Abstract
Auxins are plant hormones that mediate many aspects of plant growth and development. In higher plants, auxins are polarly transported from sites of synthesis in the shoot apex to their sites of action in the basal regions of shoots and in roots. Polar auxin transport is an important aspect of auxin functions and is mediated by cellular influx and efflux carriers. Little is known about the molecular identity of its regulatory component, the efflux carrier [Estelle, M. (1996) Current Biol. 6, 1589–1591]. Here we show that mutations in the Arabidopsis thaliana AGRAVITROPIC 1 (AGR1) gene involved in root gravitropism confer increased root-growth sensitivity to auxin and decreased sensitivity to ethylene and an auxin transport inhibitor, and cause retention of exogenously added auxin in root tip cells. We used positional cloning to show that AGR1 encodes a putative transmembrane protein whose amino acid sequence shares homologies with bacterial transporters. When expressed in Saccharomyces cerevisiae, AGR1 promotes an increased efflux of radiolabeled IAA from the cells and confers increased resistance to fluoro-IAA, a toxic IAA-derived compound. AGR1 transcripts were localized to the root distal elongation zone, a region undergoing a curvature response upon gravistimulation. We have identified several AGR1-related genes in Arabidopsis, suggesting a global role of this gene family in the control of auxin-regulated growth and developmental processes.
Plant roots typically grow downward, at a defined angle from the gravity vector. They respond to deviations from the defined growth angle (gravistimulation) by developing a curvature at the distal and central elongation zones, which eventually allows their tip to resume a normal growth pattern. The gravitropic response involves perception of the gravistimulus by the root cap columella cells, transduction of that physical information into physiological signals, transmission of the signals to the distal and central elongation zones, and curvature response (1). Physiological evidence suggests that auxin and, possibly, apoplastic Ca2+ constitute the physiological signals that are transmitted to the root distal and central elongation zones and are responsible for the curvature response (1).
Basipetal transmission of auxin is mediated by polar auxin transport machinery, which involve influx and efflux carriers. The polarity of auxin transport probably is established by a basal localization of the efflux carrier in transporting cells (2). In this report, we demonstrate that the Arabidopsis thaliana AGR1 gene encodes a component of the auxin efflux carrier that mediates the root gravitropic response.
MATERIALS AND METHODS
Plant Stocks.
The following alleles were analyzed and found to belong to the same complementation group: agr1–1, agr1–2, agr1–3 (Landsberg erecta; previously named as agr-1, agr-2, and agr-3, respectively) (3), agr1–4 (Wassilewskija; WS), agr1–5, agr1–6, agr1–7, and agr1–8 (Estland), wav6 (Landsberg; agr1–52) (4), and eir1–1 (Columbia) (5). Manipulation of plant stocks was described previously (6).
Quantification of Root Growth.
The rate of root growth was measured (6). Briefly, 4- to 5-day-old seedlings were transplanted onto 0.8% agar-based media containing one-half strength Murashige and Skoog (MS) salts and 1.5% sucrose (Sigma), with or without one of the chemicals described below. They were grown for 48 hr in a growth chamber at 22°C with a photoperiod of 16 hr and a light intensity of 80 μE m−2 s−1. Pictures were taken 0, 24, and 48 hr after seedlings were transferred to the test medium. Root length was measured by using an nih image analysis program (http://rsb.info.nih.gov/nih-image/). Reported results correspond to the growth rates measured over the 48-hr test period. However, identical results were obtained when growth rates were measured over a 24-hr period (data not shown). 1-Naphthaleneacetic acid (1-NAA), 1-aminocyclopropane-1-carboxylic acid (ACC), and 2,3,5-triiodobenzoic acid (TIBA) were obtained from Sigma and prepared as 10 mM stock solutions in diluted NaOH, 10% isopropanol, and water, respectively (7).
Auxin Efflux Assays.
Auxin efflux in root tips was measured using a modification of the procedure described earlier (8). Three-millimeter-long root tips of 20 5-day-old light-grown seedlings were excised and incubated for 2 hr in one-half MS media containing 1.5% sucrose and 3.3 × 10−8 M 3-[5(n)-3H]indolylacetic acid (3H-IAA; 21Ci/mmol; Amersham). Samples were rinsed and incubated for 2 hr in the same media without 3H-IAA. After another rinse, samples were soaked for 18 hr in 5 ml scintillation fluid and amounts of 3H-IAA present in the root tips were measured in a scintillation counter.
Root-Wave Assay.
Root waving on a tilted agar surface was examined to assess root gravitropism (4, 6).
Positional Cloning.
A mapping population segregating for polymorphic chromosomal markers in the WS and Columbia ecotypes was generated by self-pollination of F1 plants derived from a cross between homozygous agr1–4 plants (WS) and wild-type Columbia plants. F2 progeny were self-pollinated to generate F3 families. Individual F2 plants were progeny-typed to define their genetic constitution at AGR1. Two pools of 22 homozygous wild-type and 22 homozygous mutant F3 families were generated and their genomic DNAs were used in a bulked segregant analysis (9) to identify the AGR1-linked nga129 (10) and LFY3 (11) cleared amplified polymorphic sequences markers.
To fine-map AGR1, two populations of F2 plants segregating for polymorphic chromosomal markers were generated by self-pollination of F1 plants derived from crosses between homozygous agr1–4 plants (WS ecotype) and transgenic No-O lines (12) carrying a Ds element either 7 cM proximal to AGR1 (Ds389–14) or 3 cM distal to AGR1 (Ds392–13) (Fig. 2a). Individual F3 families were tested for hygromycin resistance (Ds marker) (12) and for root gravitropism. Families homozygous for agr1–4 and hemizygous for Ds, or heterozygous for AGR1 and null for Ds, were retained, because each of them carried recombined chromosome 5 with a recombination breakpoint between Ds and AGR1.
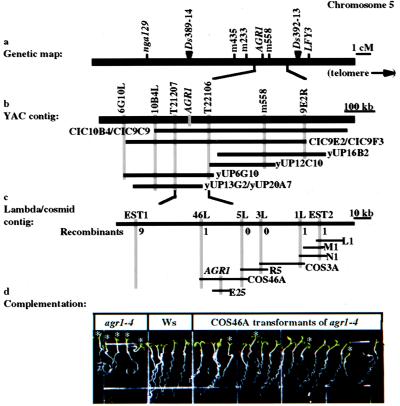
Figure 2.
Genetic and physical maps of the AGR1 region of chromosome 5. (a) Genetic map of the nga129 and LFY3 region of chromosome 5, showing the relative genetic positions of the nga129, m233, m558, and LFY3 markers, and the positions of the Ds 389–14 and Ds 392–13 insertions. (b) Mapping of AGR1 on a YAC contig of cloned genomic DNA fragments, showing the relative positions of specific RFLP markers (drawn as vertical shaded bars) and positions of individual YAC clones (drawn as horizontal black bars). (c) Fine mapping of AGR1 on a contig of cosmid (COS3A and COS46A) and lambda (L1, M1, N1, and R5) genomic clones, showing the number of recombinants identified in the mapping populations (see Materials and Methods) with recombination breakpoints between AGR1 and individual RFLP markers. Scales are in recombination units for a or kilobase pairs of DNA for b and c. (d) Wavy root-growth phenotype of segregating F2 plants (Right) derived from an agr1–4 plant (Left) transformed with the COS46A clone. Wild-type untransformed control plants are also shown (Center). Mutant seedlings are indicated by an asterisk.
Restriction fragment length polymorphism (RFLP) analysis of these two populations was carried out as described (11). Libraries for genomic lambda (13), cosmid (14), and yeast artificial chromosome (YAC; ref. 15) clones were screened by using standard procedures (16). End-fragment probes were isolated by PCR amplification (16) or by restriction-enzyme digestion.
Plant Transformation.
Genomic DNA fragments in a binary cosmid vector (COS3A and COS46A) were introduced into agr1–1, agr1–4, and agr1–5 mutants by a vacuum-infiltration transformation procedure (17). Similarly, an EcoRI subfragment of COS46A (E25) was inserted into the pBIN19 binary vector and introduced into transgenic agr1–1 and agr1–5 plants. Transgenic plants were selected on one-half MS media containing either 20 μg/ml of hygromycin (Sigma) for COS3A and COS46A transformants or 50 μg/ml of kanamycin (Sigma) for E25 transformants. Individual T2 progeny from several independent primary transformants were scored for root-waving and antibiotic-resistance phenotypes, as described (6).
Amplification and Sequencing of Genomic DNA and Full-Length cDNA.
Genomic DNA was isolated from agr1–4, agr1–5, and agr1–6 mutant and wild-type plants. The corresponding AGR1 locus was PCR-amplified to generate two overlapping fragments with the following PCR-primer pairs: (i) 5′-CTAACACGTTGGTAATGGGAATC-3′ and 5′-TGTCGTGAGGAGGAATAGAAACTT-3′, and (ii) 5′-GGAGTCAAGAAAAAGGAAAGTGG-3′ and 5′-CGCAAAACTTATCATCAGCAACTA-3′. PCRs were carried out by using the Elongase enzyme mix, according to the manufacturer’s instructions (GIBCO/BRL).
The full-length AGR1 cDNA was amplified by reverse-transcriptase–PCR, using the following primer pair: 5′-ATGATCACCGGCAAAGACATG-3′ and 5′-CACCTTTGGGTCGTATCGCC-3′, as described (16). The first-strand cDNA was synthesized from 10 μg of total Ler-root RNA by using a Superscript II RNaseH− Reverse Transcriptase (GIBCO/BRL) following manufacturer’s instructions.
The 7-kb BamHI subfragment of E25 was sequenced by using a shot-gun subcloning and sequencing approach. Genomic fragments containing the coding region of the agr1–4 allele, or overlapping with deletion breakpoints in agr1–5 and agr1–6, as well as a cDNA fragment containing the alternatively spliced exon of agr1–4 also were sequenced using a dye terminator cycle sequencing kit (Perkin–Elmer) and an Applied Biosystems sequencer.
A 900-bp-long AGR1 cDNA clone, containing 600 bp of the coding region, 265 bp of the 3′ untranslated region, and a poly(A) tail, was isolated from a cDNA library (18) using a 32P-labeled E25 probe and sequenced.
Functional Assays in Saccharomyces cerevisiae.
The pAGR1 and pANTI-AGR1 plasmids were generated by cloning a PCR-amplified full-length AGR1 cDNA into the EcoRI site of p426 GAL1 (19) (ATCC no. 87333), downstream of the GAL promoter, in the sense and antisense orientations, respectively. Both plasmids were introduced into Saccharomyces cerevisiae strain W3031A (MATa, ade2–1, his3–11/15, leu2–3/112, trp1–1, ura3–1, can1–100) as described (16). Transformants were isolated and grown on synthetic complete medium without uracil, supplemented with 2% glucose (16). Because W3031A[pAGR1] transformants did not grow in the presence of galactose, and because the GAL promoter is expressed at a low level when cells are grown on a glucose-containing medium (19), all experiments were carried out on synthetic complete medium without uracil and supplemented with 2% glucose.
To measure auxin efflux in yeast, overnight-grown cultures of W3031A[pAGR1] and W3031A[pANTI-AGR1] were centrifuged. Cells were washed and diluted in fresh medium and regrown for 10 hr. Cells (2 × 109) were pelleted, washed twice with 10 ml of water, and resuspended in 1 ml of 10 mM Mes-Tris buffer (pH 4.6) containing 2% glucose. After a 15-min preincubation at 30°C, 476 pmol of 3H-IAA (21Ci/mmol; Amersham) was added to each culture and cells were incubated for 20 min before being washed three times with 10 ml ice-cold water, resuspended in 10 ml of 10 mM Mes-Tris buffer (pH 4.6) containing 2% glucose, and incubated at 30°C. One-milliliter aliquots were removed at times 0 and 60 min. Cells were pelleted and resuspended in 100 μl of ice-cold water. Retained radioactive counts were determined by a scintillation counter.
To determine the resistance of transformed cells to 5-fluoro-indole-3-acetic-acid (F-IAA; Sigma), overnight-grown cultures of W3031A[pAGR1] and W3031A[pANTI-AGR1] were diluted in SC growth medium without uracil to a cell density of OD600 = 0.05. For each strain, five equal aliquots were prepared, and different amounts of this F-IAA stock solution (100 mM in ethanol) were added to reach final concentrations of 0, 0.01, 0.1, 1, and 3 mM, respectively. Cultures were grown at 30°C on a rotating shaker, and ODs were measured with a Milton Roy Spectrophotometer Spectronic 20D immediately after addition of F-IAA (time 0), as well as after 3, 7, 10, and 13 hr of growth. Three independent cultures were subjected to that experimental scheme. At each time point, the average OD was calculated for each culture condition, and a relative growth inhibition ratio was calculated by dividing the average OD difference between the control and the F-IAA-treated cultures by the average OD of the control culture. Three millimolar F-IAA completely inhibited the growth of both transformed cell types, while 0.01 and 0.1 mM F-IAA had no effects on the growth of either cultures (data not shown). Data are shown for cells treated with 1 mM F-IAA.
Northern Blot and Whole-Mount in Situ Hybridization Experiments.
Total RNA was isolated from plant tissues, electrophoresed, blotted, and hybridized with 32P-labeled probes following standard procedures (16).
Whole-mount in situ hybridization experiments on 3-day-old Arabidopsis seedlings were carried out as described (17) with the following modifications. A recombinant pZL1 plasmid (GIBCO/BRL) carrying the 900-bp-long AGR1 cDNA was linearized with either SalI or NotI to generate templates for in vitro synthesis of digoxigenin-labeled sense or antisense AGR1 RNA probes, following the manufacturer’s instructions (Boehringer Mannheim). The RNA probes were hydrolyzed to 150 nt, and 10 ng was used in the hybridization reactions.
RESULTS
agr1 Mutations Alter Root-Growth Sensitivity to 1-NAA, ACC, and TIBA.
Mutations in the AGR1 gene were identified and shown to confer specific defects in root gravitropism (3–5) (see Materials and Methods). Interestingly, one of these alleles (eir1–1) also was shown to mediate increased root-growth resistance to ethylene (5). The agr1–3 allele, on the other hand, was reported to confer increased root sensitivity to auxin (3) (see Materials and Methods). These observations prompted us to analyze the root-growth response to both auxin and ethylene for plants homozygous for several agr1 alleles (agr1–1 to agr1–8, eir1–1, and wav6; see Materials and Methods).
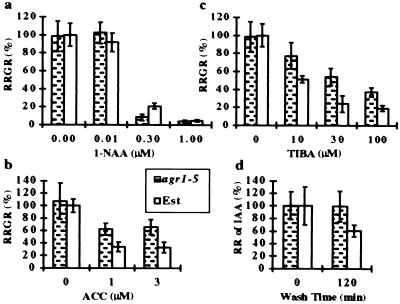
Fig. 1a shows that the growth of agr1–5 roots was inhibited more by 0.3 μM 1-NAA than that of wild-type Estland roots. The difference was highly significant (t test, P = 2 × 10−6). Both wild-type and agr1–5 mutant roots grew at comparable rates in the absence of 1-NAA or in the presence of 0.01, 1, and 3 μM 1-NAA (0.1 < P < 0.8). An increased root-growth sensitivity to 0.3 μM 1-NAA was observed in plants homozygous for all agr1 alleles tested (data not shown).
Figure 1.
Physiological analysis of agr1–5 mutant seedlings. Effects of 1-NAA (a), ACC (b), and TIBA (c) on wild-type and agr1–5 mutant root growth rates. The average root growth rate of wild-type Estland seedlings in media containing no supplements (12.3 mm for a and c, and 9.3 mm for b) was plotted as 100%. Each data point represents the average of the relative root growth rate (RRGR) for 6–16 wild-type Estland seedlings and for 10–12 agr1–5 mutant seedlings. (d) Relative retention (RR) of 3H-IAA by wild-type and agr1–5 mutant root tips. The average 3H-IAA retention at time 0 by wild-type and agr1–5 root tips (4,465 and 3,912 cpm, respectively) was plotted as 100%. Each data point represents the average of three replicates. In a–d, the SDs are represented by vertical bars. See Materials and Methods for experimental procedures.
agr1 mutations also caused decreased sensitivity to ACC, a precursor of ethylene biosynthesis. Fig. 1b shows that agr1–5 roots grew faster than wild-type roots in the presence of 1 or 3 μM ACC (P = 2 × 10−9 and 7 × 10−9, respectively). Plants homozygous for all agr1 alleles tested exhibited a similar degree of decreased sensitivity to 1 or 3 μM ACC (data not shown).
To test the possible involvement of AGR1 in the polar transport of auxin in roots, we compared the inhibitory effects of a polar auxin transport inhibitor, TIBA, on wild-type and mutant root growth (see Materials and Methods). Results shown in Fig. 1c indicate that agr1–5 mutant roots grew significantly faster than wild-type roots in the presence of 10, 30, or 100 μM TIBA (P = 7.8 × 10−4, 1.9 × 10−5, and 8.7 × 10−7, respectively). Mutant root growth was at 1.4–2.5 times the rate of wild-type root growth in the presence of TIBA for agr1–1, agr1–2, agr1–3, agr1–5, agr1–6, eir1–1, and wav6 (data not shown).
agr1 Mutants Retain More Preloaded 3H-IAA in Their Root Tips than Wild Type.
To identify possible defects in the auxin efflux activity in mutant roots, we measured the retention of preloaded 3H-IAA by wild-type and agr1–5 mutant root tips. Fig. 1d shows that 3H-IAA-pretreated agr1–5 mutant root tips retained most of the radioactive counts after a 2-hr washing period while wild-type root tips retained only 60% of the counts during that period (P = 0.1 for the experiment shown in Fig. 1d, and 2 × 10−2 and 2 × 10−4 in two other experiments).
AGR1 Encodes a Putative Transmembrane Protein with Homologies to Bacterial Transporters.
To characterize AGR1, we adopted a map-based strategy to clone the locus (Fig. 2a). AGR1 was mapped between the m233 and m558 RFLP markers (20) on chromosome 5 (Fig. 2a). Hence, a contig of genomic clones in the form of YACs overlapping with the m558 marker was identified (15) and characterized (Fig. 2b). New molecular markers (T21207, T22106, and 10B4L) were identified and ordered along the contig (Fig. 2b; Materials and Methods). The molecular marker closest to AGR1 (T22106) then was used as a probe to generate a contig of lambda and cosmid clones (Fig. 2c; Materials and Methods). Additional RFLP markers were mapped relative to AGR1 on that contig. 1L and 46L were identified as the closest markers flanking AGR1 (Fig. 2c).
AGR1 was mapped further onto the cosmid COS46A clone by a transformation–rescue approach. When introduced into agr1–1, agr1–4, and agr1–5 plants, COS46A rescued the altered root-wave phenotype associated with the mutations (Fig. 2d), while the COS3A clone did not (data not shown). Furthermore, the wild-type root-wave phenotype was found to cosegregate with the presence of the COS46A transgene in the T2 progenies of at least 16 COS46A primary transformants (data not shown). An internal 10-kb EcoRI subclone of COS46A (named E25) also was able to rescue the mutant root-wave phenotype when introduced into agr1–1 and agr1–5 mutant plants (data not shown).
A 7-kb BamHI subfragment of E25 was sequenced and a functional gene was predicted (21). The full-length cDNA was cloned and sequenced. As shown in Fig. 3a, the E25 gene consists of 9 exons and 8 introns. It encodes a hypothetical protein of 647 aa with a predicted molecular mass of 69 kDa.
Figure 3.
The Arabidopsis AGR1 gene. (a) The structure of AGR1. The exons are shown in solid boxes. The initiation (ATG) and stop (TAA) codons are also shown, with their positions indicated above the nucleotide sequence. The first nucleotide of the initiation codon was arbitrarily chosen as 1. (b) Nature of the mutations found in the agr1–4, agr1–5, and agr1–6 alleles. Exon and intron sequences are shown in upper- and lowercase letters, respectively. The exon derived from the alternative splicing of the agr1–4 transcript is underlined. Deleted nucleotides are represented by dashes. Numbers in parentheses represent nucleotides omitted because of space limitation. (c) The amino acid sequence of the AGR1 protein. Predicted transmembrane domains are boxed (22). (d) Kyte–Doollitle hydropathy plot of the AGR1 protein sequence. (e) Alignment of the amino acid sequences of AGR1 and Methanococcus MdcF (24). The AGR1 (top) and the MdcF (bottom) sequences were aligned using the nih gapped blast search program (23). Identical and functionally conserved amino acids are shown in solid and shaded boxes, respectively. Sequence gaps are represented by dashed lines.
A molecular analysis of the E25 gene in agr1–4, agr1–5, and agr1–6 plants confirmed its AGR1 identity (Fig. 3 a and b). The agr1–4 allele was found to carry a 6-bp deletion that altered the acceptor splice site of intron 6. Analysis of the corresponding cDNA sequence identified an alternative splicing of the transcribed sequence. Consequently, the agr1–4 mRNA carried a reading-frame shift and a premature stop codon (Fig. 3b). The agr1–5 allele was shown to carry a 898-bp deletion starting at the middle of the fifth intron and ending 123 bp downstream of the stop codon. As a result, a total of 128 aa should be deleted from the carboxyl end of the putative AGR1 protein, if expressed. The agr1–6 allele was shown to carry a 54-bp deletion in the seventh exon (Fig. 3b), resulting in the removal of 18 aa beginning at position 550 of the protein and the replacement of alanine-568 by serine.
The agr1–7 and agr1–8 alleles also were characterized. A deletion spanning the entire AGR1 locus and at least 0.5 cM of chromosomal DNA distal to the locus was identified in both alleles (data not shown).
Hydropathy analysis (22) of the deduced amino acid sequence of the AGR1 protein suggested that the region between positions 152 and 502 is hydrophilic, while the N and C termini are hydrophobic and predicted to form a total of 10 transmembrane domains (Fig. 3 c and d).
Database searches (23) with the AGR1 amino acid sequence identified positive matches, including two hypothetical proteins from A. thaliana (accession nos. 2829903 and 2829921, 53 and 47% amino acid sequence identities, respectively), YwkB-homolog protein from Methanococcus jannaschii (accession no. F64428, 23% identity), and MdcF protein from Klebsiella pneumoniae (accession no. 2240016). The homologous sequences from A. thaliana and M. jannaschii were identified by genome sequencing projects, and their functions are not known. Furthermore, the 319-aa-long MdcF protein of K. pneumoniae, which shares 23% identical and another 21% functionally conserved amino acids with AGR1 over its entire sequence (Fig. 3e), was predicted to be a malonate transporter. The MdcF protein was shown to share sequence homologies with a number of transporters (24) and with the hypothetical transmembrane YwkB protein of Bacillus subtilis (GenBank accession no. 1176957).
AGR1-Expressing S. cerevisiae Cells Retain Higher Levels of Preloaded 3H-IAA Than Control Cells.
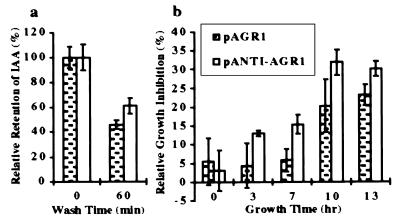
To test whether the AGR1 protein mediates the efflux of auxin from cells, we tested its ability to promote 3H-IAA release from loaded S. cerevisiae cells. Results shown in Fig. 4a indicated that AGR1-expressing yeast cells retained less radioactivity after a 1-hr washing period than control cells expressing an antisense AGR1. Furthermore, AGR1-expressing yeast cells were more resistant to 5-fluoro-IAA, a toxic IAA derivative, than control cells (Fig. 4b).
Figure 4.
AGR1 mediates auxin transport in S. cerevisiae. (a) Relative retention of 3H-IAA by yeast cells transformed with pAGR1 (dotted bar) or with pANTI-AGR1 (open bar), after 0 and 60 min in the wash solution (see Materials and Methods). Total number of counts at time 0 (470 cpm for W3031A[pAGR1] and 752 cpm for W3031A[pANTI-AGR1]) are plotted as 100%. Each data point represents an average of two measurements. The vertical bars represent the range of the measurements. (b) Relative growth inhibition by 1 mM F-IAA of yeast cells transformed with pAGR1 (dotted bar) or with pANTI-AGR1 (open bar) after 0, 3, 7, 10, and 13 hr of treatment (see Materials and Methods). Each point represents an average of three independent measurements. SDs are shown by vertical bars.
AGR1 Is Expressed in the Distal and Central Elongation Zones of A. thaliana.
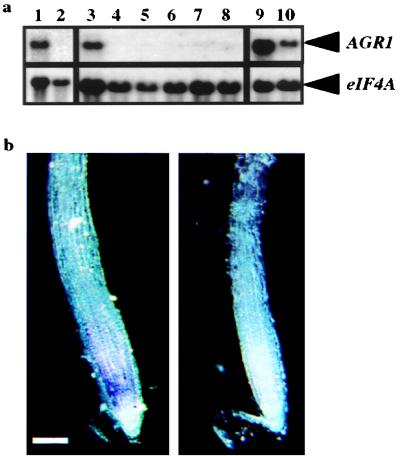
A combination of Northern blot analysis and in situ hybridization experiments were used to define the pattern of AGR1 expression in Arabidopsis seedlings and plants. Fig. 5a shows that the AGR1 gene is expressed specifically in roots of both 7-day-old seedlings and 24-day-old plants (Fig. 5a, lanes 1 and 3). No AGR1 transcripts were detected in hypocotyls and cotyledons of young seedlings (Fig. 5a, lane 2) or in rosette leaves, cauline leaves, inflorescence stems, flowers, or siliques of mature plants (Fig. 5a, lanes 4–8). On the other hand, AGR1 transcripts were found in root, hypocotyl, and cotyledon tissues of 5-day-old etiolated seedlings (Fig. 5a, lanes 9 and 10). When compared with the expression level of a control eIF4A gene, the steady-state level of AGR1 transcripts was found to be 10 times higher in roots than in hypocotyl and cotyledon tissues of etiolated seedlings (Fig. 5a, lanes 9 and 10).
Figure 5.
The pattern of AGR1 gene expression. (a) Northern blot analysis of total RNA extracted from tissues of 7-day-old seedlings (lanes 1 and 2) or 24-day-old (lane 3 to 8) or 5-day-old etiolated (lanes 9 and 10) plants. Tissues from roots (lanes 1, 3, and 9), hypocotyls and cotyledons (lanes 2 and 10), rosette leaves (lane 4), cauline leaves (lane 5), inflorescence stems (lane 6), flowers (lane 7), and siliques (lane 8) were analyzed. A 900-bp cDNA corresponding to the 3′ end of AGR1 was used as the probe (see Materials and Methods). After exposure, the blots were stripped and rehybridized with an eIF4A probe to control for loading differences. (b) Whole-mount in situ hybridization analysis of 3-day-old wild-type root tips, using digoxygenin-labeled antisense (Left) and sense (Right) RNA probes derived from in vitro transcription of the 900-bp-long AGR1 cDNA template (see Materials and Methods). Photos were taken under a dark-field microscope. (Bar = 150 μm.)
Whole-mount in situ hybridization using an antisense AGR1 RNA probe revealed the existence of AGR1 transcripts in cells of the distal elongation zone (Fig. 5b Left). The hybridization signal intensity decreased from the tip of the root to the mature zone (Fig. 5b Left). Control hybridization experiments using either a sense AGR1 RNA probe on wild-type seedlings (Fig. 5b Right) or an antisense AGR1 RNA probe on agr1–7 seedlings (data not shown) indicated that the signal found in Fig. 5b (Left) was AGR1-specific.
DISCUSSION
All agr1 alleles conferred altered root gravitropism, increased root-growth sensitivity to high concentrations of 1-NAA, and decreased root-growth sensitivity to ACC, a precursor of ethylene biosynthesis. Most alleles also conferred a decreased root-growth sensitivity to TIBA, a specific inhibitor of polar auxin transport (Fig. 1). The only exception was agr1–4, which exhibited a wild-type response to TIBA (data not shown). This phenotypic difference between agr1–4 and other agr1 alleles may reflect functional differences among agr1 alleles or a difference in phenotypic expression of agr1 mutations in different ecotype backgrounds.
These phenotypes suggest that AGR1 plays an important role in the efflux phase of polar auxin transport in roots. Indeed, TIBA is a specific inhibitor of the auxin efflux carrier in plants (2). Furthermore, ethylene is known to promote a reduction in both the abundance of a membrane-associated regulatory subunit of the auxin efflux carrier and the auxin transport activity (2). At the same time, a defect in the auxin efflux carrier is expected to result in an increased sensitivity to auxin, consistent with our observation (Fig. 1a). Indeed, mutant root cells are not expected to export as efficiently as wild-type cells an excessive amount of inhibitory auxin taken up from the medium. Interestingly, the latter phenotype is opposite from the phenotype exhibited by aux1 roots, which carry a defective auxin influx carrier (17).
An involvement of AGR1 in the efflux phase of polar auxin transport in roots is supported by the fact that preloaded 3H-IAA is retained more efficiently by agr1 root tips than by wild-type root tips. However, we have not formally excluded possible changes in auxin metabolism or possible differential effects of wounding on 3H-IAA retention by wild-type and mutant root tips (Fig. 1d). It is interesting to note that agr1–5 mutant root tips did not retain more 3H-IAA than wild-type root tips immediately after loading. This surprising result may be explained by the fact that AGR1 is expressed only in limited and specific regions of the root tips. Alternatively, it is possible that more complex processes feedback-regulate the intake, or modify the transport, of exogenously added auxin in different regions of the root tip.
AGR1 involvement in polar auxin transport is demonstrated by the fact that AGR1 expression in S. cerevisiae results in increased resistance to F-IAA, a toxic IAA-derived compound, and an enhanced cellular efflux of preloaded 3H-IAA (Fig. 4). That model also is in complete agreement with the fact that AGR1 is expressed in the distal and central elongation zones of Arabidopsis root tips (Fig. 5) and encodes a putative transmembrane protein with homologies to bacterial transporters (Fig. 3).
In addition to being specifically expressed in the root distal and central elongation zones of light- and dark-grown seedlings, AGR1 is also expressed in the hypocotyl and cotyledon tissues of young etiolated seedlings (Fig. 5). The latter observation is not surprising since agr1 mutations cause a transient randomization of hypocotyl tip angles in etiolated seedlings early after germination (E.R. and Kenneth L. Poff, unpublished results).
While our work was being reviewed, an independent paper (25) reported on the physiological characterization and molecular cloning of EIR1/AGR1. Similarly, these authors observed an increased root-growth resistance to ACC and TIBA of eir1 mutants compared with wild type. However, they reported no differences in root-growth sensitivities to 1-NAA. The inconsistency may result from differences in experimental procedures and/or in alleles tested. Luschnig et al. (25) reported that EIR1-expressing yeast cells were more resistant to F-IAA and that eir1 mutations affected the gravitropically regulated PIG4∷GUS reporter gene expression. They concluded that AGR1 encodes a protein involved in auxin transport in roots. Our in situ localization of AGR1 transcripts in the distal and central elongation zones of the root tip (Fig. 5) provides further evidence to support this model. In addition, our direct analysis of 3H-IAA retention in root tips (Fig. 1) and, more importantly, in AGR1-expressing yeast cells (Fig. 4) demonstrates that AGR1 is indeed an important component of the auxin efflux carrier in root tips.
Localization of the polar auxin transport machinery in the root distal and central elongation zones is consistent with the fountain model of gravitropism, which hypothesizes that an auxin gradient established by lateral auxin transport across a gravistimulated root cap is transported basipetally to the distal and central elongation zones, where it differentially regulates cellular elongation on opposite flanks (reviewed in ref. 1). It is possible that AGR1 facilitates the transport of the gradient from the cap to more basal regions of the root. Mutations in AGR1 would simply affect the maximal flow of auxin from the cap to the distal and central elongation zones. Hence, the auxin flow required on the bottom side of a gravistimulated root would be blocked in agr1 mutants. This model is in complete agreement with the fact that agr1 mutations promote an increased root-growth sensitivity to only high concentrations of 1-NAA (Fig. 1a). It also predicts that another mechanism, possibly mediated by other members of the AGR1 gene family, may be responsible for the establishment of an auxin gradient across the root cap in response to gravistimulation.
Alternatively, it is also possible that AGR1 is more directly involved in creating the functional gradient of auxin across a gravistimulated root tip. AGR1 activity could be directly regulated by gravity signals, promoting a differential flow of auxin on opposite flanks of a gravistimulated root (26). It is worth noting that fast changes in proton fluxes have been detected in proximity of the distal elongation zone in response to gravistimulation (27–29), indicating that this region can quickly respond to changes in root-tip orientation within the gravity field.
Mutations in the AGR1 gene result in a specific defect in root gravitropism, without dramatic pleiotropic phenotypes. Yet, polar auxin transport has been implicated in the regulation of many aspects of plant development, such as vascular tissue differentiation (30), root hair formation, lateral root initiation (31), pattern formation during embryogenesis (32), and development of inflorescence axes, flowers, and leaves (7). This apparent discrepancy may reflect some genetic redundancies in the control of polar auxin transport in plants. Genetic redundancy may also explain why AGR1 mutations do not result in complete root-growth resistance to TIBA (Fig. 1b). The identification of at least two A. thaliana AGR1 paralogs and of several A. thaliana expressed sequence tags potentially encoding AGR1-like proteins (R.C. and P.H.M., unpublished data) supports this hypothesis. A mutational analysis of these paralogs likely will provide further evidence to support a regulatory role of auxin efflux in the development of auxin gradients along and across plant organs and to define the role of such gradients in the regulation of plant growth and development.
Acknowledgments
We thank T. Sullivan and M. Bennett for discussions; K. Carroll, W. Alphin, and F. Garlick for assistance; W. Sinclair, A. Trewavas, E. Maher, J. Ecker, and K. Okada for providing agr1 alleles; N. Fedoroff, P. Green, and C. Kung for providing the Ds389-14 and Ds392-13 lines, the eIF4A probe, and strain W3031A, respectively; and the Arabidopsis Biological Resource Center for providing DNA stocks and libraries. This work was supported in part by grants from the National Institutes of Health (R01-GM 48053) and the National Aeronautics and Space Administration (NASA) (NAG5-4596), and by a Packard Fellowship (to P.H.M.). R.C. was supported by NASA Grant NAG5-4596; P.H. was supported by Belgian Fonds National de la Recherche Scientifique and European Molecular Biology Organization fellowships; J.S. was supported by National Science Foundation (NSF)/Department of Energy (DOE)/U.S. Department of Agriculture (USDA) (92-2033) and National Institutes of Health (5-T32-6M07133) grants; and E.R. was supported by a NSF/DOE/USDA grant (92-2033). This is paper no. 3528 of the Laboratory of Genetics.
ABBREVIATIONS
- ACC
1-aminocyclopropane-1-carboxylic acid
- F-IAA
5-fluoro-1-indole-3-acetic acid
- IAA
indole-3-acetic acid
- 1-NAA
1-naphthaleneacetic acid
- TIBA
2,3,5-triiodobenzoic acid
- WS
Wassilewskija
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF087459).
References
- 1.Masson P H. BioEssays. 1995;17:119–127. doi: 10.1002/bies.950170207. [DOI] [PubMed] [Google Scholar]
- 2.Lomax T L, Muday G K, Rubery P H. In: Plant Hormones-Physiology, Biochemistry, and Molecular Biology. Davies P J, editor. Dordrecht, The Netherlands: Kluwer; 1995. pp. 509–530. [Google Scholar]
- 3.Bell C J, Maher E P. Mol Gen Genet. 1990;220:289–293. [Google Scholar]
- 4.Okada K, Shimura Y. Science. 1990;250:274–276. doi: 10.1126/science.250.4978.274. [DOI] [PubMed] [Google Scholar]
- 5.Roman G, Lubarsky B, Kieber J J, Rothenburg M, Ecker J R. Genetics. 1995;139:1393–1409. doi: 10.1093/genetics/139.3.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rutherford R, Masson P. Plant Physiol. 1996;111:987–998. doi: 10.1104/pp.111.4.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Okada K, Ueda J, Komaki M K, Bell C J, Shimura Y. Plant Cell. 1991;3:677–684. doi: 10.1105/tpc.3.7.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garbers C, DeLong A, Deruere J, Bernasconi P, Soll D. EMBO J. 1996;15:2115–2124. [PMC free article] [PubMed] [Google Scholar]
- 9.Michelmore R W, Paran I, Kesseli R V. Proc Natl Acad Sci USA. 1991;88:9828–9832. doi: 10.1073/pnas.88.21.9828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bell C J, Ecker J R. Genomics. 1994;19:137–144. doi: 10.1006/geno.1994.1023. [DOI] [PubMed] [Google Scholar]
- 11.Konieczny A, Ausubel F M. Plant J. 1993;4:403–410. doi: 10.1046/j.1365-313x.1993.04020403.x. [DOI] [PubMed] [Google Scholar]
- 12.Smith D, Yanai Y, Liu Y G, Ishiguro S, Okada K, Shibata D, Whittier R F, Fedoroff N V. Plant J. 1996;10:721–732. doi: 10.1046/j.1365-313x.1996.10040721.x. [DOI] [PubMed] [Google Scholar]
- 13.Reerie W G, Feldmann K A, Marks M D. Genes Dev. 1992;8:1388–1399. doi: 10.1101/gad.8.12.1388. [DOI] [PubMed] [Google Scholar]
- 14.Schulz B, Bennett M J, Dilkes B P, Feldmann K A. Plant Molecular Biology K3. Dordrecht, the Netherlands: Kluwer; 1994. [Google Scholar]
- 15.Schmidt R, Love K, West J, Lenehan Z, Dean C. Plant J. 1997;11:563–572. doi: 10.1046/j.1365-313x.1997.11030563.x. [DOI] [PubMed] [Google Scholar]
- 16.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1994. [Google Scholar]
- 17.Bennett M J, Marchant A, Green H G, May S T, Ward S P, Millner P A, Walker A R, Schulz B, Feldmann K A. Science. 1996;273:948–950. doi: 10.1126/science.273.5277.948. [DOI] [PubMed] [Google Scholar]
- 18.Newman T, deBruijn F J, Green P, Keegstra K, Kende H, McIntosh L, Ohlrogge J, Raikhel N, Somerville S, Thomashow M, et al. Plant Physiol. 1994;106:1241–1255. doi: 10.1104/pp.106.4.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mumberg D, Mueller R, Funk M. Nucleic Acids Res. 1994;22:5767–5768. doi: 10.1093/nar/22.25.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang C, Bowman J L, DeJohn A W, Lander E S, Meyerowitz E M. Proc Natl Acad Sci USA. 1988;85:6856–6860. doi: 10.1073/pnas.85.18.6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hebsgaard S M, Korning P G, Tolstrup N, Engelbrecht J, Rouze P, Brunak S. Nucleic Acids Res. 1996;24:3439–3452. doi: 10.1093/nar/24.17.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hofmann K, Stoffel W. Biol Chem Hoppe-Seyler. 1993;347:166. doi: 10.1515/bchm3.1992.373.1.187. [DOI] [PubMed] [Google Scholar]
- 23.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang J, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoenke S, Schmid M, Dimroth P. Eur J Biochem. 1997;246:530–538. doi: 10.1111/j.1432-1033.1997.00530.x. [DOI] [PubMed] [Google Scholar]
- 25.Luschnig C, Gaxiola R A, Grisafi P, Fink G R. Genes Dev. 1998;12:2175–2187. doi: 10.1101/gad.12.14.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sachs T. Adv Bot Res. 1981;9:152–265. [Google Scholar]
- 27.Mulkey T J, Evans M L. Science. 1981;212:70–71. doi: 10.1126/science.212.4490.70. [DOI] [PubMed] [Google Scholar]
- 28.Bjokmann T, Leopold A C. Plant Physiol. 1987;83:841–846. doi: 10.1104/pp.84.3.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zieschang H E, Köhler K, Sievers A. Planta. 1993;190:546–554. [Google Scholar]
- 30.Hardtke C S, Berleth T. EMBO J. 1998;17:1405–1411. doi: 10.1093/emboj/17.5.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Celenza J L J, Grisafi P L, Fink G R. Genes Dev. 1995;9:2131–2142. doi: 10.1101/gad.9.17.2131. [DOI] [PubMed] [Google Scholar]
- 32.Hadfi K, Speth V, Neuhaus G. Development (Cambridge, U.K.) 1998;125:879–887. doi: 10.1242/dev.125.5.879. [DOI] [PubMed] [Google Scholar]