Abstract
Many homeobox genes control essential developmental processes in animals and plants. In this report, we describe the first cDNA corresponding to a homeobox gene isolated from a gymnosperm, the HBK1 gene from the conifer Picea abies (L.) Karst (Norway spruce). The sequence shows distinct similarities specifically to the KNOX (knotted-like homeobox) class of homeobox genes known from different angiosperm plants. The deduced amino acid sequence of HBK1 is strikingly similar within the homeodomain (84% identical) to the maize gene Knotted1 (Kn1), which acts to regulate cell differentiation in the shoot meristem. This similarity suggested that the phylogenetic association of HBK1 with the KNOX genes might be coupled to a conservation of gene function. In support of this suggestion, we have found HBK1 to be expressed in the apical meristem in the central population of nondifferentiated stem cells, but not in organ primordia developing at the flanks of the meristem. This pattern of expression is similar to that of Kn1 in the maize meristem. We show further that HBK1, when expressed ectopically in transgenic Arabidopsis plants, causes aberrations in leaf development that are similar to the effects of ectopic expression of angiosperm KNOX genes on Arabidopsis development. Taken together, these data suggest that HBK1 has a role, similar to the KNOX genes in angiosperms, in the control of cellular differentiation in the apical meristem of spruce. The data also indicate that KNOX-gene regulation of vegetative development is an ancient feature of seed plants that was present in the last common ancestor of conifers and angiosperms.
The presence of homeobox genes in plants, first documented in the early 1990s (1–5), has provoked considerable interest among developmental biologists, because homeobox genes in animals encode transcription factors that are essential determinants of development. Even though the plant homeobox genes identified to date do not appear to be closely related to their animal counterparts, but instead form several different plant-specific classes within the homeobox superfamily of genes, there is evidence that some of the plant homeobox genes have essential roles also in the control of developmental processes. This is the case, for example, for the maize gene Knotted1 (Kn1), first identified by a gain-of-function mutation that causes alterations in the timing and position of cell differentiation in the developing leaf (2, 6). In wild-type plants, the KN1 protein was detected in the central population of nondifferentiated stem cells in vegetative and floral meristems, but not in the differentiating primordia that develop on the flanks of the meristem or in leaves. This protein distribution suggested a role for Kn1 in the control of cell differentiation in the meristem (7). Recently, Kn1 loss-of-function mutants have been isolated that reveal a role for Kn1 in meristem maintenance (8). In Arabidopsis, the closely related gene Shoot Meristemless (STM) has an essential role in the formation and/or maintenance of the meristem (9). STM is expressed in the cells that give rise to the shoot meristem during early embryogenesis and later in development it is expressed in the vegetative meristem, in a pattern similar to that of Kn1 expression.
Kn1 and STM belong to the apparently plant-specific knotted-like homeobox (KNOX) class of homeobox genes, which in turn is part of the TALE (three amino acid loop extension) superclass of atypical homeobox genes (10, 11). TALE includes genes from animals and fungi as well as plants; e.g., the human PRL and the Saccharomyces pombe MATi both have roles in the specification of cell fate in their respective systems (12–14). Two subclasses of KNOX genes are distinguished by sequence criteria in angiosperm plants. Class I genes differ also from the less well-characterized class II genes in expression and by functional criteria. The class I genes studied are expressed in meristematic tissue, mainly in apical meristems. Class II genes, on the other hand, are expressed in most organs and tissues with high abundance in a tissue or organ specific for each gene and are not expressed in meristematic tissues (15–24). Further, all class I genes analyzed give rise to similar and distinct phenotypic effects when ectopically expressed in transgenic plants, i.e., perturbations in the development of leaves leading to morphological defects such as asymmetry over the midrib, lobing, and wrinkling (17, 22, 25). No developmental defects have been recorded in plants expressing a class II gene ectopically (21).
In this report, we address the question of the phylogenetic origin of this apparently plant-specific class of developmental regulators. We have cloned the cDNA corresponding to a homeobox gene, HBK1 (homeobox of KNOX class), from the conifer Picea abies (Norway spruce), which, by sequence, expression, and functional criteria is a member of the KNOX class I. These results suggest a role for HBK1 in the control of meristem function in spruce. Conifers are thought to have split from the lineage leading to the angiosperms more than 300 million years ago (26). Our data indicate that the origin of the KNOX class and the distinction between the KNOX class I and class II genes predate this event and demonstrate that the KNOX genes were active in the control of meristem function in the last common ancestor of angiosperms and conifers.
MATERIALS AND METHODS
Screening of cDNA Libraries.
Clones (200,000) of a cDNA library constructed from spruce embryo RNA (27) were screened with a BamHI-XhoI fragment of the Knotted1 cDNA clone pBSKN1 containing the homeobox but excluding the 5′ end of the clone encoding the histidine-rich region. A partial cDNA was isolated and used subsequently to isolate a full-length cDNA by the screening of 200,000 clones of a spruce female strobilus cDNA library (28). Labeling, hybridization, and washing were performed as described (27). Positive clones were subcloned into the EcoRI site of pBS SK + (Stratagene). HBK1 has an internal EcoRI site at position 1583 and the two fragments were subcloned separately (clones phbk1–5′ and phbk1–3′).
Sequence Analysis.
By using deletions and synthetic oligonucleotide primers, both strands of the clones phbk1–5′ and phbk1–3′ were sequenced by the chain termination method by using Sequenase DNA Polymerase (United States Biochemical) and double-stranded plasmid DNA as a template.
Transgenic Plants.
An HBK1 clone was reconstituted from phbk1–5′ and phbk1–3′ and cloned subsequently downstream of the cauliflower mosaic virus 35S promoter in pBINHyg-TX (29). This construct was introduced into the Agrobacterium tumefaciens strain C58∷pGV2260 by use of triparental mating. The resulting Agrobacterium strain was used to transform Arabidopsis thaliana ecotype Wassilewskija by an infiltration protocol (30) with minor modifications. Transformants were selected on 50 μg ml−1 hygromycin. The phenotype of T2 plants grown under long-day conditions at 20–22°C was studied.
In Situ Hybridization.
The in situ hybridizations were performed essentially as described (31). Branches with resting vegetative buds were collected from 20-year-old spruce trees and were kept under greenhouse conditions for 6 days to resume growth. Buds were dissected and fixed subsequently and were embedded and sectioned as described (32). A 3′ noncoding subclone of HBK1 was used as template in in vitro transcription incorporating 33P-labeled UTP (Amersham). The essential changes in the protocol are the introduction of a prehybridization step and the addition of DTT at 0.1 M and 0.012 M in the hybridization and wash solutions, respectively. Slides were dehydrated and coated with Kodak NBT2 emulsion and were exposed for 4 weeks. Sections were stained in 20% Gill’s hematoxylin (BDH).
RESULTS AND DISCUSSION
A Homeobox Gene of the KNOX Class I Type from Norway Spruce.
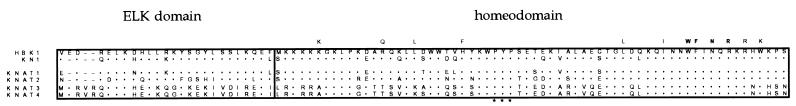
A spruce cDNA library constructed from zygotic embryos, induced to form adventitious meristems by a treatment with the hormone cytokinin (27), was screened with a fragment of the maize Kn1 cDNA. Plaques (200,000) were screened and four clones representing two different sequences were isolated. A full-length cDNA clone corresponding to one of these sequences was isolated subsequently from a second cDNA library, constructed from young female strobilus of spruce (28) by use of one of the spruce clones. The 1,988-nt-long cDNA contains one ORF with two potential start codons at positions 453 and 477, respectively. The spruce protein sequence deduced from these ORFs contains 442 or 434 amino acids, giving the putative protein a molecular weight of 49,484 or 48,588, respectively. A comparison of this sequence with that of a range of homeodomain proteins from different organisms revealed the presence in the spruce protein of a homeodomain with distinct similarities specifically to the KN1 homeodomain. As shown in Fig. 1, the spruce protein contains the four amino acid residues invariant among all homeodomains studied (33, 34). These amino acids make DNA contact and specify the DNA-binding properties of the homeodomain (35–37). Four of the eight amino acids that are highly conserved in most homeodomain classes are present in the spruce protein. In the remaining four highly conserved positions, the spruce protein differs from the consensus homeodomain sequence but is identical to KN1. The presence of a conserved proline-tyrosin-proline insertion in a position between helix 1 and helix 2 of the homeodomain distinguishes the spruce protein, like KN1, as a member of the TALE class of atypical homeodomain proteins (10, 11). The spruce and maize sequences show an overall identity of 46% and, across an 88-amino-acid-long region containing the homeodomain (Fig. 1), the degree of sequence identity is 84%. For comparison, the corresponding identity scores between KN1 and its related sequences from other angiosperms are 84% and 89% for the Arabidopsis STM and KNAT1 proteins, respectively, 83% for soybean SHB1, and 74% for maize KNOX10 (15, 17, 18, 38). In addition to the homeodomain, this conserved region also contains a second sequence element identified as conserved between KN1 and related angiosperm proteins, but absent from TALE proteins of nonplant origin, i.e., the ELK-domain, N-terminal to the homeodomain (2). This protein motif is thought to mediate protein–protein interactions (2, 11). The presence of an ELK domain in the conifer protein indicates that the ELK-homeodomain proteins constitute a distinct class of homeodomain proteins present in all seed plants. The sequence similarity between the proteins extends toward their N termini into the KNOX domain, common to all known TALE proteins (11). Thus, sequence data demonstrate that the spruce gene is a member of the KNOX class of homeobox genes, and hence we name it HBK1 (homoebox of KNOX class) to indicate this fact.
Figure 1.
Alignment of the amino acid sequences deduced from spruce HBK1, maize Kn1, and Arabidopsis KNAT1–4 genes. Kn1, KNAT1, and KNAT2 represent KNOX class I genes and KNAT3 and KNAT4 represent KNOX class II genes. ELK and the homeodomain regions are boxed. The three amino acids inserted in all TALE homeodomains are indicated by asterisks. Amino acids conserved among most homeodomains are indicated above the sequence, and invariant amino acids are shown in bold.
HBK1 is more similar to KNOX proteins belonging to class I, as represented by KN1, KNAT1, and KNAT2, than to the class II proteins, represented by KNAT3 and KNAT4, over the ELK-homeodomain region (Fig. 1). HBK1 lacks a 2 amino acid insertion in the ELK domain typical for class II proteins and shows a distinctly higher degree of sequence identity to the class I proteins within both the ELK domain and the homeodomain. This relatedness of HBK1 with the KNOX class I genes is consistent with phylogenetic analyses carried out (unpublished results; personal communication, N. Sinha and G. Bharathan). The presence of HBK1 in spruce indicates that the ELK- homeodomain encoding genes are ancient and were present in the last common ancestor of conifers and angiosperms. Provided that the rate of sequence divergence has not been dramatically different between the class I and II genes, our data imply also that the gene duplication giving rise to the separate gene lineages must have occurred before the split between the conifer and angiosperm lineages.
The high degree of sequence conservation, in spite of the long evolutionary distance between spruce and angiosperms, implies that the selection pressure to maintain the sequence, imposed by functional constraints, must be high. Consequently, the molecular function of these proteins may have been conserved between conifers and angiosperms.
HBK1 Is Expressed in the Vegetative Spruce Meristem.
KNOX class I genes in maize and Arabidopsis show distinct transcript patterns in the vegetative and inflorescence meristems, as well as in floral meristems (16–18). We therefore examined spruce meristems for the presence of HBK1 transcript by use of in situ hybridization. Sections of vegetative buds were hybridized to a 3′ noncoding HBK1-specific probe. In both longitudinal and transverse sections, HBK1 transcript was detected at low levels in the central zone of the meristem (Fig. 2A and C), whereas no expression was detectable in the developing needle primordia surrounding the meristem (Fig. 2A). Down-regulation of HBK1 in needle primordia is apparent at a very early stage of development (arrows in Fig. 2A). This pattern is similar to the expression of the angiosperm class I genes Kn1, STM, and Knat1 in vegetative meristems. The isolation of the spruce cDNA from a female strobilus library indicates that HBK1 is active also in reproductive meristems, in agreement with the expression of the angiosperm genes in inflorescence and floral meristems. Our expression data is in support of HBK1 being a KNOX class I gene and indicates that HBK1, like Kn1 and STM in angiosperms, has its primary role in the shoot apical meristem.
Figure 2.
Distribution of HBK1 transcript in vegetative buds of Norway spruce. (A) Longitudinal section through the vegetative bud hybridized with an antisense probe. Arrows point to the youngest needle primordia. (B) Longitudinal section through the vegetative bud hybridized with a sense probe. Structures indicated are meristem (M), needle primordia (NP), and pith (P). (C) Transverse section through the vegetative bud hybridized with an antisense probe. Structures indicated are meristem (M) and needle primordia (NP). Pictures are bright-field/dark-field double exposures. For dark-field exposures, epiillumination and a yellow filter were used to visualize the silver grains in yellow. The yellow stain in cells of the pith and older needle primordia originates from light reflected by cell walls and phenolic compounds contained in vacuoles. Note the similarity of this pattern in A and B.
Ectopic Expression of HKB1 in Arabidopsis Results in Altered Leaf Morphology.
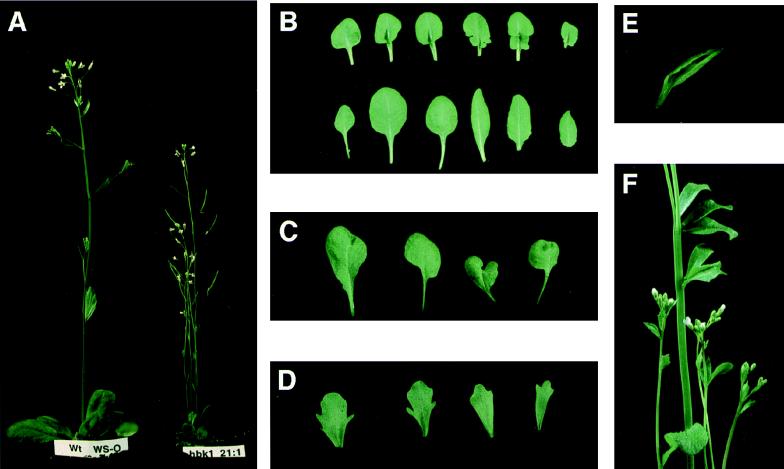
To assess the function of the spruce gene in comparison to that of angiosperm KNOX genes, we constructed transgenic Arabidopsis plants expressing HBK1 ectopically under the control of the Cauliflower Mosaic Virus 35S promoter. The phenotypic properties of the resulting transformant lines were analyzed in the T2 generation. Eleven independent transformant lines shared the same overall phenotypic aberrations, although the degree to which the plants deviated from wild type differed both within and between the lines. The rosette leaves of all lines developed a leaf shape that differed from wild type, which develops symmetrical spatulate leaves with slight serrations in the leaf margins. Transgenic leaves often had an asymmetrical outline over the midrib (Fig. 3B) or, in some cases, were deeply lobed at the base of the leaf (Fig. 3C). In severely affected lines, similar effects were seen also on cauline leaves (Fig. 3D). Differential growth of the leaves often gave rise to wrinkling. A few lines developed twin leaves, i.e., leaves that had two leaf blades sharing a midvein, in some cases with the midvein split toward the tip of the leaf (Fig. 3E). All but four of the lines showed different extents of dwarfing, illustrated in Fig. 3A. Populations from several lines included plants with stem fasciation (Fig. 3F).
Figure 3.
Characteristics of the phenotype of transgenic Arabidopsis plants expressing the spruce HBK1 gene. (A) A wild-type Wassiljewska (Left) and a dwarfed transgenic plant carrying the HBK1 gene (Right). (B) (Upper) Lobed rosette leaves from a transgenic plant carrying the HBK1 gene. (Lower) Rosette leaves from a wild-type Wassiljewska plant. (C) Misshaped rosette leaves from a transgenic plant carrying the HBK1 gene. (D) Lobed cauline leaves from a transgenic plant carrying the HBK1 gene. (E) A “twin” rosette leaf from a transgenic plant carrying the HBK1 gene. (F) An inflorescence of a plant carrying the HBK1 gene showing stem fasciation.
The phenotypic effects are very similar to the effects in Arabidopsis plants expressing KNOX class I genes from angiosperms. In addition to the defects in leaf development, ectopic expression of the rice gene OSH1 and high-level ectopic expression of Kn1 and Knat1 induce a reduction in size and shape defects in floral organs as well as the formation of ectopic meristems in transgenic Arabidopsis (17, 22, 25). We have not observed any defects in flower development or ectopic meristem formation in Arabidopsis plants expressing HBK1. This may be due to a difference in gene product function, expression levels, or growth conditions. Similarly, ectopic meristems have not been found in plants ectopically expressing Knat2 (25). In contrast to the effect of ectopic expression of class I genes, the class II gene Knat3 does not affect development in Arabidopsis (21).
Several lines of evidence presented in this report support the notion that HBK1 is phylogenetically related to the KNOX class I genes, and that the function of this gene class has been conserved broadly throughout the evolution of seed plants. First, HBK1 is highly similar in sequence to the KNOX class I genes, especially in parts of the gene that encode known functional domains of the KNOX proteins. Second, the expression patterns in the vegetative meristems are very similar between the genes. Third, HBK1 and the class I angiosperm genes cause very similar developmental defects when ectopically expressed in Arabidopsis. These data indicate that the regulation of gene expression in the meristem by homeodomain proteins represents a general mechanism for the control of development that has been conserved in the evolution of seed plants.
These findings are interesting because the understanding of developmental control mechanisms in conifers is very limited, whereas the ecological and economical importance of vegetative growth of this plant group is considerable. The conservation of gene sequence and function is also interesting from an evolutionary perspective, because little information is available from nonangiosperm plants on the molecular networks that control development at the genetic level. Two interesting parallels exist. Firstly, in the MADS-box class of developmental control genes, one subclass of genes, the C class, which functions in stamen and pistil development in angiosperms, is represented by at least one gene in a conifer (28), and the functional properties are conserved at least in part between conifers and angiosperms (39). Secondly, a gene from pine, Needly (40), is homologous to, and can functionally substitute for, the Leafy gene in Arabidopsis, a central component in the genetic control system for the transition from vegetative to reproductive development. These three cases indicate that the central mechanisms by which angiosperms and conifers control the progression through the life cycle are of common origin, and that similarities in molecular detail may be quite extensive. These results have implications for research and biotechnology on conifers. Arabidopsis should be a useful model plant also for work on at least some aspects of conifer biology as well as for evolutionary biology, in that molecular genetic information on developmental control mechanisms may be used to trace the genetic basis for morphological diversification among a wide range of plants. In this latter respect, the KNOX class I genes may be of special interest. For example, an extension of research on KNOX genes into plants originating from the deeper branches of the phylogenetic tree, the clubmosses, ferns, and mosses, might help elucidate the origin of the leaf.
Acknowledgments
We thank Dr. Sarah Hake for kindly providing the Kn1 cDNA and Mats Nilsson for his work with cloning and sequencing. We also thank two anonymous reviewers for critical comments on the manuscript. This work was supported by grants from the Swedish Natural Science Research Council and the Swedish Foundation for Strategic Research.
ABBREVIATIONS
- TALE
a three amino acid loop extension
- KNOX
knotted-like homeobox
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF063248).
References
- 1.Ruberti I, Sessa G, Lucchetti S, Morelli G. EMBO J. 1991;10:1787–1791. doi: 10.1002/j.1460-2075.1991.tb07703.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vollbrecht E, Veit B, Sinha N, Hake S. Nature (London) 1991;350:241–243. doi: 10.1038/350241a0. [DOI] [PubMed] [Google Scholar]
- 3.Bellman R, Werr W. EMBO J. 1992;11:367–374. doi: 10.1002/j.1460-2075.1992.tb05415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mattsson J, Söderman E, Svenson M, Borkird C, Engström P. Plant Mol Biol. 1992;18:1019–1022. doi: 10.1007/BF00019223. [DOI] [PubMed] [Google Scholar]
- 5.Schena M, Davis R W. Proc Natl Acad Sci USA. 1992;89:3894–3898. doi: 10.1073/pnas.89.9.3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hake S, Vollbrecht E, Freeling M. EMBO J. 1989;8:15–22. doi: 10.1002/j.1460-2075.1989.tb03343.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith L G, Greene B, Veit B, Hake S. Development (Cambridge, UK) 1992;116:21–30. doi: 10.1242/dev.116.1.21. [DOI] [PubMed] [Google Scholar]
- 8.Kerstetter R A, Laudencia-Chingcuanco D, Smith L G, Hake S. Development (Cambridge, UK) 1997;124:3045–3054. doi: 10.1242/dev.124.16.3045. [DOI] [PubMed] [Google Scholar]
- 9.Barton M K, Poethig R S. Development (Cambridge, UK) 1993;119:823–831. [Google Scholar]
- 10.Bertolino E, Reimund B, Wildt-Perinic D, Clerc R G. J Biol Chem. 1995;270:31178–31188. doi: 10.1074/jbc.270.52.31178. [DOI] [PubMed] [Google Scholar]
- 11.Bürglin T R. Nucl Acids Res. 1997;25:4173–4180. doi: 10.1093/nar/25.21.4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kamps M P, Murre C, Sun X-H, Baltimore D. Cell. 1990;60:547–555. doi: 10.1016/0092-8674(90)90658-2. [DOI] [PubMed] [Google Scholar]
- 13.Nourse J, Mellentin J D, Galili N, Wilkinson J, Stanbridge E, Smith S D, Cleary M L. Cell. 1990;61:1225–1236. doi: 10.1016/0092-8674(90)90657-z. [DOI] [PubMed] [Google Scholar]
- 14.Kelly M, Burke J, Smith M, Klar A, Beach D. EMBO J. 1988;7:1537–1547. doi: 10.1002/j.1460-2075.1988.tb02973.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kersetter R, Vollbrecht E, Lowe B, Veit B, Yamaguchi J, Hake S. Plant Cell. 1994;6:1877–1887. doi: 10.1105/tpc.6.12.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jackson D, Veit B, Hake S. Development (Cambridge, UK) 1994;120:405–413. [Google Scholar]
- 17.Lincoln C, Long J, Yamaguchi J, Serikawa K, Hake S. Plant Cell. 1994;6:1859–1876. doi: 10.1105/tpc.6.12.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Long J A, Moan E I, Medford J I, Barton M K. Nature (London) 1996;379:66–69. doi: 10.1038/379066a0. [DOI] [PubMed] [Google Scholar]
- 19.Dockx J, Quaedvlieg N, Keultjes G, Kock P, Weisbeek P, Smeekens S. Plant Mol Biol. 1995;28:723–737. doi: 10.1007/BF00021196. [DOI] [PubMed] [Google Scholar]
- 20.Serikawa K A A, Martinez-Laborda A, Zambryzki P. Plant Mol Biol. 1996;32:673–683. doi: 10.1007/BF00020208. [DOI] [PubMed] [Google Scholar]
- 21.Serikawa K A, Martinz-Laborda A, Kim H, Zambryski P C. Plant J. 1997;11:853–861. doi: 10.1046/j.1365-313x.1997.11040853.x. [DOI] [PubMed] [Google Scholar]
- 22.Matsuoka M, Ichikawa H, Saito A, Tada Y, Fujimura T, Kano-Murakami Y. Plant Cell. 1993;5:1039–1048. doi: 10.1105/tpc.5.9.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Janssen B-J, Williams A, Chen J-J, Mathern J, Hake S, Sinha N. Plant Mol Biol. 1998;36:417–425. doi: 10.1023/a:1005925508579. [DOI] [PubMed] [Google Scholar]
- 24.Watillon B, Kettmann R, Boxus P, Burny A. Plant Mol Biol. 1997;33:757–763. doi: 10.1023/a:1005708700636. [DOI] [PubMed] [Google Scholar]
- 25.Chuck G, Lincoln C, Hake S. Plant Cell. 1996;8:1277–1289. doi: 10.1105/tpc.8.8.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stewart W N, Rothwell G W. Paleobotany and the Evolution of Plants. Cambridge, U.K.: Cambridge Univ. Press; 1993. [Google Scholar]
- 27.Sundås A, Tandre K, Holmstedt E, Engström P. Plant Mol Biol. 1992;18:713–724. doi: 10.1007/BF00020013. [DOI] [PubMed] [Google Scholar]
- 28.Tandre K, Albert V A, Sundås A, Engström P. Plant Mol Biol. 1995;27:69–78. doi: 10.1007/BF00019179. [DOI] [PubMed] [Google Scholar]
- 29.Becker D. Nucleic Acids Res. 1991;18:203. doi: 10.1093/nar/18.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bechtold N, Ellis J, Pelletier G. C R Acad Sci. 1993;316:1194–1199. [Google Scholar]
- 31.Jackson D P. In: Molecular Plant Pathology: A Practical Approach. Bowles D J, Gurr S J, McPhereson M, editors. Oxford: Oxford Univ. Press; 1991. pp. 163–174. [Google Scholar]
- 32.Stabel P, Sundås A, Engström P. Planta. 1991;183:520–527. doi: 10.1007/BF00194273. [DOI] [PubMed] [Google Scholar]
- 33.Scott M P, Tamkun J W, Hartzell G W., III Biochim Biophys Acta. 1989;989:25–48. doi: 10.1016/0304-419x(89)90033-4. [DOI] [PubMed] [Google Scholar]
- 34.Bürglin T A. In: Guidebook to Homeobox Genes. Dudoule D, editor. Oxford: Oxford Univ. Press; 1994. [Google Scholar]
- 35.Kissinger C R, Beishan L, Martin-Blanco L, Kornberg T B, Pabo C O. Cell. 1990;63:579–590. doi: 10.1016/0092-8674(90)90453-l. [DOI] [PubMed] [Google Scholar]
- 36.Otting G, Qian Y Q, Billeter M, Müller M, Affolter M, Gehring W, Wüthrich K. EMBO J. 1990;9:3085–3092. doi: 10.1002/j.1460-2075.1990.tb07505.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wolberger C, Vershon A K, Liu B, Johnson A D, Pabo C O. Cell. 1991;67:517–528. doi: 10.1016/0092-8674(91)90526-5. [DOI] [PubMed] [Google Scholar]
- 38.Ma H, McMullen M D, Finer J J. Plant Mol Biol. 1994;24:465–473. doi: 10.1007/BF00024114. [DOI] [PubMed] [Google Scholar]
- 39.Tandre K, Svenson M, Svensson M E, Engström P. Plant J. 1998;15:615–623. doi: 10.1046/j.1365-313x.1998.00236.x. [DOI] [PubMed] [Google Scholar]
- 40.Mouradov A, Glassick T, Hamdorf B, Murphy L, Fowler B, Marla S, Teasdale D. Proc Natl Acad Sci USA. 1998;95:6537–6542. doi: 10.1073/pnas.95.11.6537. [DOI] [PMC free article] [PubMed] [Google Scholar]