of 50 leaves examined (Fig. 2C). In contrast, inoculated NahG-expressing plants and npr1–1 mutants revealed the presence of intercellular growing hyphae and oospores in all of 50 examined leaves (Fig. 2C).
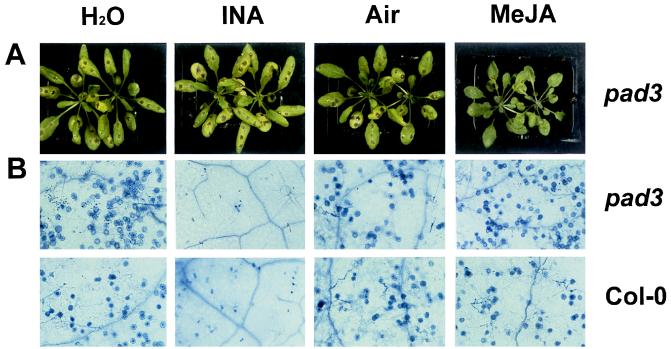
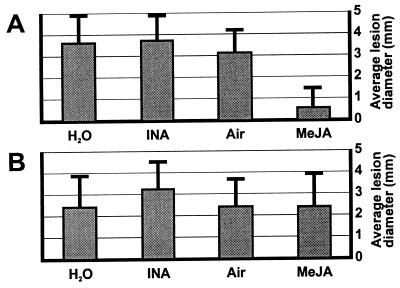
The discovery that a JA-dependent defense-response pathway plays a role in resistance raises the expectation that JA-related compounds may be used to boost the resistance level of plants against particular pathogens. A benzothiadiazole-type agrochemical that mimics the action of SA has been described recently (18, 19) and is currently commercially available for protection of crops via its ability to activate endogenous SA-dependent defense responses. Interestingly, Alternaria species have been reported not to be controlled by this compound (18, 19). It therefore seemed worthwhile to test whether MeJA could increase the resistance of Arabidopsis to A. brassicicola. For this purpose, various Arabidopsis genotypes were screened for their susceptibility to different A. brassicicola strains to identify a combination leading to a true infectious disease. An Arabidopsis mutant, pad3, which is affected in its ability to produce the antimicrobial secondary metabolite camalexin (20), was found to be susceptible to A. brassicicola strain MUCL20297. The pad3 mutant was, unlike coi1–1, still able to induce PDF1.2 in response to fungal infection (B.P.H.J.T., unpublished results). Hence, the pad3 mutation causes susceptibility to A. brassicicola because of a defect in yet another defense response different from the pathways leading to AMP induction. Untreated pad3 mutants developed lesions with an average diameter of 3.5 mm 6 days after inoculation with A. brassicicola (Figs. 4A and 5A). When pad3 mutants had been incubated for 48 hr in an atmosphere containing 150 nM gaseous MeJA before inoculation, the average diameter of lesions formed 6 days after infection was reduced to 0.6 mm (Figs. 4A and 5A). When coi1–1 mutants were pretreated with MeJA, however, no reduction in average lesion size was observed (Fig. 5B), indicating that a functional JA signal-transduction pathway is essential for MeJA-mediated protection against A. brassicicola. Pretreatment of pad3 mutants by spraying with 2,6-dichloroisonicotinic acid (INA), a functional analog of SA, did not result in reduced lesion development compared with the water-treated pad3 plants (Figs. 4A and 5A). In contrast, spraying Arabidopsis plants with INA very efficiently protected Arabidopsis Col-0 plants against infection by P. parasitica strain Noco, as shown before (5, 19), whereas pretreatment by exposure to 150 nM gaseous MeJA before inoculation did not confer any protection from this particular pathogen (Fig. 4B).
Figure 4.
Protective effect of exogenously applied MeJA and INA after infection by A. brassicicola and P. parasitica. (A) Arabidopsis mutant pad3 was infected with A. brassicicola. Fourty-eight hr before inoculation, separate sets of plants were treated as follows: sprayed with water on the leaves, H2O; sprayed with 1 mg/ml INA wettable powder with 25% active ingredient, INA; untreated in an airtight translucent container, Air; or treated with 150 nM gaseous MeJA in an airtight translucent container, MeJA. Observations were made 6 days after inoculation. (B) Arabidopsis mutant (pad3) and wild-type (Col-0) were infected with P. parasitica strain Noco. Treatments were as described for A. Six days after inoculation, plants were stained with lactophenol-trypan blue (32). Representative microscopy pictures of leaves are shown. Oospores can be seen on plants pretreated with water, air, and MeJA. Plants treated with INA only show signs of local hypersensitive reaction.
Figure 5.
Quantification of the protective effect of exogenously applied MeJA and INA on infection by A. brassicicola. (A) Average lesion diameter with SDs from measurements of 30 lesions on pad3 mutants subjected to the indicated pretreatments, as in the legend to Fig. 4A. Experiments were performed three times with similar results. (B) Average lesion diameter with SDs from measurements of 30 lesions on coi1–1 mutants subjected to the indicated pretreatments. Experiments were performed twice with similar results.
DISCUSSION
It has been known for a long time that infection attempts of microbial pathogens on plants trigger a complex set of defense responses requiring activation of distinct signaling pathways (21, 22). In Arabidopsis, induction of the antimicrobial protein genes PR-1, PR-2, and PR-5 follows an SA-dependent pathway (3, 5, 7), whereas the induction of the plant defensin PDF1.2, the basic chitinase PR-3, and the hevein-like protein PR-4 depends on a pathway involving at least JA as a signal molecule. The availability of different Arabidopsis mutants affected in these response pathways allows for assessment of the significance of these pathways in resistance against pathogens.
We have shown that coi1–1, a MeJA-insensitive mutant unable to induce PDF1.2, PR-3, and PR-4 on pathogen challenge, is more susceptible than wild-type plants to infection by the fungal pathogens A. brassicicola and B. cinerea, but not P. parasitica. In contrast, the Arabidopsis genotypes NahG and npr1, which are blocked in their response to SA and fail to induce PR-1 on pathogen challenge, are as resistant as wild-type plants to A. brassicicola and B. cinerea, whereas they are more susceptible to P. parasitica. Hence, the distinct defense-response pathways appear to be specifically efficient against distinct pathogens. Our data, however, do not prove that the efficacy is determined by the particular set of antimicrobial proteins that we have identified, as many yet-unknown effector molecules may also be involved.
We have also shown that it is possible to boost the resistance level of Arabidopsis plants against A. brassicicola by exogenous application of MeJA but not by INA, a functional analog of SA. On the other hand, INA, but not MeJA, was effective in protecting Arabidopsis plants against P. parasitica. Hence, it may be feasible to develop JA-mimicking agrochemicals that protect crops against particular economically important pathogens, and the activity of such compounds may be complementary to that of SA-mimicking agrochemicals such as INA and benzothiadiazoles.
The potential of jasmonates to protect plants from infection by pathogens has been shown before for the interactions between potato and Phytophthora infestans (23), between cotton and Verticillium dahliae (24), and between Arabidopsis and Pythium mastophorum (25). On the other hand, MeJA failed to protect wheat against the powdery mildew fungus Erysiphe graminis (26), whereas SA-mimicking compounds such as INA and benzothiadiazole conferred effective protection (18, 26). We have shown that at least protection of Arabidopsis against A. brassicicola infection is caused by activation of responses that require a functional JA signal-transduction pathway, as the protection was observed for the pad3 mutant but not for the JA-insensitive coi1–1 mutant. Hence, a direct effect of MeJA on development of A. brassicicola can be excluded in our experimental system. In agreement with this notion, we could not observe any detrimental effect of MeJA on growth or development of this fungus in vitro at a MeJA concentration up to 50 μM. The dependency of MeJA-mediated protection on a functional COI1 gene product, and hence on a functional jasmonate response pathway, was also shown in the case of Arabidopsis challenged with Pythium mastophorum (25).
The multifaceted character of the inducible defense system of Arabidopsis bears strong resemblance to that of the fruitfly Drosophila melanogaster. In Drosophila, pathogen-induced accumulation of the antifungal peptide drosomycin is controlled by an NF-κB/IκB-like signaling pathway, whereas expression of various genes encoding antibacterial peptides is under control of the unknown gene IMD (27, 28). Interestingly, drosomycin shows striking sequence and structural homology to the Arabidopsis plant defensin PDF1.2 (29, 30). Mutants in the NF-κB/IκB-like signaling pathway are highly susceptible to Aspergillus flavus infection but not to E. coli infection, whereas the opposite is true for imd mutants (27, 28). The role of oxylipin-type molecules in the induction of antimicrobial peptides in insects has not yet been investigated. However, it has been shown that treatment of insect larvae with inhibitors of oxylipin biosynthesis severely reduces their ability to clear bacterial pathogens from their hemolymph, leading to increased mortality (31). Hence, oxylipin-type molecules appear to be fundamental in the pathophysiology of both plants and animals.
Acknowledgments
We are indebted to Drs. J. Turner, X. Dong, J. Ryals, and J. Glazebrook for providing the Arabidopsis genotypes coi1–1, npr1–1, NahG, and pad3, respectively. We also thank Dr. W. Schäfer for providing the pGUS5 plasmid and Y. Tadesse for assistance with biolistic transformation of A. brassicicola. This work was supported in part by a grant from the Fonds voor Wetenschappelijk Onderzoek-Vlaanderen. B.P.H.J.T. is Research Assistant with this fund and I.A.M.A.P. is recipient of a predoctoral grant of the ‘Vlaams Instituut voor de Bevordering van het Wetenschappelijk Technologisch Onderzoek in de Industrie’.
ABBREVIATIONS
- AMP
antimicrobial protein/peptide
- JA
jasmonic acid
- MeJA
methyl jasmonate
- SA
salicylic acid
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Dürner J, Shah J, Klessig D F. Trends Plant Sci. 1997;2:266–274. [Google Scholar]
- 2.Ryals J A, Neuenschwander U H, Willits M G, Molina A, Steiner H-Y, Hunt M D. Plant Cell. 1996;8:1809–1819. doi: 10.1105/tpc.8.10.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Uknes S, Mauch-Mani B, Moyer M, Potter S, Williams S, Dincher S, Chandler D, Slusarenko A, Ward E R, Ryals J A. Plant Cell. 1992;4:645–656. doi: 10.1105/tpc.4.6.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Uknes S, Winter A, Delaney T, Vernooij B, Morse A, Friedrich L, Nye G, Potter S, Ward E, Ryals J A. Mol Plant–Microbe Interact. 1993;6:692–698. [Google Scholar]
- 5.Delaney T, Uknes S, Vernooij B, Friedrich L, Weymann K, Negrotto D, Gaffney T, Gut-Rella M, Kessmann H, Ward E, et al. Science. 1994;266:1247–1250. doi: 10.1126/science.266.5188.1247. [DOI] [PubMed] [Google Scholar]
- 6.Cao H, Glazebrook J, Clarke J D, Volko S, Dong X. Cell. 1997;88:57–63. doi: 10.1016/s0092-8674(00)81858-9. [DOI] [PubMed] [Google Scholar]
- 7.Cao H, Bowling S A, Gordon A S, Dong X. Plant Cell. 1994;6:1583–1592. doi: 10.1105/tpc.6.11.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hunt M D, Delaney T P, Dietrich R A, Weymann K B, Dangl J L, Ryals J A. Mol Plant–Microbe Interact. 1997;10:531–536. doi: 10.1094/MPMI.1997.10.5.531. [DOI] [PubMed] [Google Scholar]
- 9.Penninckx I A M A, Eggermont K, Terras F R G, Thomma B P H J, De Samblanx G W, Buchala A, Métraux J-P, Manners J M, Broekaert W F. Plant Cell. 1996;8:2309–2323. doi: 10.1105/tpc.8.12.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xie D-X, Feys B F, James S, Nieto-Rostro M, Turner J G. Science. 1998;280:1091–1094. doi: 10.1126/science.280.5366.1091. [DOI] [PubMed] [Google Scholar]
- 11.Broekaert W F, Terras F R G, Cammue B P A, Vanderleyden J. FEMS Microbiol Lett. 1990;69:55–60. [Google Scholar]
- 12.Mönke E, Schäfer W. Mol Gen Genet. 1993;241:73–80. doi: 10.1007/BF00280203. [DOI] [PubMed] [Google Scholar]
- 13.Eggermont K, Goderis I J, Broekaert W F. Plant Mol Biol Rep. 1996;14:273–279. [Google Scholar]
- 14.Samac D A, Hironaka C M, Yallaly P E, Shah D M. Plant Physiol. 1990;93:907–914. doi: 10.1104/pp.93.3.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Potter S, Uknes S, Lawton K, Winter A M, Chandler D, DiMaio J, Novitzky R, Ward E, Ryals J. Mol Plant–Microbe Interact. 1993;6:680–685. doi: 10.1094/mpmi-6-680. [DOI] [PubMed] [Google Scholar]
- 16.Ponstein A S, Bres-Vloemans S A, Sela-Buurlage M B, van den Elzen P J M, Melchers L S, Cornelissen B J C. Plant Physiol. 1994;104:109–118. doi: 10.1104/pp.104.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Verburg J G, Huynh Q K. Plant Physiol. 1991;95:450–455. doi: 10.1104/pp.95.2.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gorlach J, Volrath S, Knauf-Beiter G, Hengy G, Beckhove U, Kogel K-H, Oostendorp M, Staub T, Ward E, Kessmann H, et al. Plant Cell. 1996;8:629–643. doi: 10.1105/tpc.8.4.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lawton K, Friedrich L, Hunt M, Weymann K, Delaney T, Kessmann H, Staub T, Ryals J. Plant J. 1996;10:71–82. doi: 10.1046/j.1365-313x.1996.10010071.x. [DOI] [PubMed] [Google Scholar]
- 20.Glazebrook J, Ausubel F M. Proc Natl Acad Sci USA. 1994;91:8955–8959. doi: 10.1073/pnas.91.19.8955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beffa R, Szell M, Pay A, Vögeli-Lange R, Metraux J-P, Neuhaus G, Meins F, Jr, Nagy F. EMBO J. 1995;14:5753–5761. doi: 10.1002/j.1460-2075.1995.tb00264.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dorey S, Baillieul F, Pierrel M-A, Saindrenan P, Fritig B, Kauffmann S. Mol Plant–Microbe Interact. 1997;10:646–655. [Google Scholar]
- 23.Cohen Y, Gisi U, Niderman T. Phytopathology. 1993;83:1054–1062. [Google Scholar]
- 24.Li J, Zingen-Sell I, Buchenauer H. J Plant Dis Prot. 1996;103:288–299. [Google Scholar]
- 25.Vijayan P, Shockey J, Lévesque C A, Cook R J, Browse J. Proc Natl Acad Sci USA. 1998;95:7209–7214. doi: 10.1073/pnas.95.12.7209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kogel K H, Beckhove U, Dreschers J, Munch S, Romme Y. Plant Physiol. 1994;106:1269–1277. doi: 10.1104/pp.106.4.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lemaitre B, Kromer-Metzger E, Michaut L, Nicolas E, Meister M, Georgel P, Reichhart J M, Hoffmann J A. Proc Natl Acad Sci USA. 1995;92:9465–9469. doi: 10.1073/pnas.92.21.9465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lemaitre B, Nicolas E, Michaut L, Reichhart J M, Hoffmann J A. Cell. 1996;86:973–983. doi: 10.1016/s0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- 29.Fehlbaum P, Bulet P, Michaut L, Lageux M, Broekaert W F, Hétru C, Hoffmann J A. J Biol Chem. 1994;269:33159–33163. [PubMed] [Google Scholar]
- 30.Landon C, Sodano P, Hétru C, Hoffmann C, Ptak M. Protein Sci. 1997;6:1878–1884. doi: 10.1002/pro.5560060908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stanley-Samuelson D W, Jensen E, Nickerson K W, Tiebel K, Ogg C L, Howard R W. Proc Natl Acad Sci USA. 1991;88:1064–1068. doi: 10.1073/pnas.88.3.1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Keogh R C, Deverall B J, McLeod S. Trans Br Mycol Soc. 1980;74:329–333. [Google Scholar]