Abstract
Background
Genes involved in male reproduction are often the targets of natural and/or sexual selection. SCML1 is a recently identified X-linked gene with preferential expression in testis. To test whether SCML1 is the target of selection in primates, we sequenced and compared the coding region of SCML1 in major primate lineages, and we observed the signature of positive selection in primates.
Results
We analyzed the molecular evolutionary pattern of SCML1 in diverse primate species, and we observed a strong signature of adaptive evolution which is caused by Darwinian positive selection. When compared with the paralogous genes (SCML2 and SCMH1) of the same family, SCML1 evolved rapidly in primates, which is consistent with the proposed adaptive evolution, suggesting functional modification after gene duplication. Gene expression analysis in rhesus macaques shows that during male sexual maturation, there is a significant expression change in testis, implying that SCML1 likely plays a role in testis development and spermatogenesis. The immunohistochemical data indicates that SCML1 is preferentially expressed in germ stem cells of testis, therefore likely involved in spermatogenesis.
Conclusion
The adaptive evolution of SCML1 in primates provides a new case in understanding the evolutionary process of genes involved in primate male reproduction.
Background
Proteins involved in sexual reproduction often evolve rapidly due to positive selection [1-4]. Although the selective forces are unclear, a variety of hypotheses have been proposed including mate choice, intra-sexual competition and sexual conflict, which are different forms of sexual selection. The rapid evolution of these proteins may contribute to several important biological aspects such as reproduction and speciation. It has long been recognized that gene duplication is a major source of genomic novelties. Therefore, the newly duplicated genes involved in reproduction are likely the targets of natural and/or sexual selection.
Using exon trapping, van de Vosse et al [5] identified a novel gene in human located on Xp22, named as SCM-like-1 (SCML1), which is similar with the Scm gene in Drosophila. In the human genome, SCML1 spans 18 kb and contains 8 exons. Northern blot analysis detected a major SCML1 transcript of approximately 3-kb in all human adult and fetal tissues tested [5].
SCML1 gene is a polycomb group (PcG) gene. Most of the PcG genes are expressed throughout embryonic, larval and pupal development, and are required continuously to maintain restricted homeotic expression in Drosophila. [6-10]. Most mammalian PcG genes have Drosophila homologs [11,12]. Compared to Drosophila, the mammalian PcG genes have acquired novel functions during evolution because PcG knockout mice exhibit numerous phenotypes including hematopoietic defects, neural crest defects, cardiac anomalies, and sex reversal [12,13]. SCML1 is likely a recently duplicated gene during mammalian evolution due to the absence of orthologs in Drosophila, zebrafish and chicken.
In the SCM family, there are other two genes, SCML2 and SCMH1, which have orthologs in all vertebrate species and are located on chromosome Xp22 [14] and chromosome 1p34 [15] respectively. SCMH1 is a core component of polycomb repressive complex 1 (PRC1) [16-18] which is involved in the maintenance of repression and can block chromatin remodeling[17], and it plays an important role in regulation of homeotic genes in embryogenesis[19]. SCML2 is also involved in PRC1's regulation[20]. A recent study showed that SCML2 is over-expressed in acute myeloid leukaemia, suggesting its role in differentiation and cell cycle regulation[21]. As SCML2 and SCMH1 are the ancient copies in the SCM family, they would serve as the ideal reference genes when dissecting the molecular evolution of SCML1 in primates.
Through a genome-wide comparison, we have identified 34 candidate genes including SCML1 that showed rapid nonsynonymous sequence divergence between human and chimpanzee [22], therefore an implication of adaptive evolution of these genes during primate evolution. To test whether SCML1 is the target of selection in primates, we sequenced and compared the coding region of SCML1 in major primate lineages, and we observed the signature of positive selection.
Methods
DNA samples
The major lineages of primates were sampled, including three great ape species (chimpanzee-Pan troglodytes, gorilla-Gorilla gorilla and orangutan-Pongo pygmaeus), two lesser ape species (white-browed gibbon-Bunopithecus hoolock and white-cheeked gibbon-Nomascus leucogenys), two Old World monkey species (rhesus macaque-Macaca mulatta and Yunnan snub-nosed monkey-Rhinopithecus bieti) and one New World monkey species (common marmoset-Callithrix jacchus). The common ancestor of the tested primate species can be traced back to about 45 million years ago [23]. All the DNA samples were from collection in Kunming Cell Bank of CAS and Kunming Blood Center in China.
PCR and sequencing
All the samples were sequenced for the full length coding region of SCML1. Primers for all the primates were designed by aligning the published sequences of human (Esembl ID: ENSG00000047634) and macaque (Ensembl ID: ENSMMUG00000012899, Ensemble genome browser [24]). The primer sequences are listed [see Additional file 1].
PCRs were performed with rTaq under conditions recommended by the manufacturer (TAKARA Company). Sequencing was performed in both directions with forward and reverse primers using the BigDye terminator sequencing kit on an ABI 3130 automated sequencer. There are 8 exons in SCML1 gene, and the first exon is non-translational, therefore, not sequenced in this study. Overlapping chromatogram files retrieved from the sequencer were analyzed and edited using the SeqMan program in the Lasergene software package (DNASTAR Inc).
Sequence analysis
The DNA sequences were aligned with the CLUSTALW program implanted in Mega [25,26] and checked manually. There are several in-dels (do not change the reading frame) in the coding region of common marmoset, and those sites were removed in the sequence analysis. The known phylogeny of primate species was used[23,27]. The ancestral sequences were inferred by PAML 3.15 [28]. The synonymous (ds) and nonsynonymous (dN) substitution rates of each branch were calculated with the use of the maximum likelihood method under the free-ratio model [28].
Test of selection
Positive selection can be inferred from a higher proportion of nonsynonymous than synonymous substitutions per site (dN/dS > 1). To detect specific amino acid sites under positive selection, we applied the site models in the codeml program of the PAML package. Using this set of models, we obtained the log likelihood estimates (L) of a tree topology under models that impose alternative assumptions in terms of rate variation (ω = dN/dS) over different codon sites [29,30]. The model M0 was used to evaluate the general sequence substitution pattern of SCML1 in primates assuming a constant ω ratio across codon sites. M0 estimates the overall ω for the data. The M1a model estimates single parameter, p0, with ω0 = 0, and the remaining sites with frequency p1 (p1 = 1-p0) assuming ω1 = 1. We first compared model M0 with M1a to determine which model is more realistic for the data and M1a tuned out to be the better one. Then we compared model M2a (selection) and M1a (nearly neutral) to test if invoking of positive selection in model M2a would better explain the data [31,32]. It was suggested that under certain scenarios, a beta distribution of ω is more realistic, therefore, we also conducted the selection test by comparing model M7 and M8, in which a beta distribution of ωwas assumed. We also conducted a more stringent test by comparing M8 with M8a. The LRTs between nested models were conducted by comparing twice the difference of the log-likelihood values (2ΔL) between two models [32]. If the log likelihood test suggests the presence of sites under positive selection, we then identified these sites by using a Bayesian method to estimate posterior probabilities (P) [33].
Comparative evolutionary analysis among SCML1, SCML2 and SCMH1
Sequences of SCML2 and SCMH1 genes were obtained using BLAST (GenBank and Ensembl) for five primate species including human, chimpanzee, orangutan, rhesus macaque and common marmoset. The sequence IDs are: SCML2, Homo sapiens (ENST00000398048), Macaca mulatta (ENSMMUG00000005084), Pan troglodytess (ENSPTRG00000021710); SCMH1, Homo sapiens (ENST00000326197), Pan troglodytess (ENSPTRG00000000601), Macaca mulatta (ENSMMUG00000017104). With the use of human SCML2 and SCMH1, we searched the genomes of orangutan and common marmoset with Blastn and obtained the coding sequences of these two genes[34].
Protein sequences were aligned with the CLUSTALW program implanted in Mega4 [26] and the ω calculation was conducted using the codeml program of PAML3.15 [32]. The ratios of dN and dS were estimated by using PAML3.15, and the Z test(data not show) was used to evaluate the ratio difference between each branches [26]. Similar neutrality tests described above were used in comparing the evolutionary patterns among the three genes.
RT-PCR analysis RNAs were extracted using the Tri-Reagent kit based on the manufacturer's specifications (Invitrogen Inc.). For gene expression analysis of rhesus macaques during development, a total of 20 testis samples were analyzed including ten 1–2 year old monkeys (sexually immature) and ten 7–8 years old monkeys (sexually matured). The T test was used for statistical evaluation of expression difference.
For tissue expression analysis in rhesus macaques, a total of 12 tissue types (1–2 year old male macaques) were analyzed including brain, cartilage, heart, large intestine, small intestine, liver, lung, muscle, pancreas, spleen, stomach and testis. All the rhesus macaque tissue samples were collected from the Kunming Primate Research Center, Chinese Academy of Sciences.
For real-time quantitative RT-PCR analysis, cDNAs were synthesized with SuperScript™ III (Invitrogen) from 5 μg of total RNA in a total volume of 20 μl with oligo(dT) primer in accordance with the manufacturer's instructions. SYRB Green I-based real-time PCR was carried out using the DNA Engine Opticon® 2 Continuous Fluorescence Detection System (MJ, BioRad). After an initial denature step for 5 min at 94°C, conditions for cycling are 40 cycles of 20 sec at 94°C, 20 sec at 58°C, 20 sec at 72°C. At the end of the PCR cycles, a melting curve was generated to identify specificity of the PCR product. For each run, serial dilutions of rhesus macaque GAPDH (glyceraldehyde-3-phosphate dehydrogenase) plasmids were used as standards for quantitative measurement of the amount of amplified DNA. In addition, for normalization of each sample, mGAPDH primers were used to measure the amount of GAPDH cDNA. All samples were run in triplicates and the data were presented as a ratio of SCML1/GAPDH. The ΔCt values were calculated and then converted into the linear-scale expression levels. Oligonucleotides were obtained from Invitrogen. Negative controls were performed with water as template. The primer sequences are:
GAPDH F primer 5'ACTTCAACAGCGACACCCACTC3'
GAPDH R primer 5'CCCTGTTGCTGTAGCCAAATTC3'
SCML1 F primer 5'CTCCTACCCTGAAAGTTATAGCC3'
SCML1 R primer 5'TCTGAGGGATGCACTGGAC3'
Immunohistochemical analysis
The liquid nitrogen stored tissue was sectioned (10 μm) using a HM550 tissue processor (Microm). The frozen section slides were stored at -80°C in a sealed slide box. Sections were stained using the standard immunohistochemical method. The mouse monoclonal antibodies generated using human SCML1 protein (dilution 1:100, Abnova) and the goat anti-mouse IgG antibody (dilution 1:200, Bethyl) were used following the manufacturer's instruction. The negative control used is the buffer-only samples with no mouse antibodies. Immuno-reactivity was visualized by using 0.025% 3.3'-diaminobenzidine tetrachloride/0.001% H2O2. These slides were washed with phosphate-buffered saline (pH 7.4). The sections were counterstained with hematoxylin for a few seconds.
Results
Sequence substitution pattern of SCML1 in primates
A total of nine primate species were sequenced covering the complete 990–1,002 bp coding region of SCML1. There are 215 sites (21.98%, 215/978, in-dels were not counted) showing sequence substitutions in the nine primate species tested. When translated into protein sequences (329–333 amino acids, Figure 1), there are 129 sites (38.51%) having substitutions, an implication of rapid sequence changes during primate evolution. For example, the protein sequence substitution rates (measured by dN) of SCML1 between human and Old World monkeys are relatively fast (human vs. rhesus monkey, 0.045; human vs. Yunnan snub-nosed monkey, 0.055) among those male reproduction associated genes in primates [35].
Figure 1.
Protein sequence alignment of SCML1 in human and nonhuman primates. The SAM domains are indicated. The SAM domain is 64 amino acids in length. Sterile alpha motif (SAM) domains are known to exhibit diverse protein-protein interaction modes[57] (Hum: human – Homo sapiens, Chi: Pan troglodytes, Gor: Gorilla gorilla, Ora: Pongo pygmaeus, HLG: white-browed gibbon – Bunopithecus hoolock, WCG: white-cheeked gibbon – Nomascus leucogenys, Mac: rhesus monkeys – Macaca mulatta, and SCM: Yunnan snub-nosed monkey – Rhinopithecus bieti, Mar: common marmoset – Callithrix jacchus.) The sites under positive selection are highlighted including 23N, 95S, 153L, 201T, 270L (P > 95%, labeled with *) and 92H, 242G (P > 99%, labeled with ◆). The sequence IDs are Hum: EU370780, Chi: EU370781, Gor: EU370782, Ora: EU370783, WCG: EU370784, HLG: EU370785, Mac: EU370786 and SCM: EU370787. The marmoset's ortholog of SCML1 was obtained through blast search [34].
Test of selection on SCML1 in primates
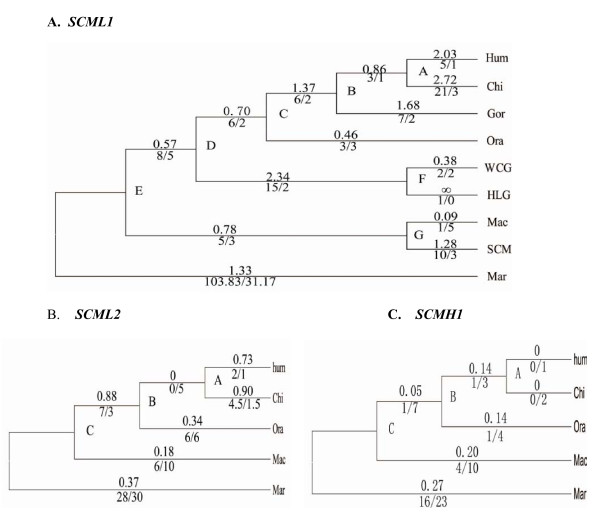
We calculated the dN/dS ratio (also called ω), which measures the rate of protein evolution as scaled to mutation rate for all the branches. We also obtained the numbers of nonsynonymous and synonymous substitutions for each branch (Figure 2A) [36]. As shown in Figure 2A, most of the primate lineages have large ω values (ω > 1) except for orangutan and rhesus macaque, again suggesting rapid amino acid changes during primate evolution.
Figure 2.
Molecular evolution of SCML1, SCML2 and SCMH reconstructed over the known phylogenetic tree of primates. A) The ω values of different lineages in primates were calculated using the SCML1 coding sequences. The phylogeny was drawn roughly to the scale of evolutionary time. ∞ refers to ω value with dS being zero. B, C) The phylogenetic trees of SCML2 and SCMH1. The ω values are shown above each branch. The numbers of nonsynonymous vs. synonymous substitutions (N/S) are shown below each branch.
The rapid protein sequence evolution can be explained either by Darwinian positive selection or by relaxation of negative (or purifying) selection. To test the hypothesis that they could be under Darwinian positive selection, we conducted the analysis for positive selection at individual amino acid sites using maximum likelihood models by estimating ω values[28,29,37]. The results are presented in Table 1. We first conducted the analysis using M0. In the M0 analysis, the log likelihood L is -2619.84, and the estimated ω = 1.169, implying that there are varied evolutionary forces acting on the amino acid sites of SCML1 (neutral, negative selection and/or positive selection). Model M1a (nearly neutral) assumes two site classes in the sequence (0 < ω < 1 and ω1 = 1 fixed), and is significantly better than M0 (2ΔL = 13.26, P = 0.0002). Therefore, we use M1a as the null hypothesis in detecting selection.
Table 1.
Neutrality tests of SCML1 in primates using maximum likelihood estimates (site-model)
| Model | lnL | Estimates of parameters | 2ΔlnL | Positively selected sites |
|---|---|---|---|---|
| M0: | -2619.84 | ω = 1.169 | None | |
| M1a | -2613.21 | p0 = 0.278 p1 = 0.722 | 13.26(1)** | Not allowed |
| M2a | -2599.55 | p0 = 0.217 p1 = 0.660 p2 = 0.123 ω2 = 5.25 |
26.52(2)** | 23N 153L 201T (95 =< P < 99%) 92H 242G (P > 99%) |
| M7 | -2616.5 | p = 1.096 q = 0.005 p0 = 0.892 p = 0.020 |
Not allowed | |
| M8 | -2599.65 | q = 0.005(p1 = 0.108) ω = 5.78 |
33.9(3)** | 3N 95S 153L 201T 270L (P > 95%) 92H 242G (P > 99%) |
| M8a | -2613.21 | p0 = 0.278 p = 0.005 q = 1.728 (p1 = 0.722) ω = 1.0 |
27.3(4)** | Not allowed |
Note: * refers to P < 0.05. ** refers to P < 0.01; (1): M1a versus M0, (2): M2a versus M1a, (3): M8 versus M7 (4): M8 versus M8a. The proportions of sites under positive selection and selective constraint are p1 and p0 respectively. The parameters p and q are used for the beta distribution B(p, q).
We next compared M1a and M2a (selection model), and M2a fits the data significantly better than M1a (2ΔL = 26.52 P < 0.0001), a strong signature of positive selection on SCML1 in primates. M2a suggests that 12.3% of the sites are under positive selection with ω2 = 5.25. In addition, to avoid the potential bias caused by the assumed substitution pattern in M1a and M2a, we also conducted the selection test by comparing M8 (selection model) and M7 (neutral model), in which a beta distribution for ω over sites was assumed. M8 provides significantly better fit to the data than M7(2ΔL = 33.9, P < 0.0001), again suggesting positive selection on SCML1 in primates. M8 suggests that 10.8% of the sites were under positive selection with ω = 5.78. Interestingly, M8 demonstrates a U-shaped distribution of beta values, suggesting that most sites are either highly conserved with dN/dS close to 0 or nearly neutral with dN/dS = 1, and only a small percentage of the sites were under positive selection. A more stringent test comparing M8 and M8a also supports the proposed positive selection (2ΔL = 27.3, P < 0.0001). The positively selected sites are shown in Table 1 and Figure 1[33,38]. Collectively, all the tests on selection can be better explained by the evolutionary model that invokes positive selection in primates.
Evolutionary pattern comparison between SCML1, SCML2 and SCMH1
As SCML1 is likely a recent duplication during mammalian evolution, we compared the evolutionary patterns between SCML1 and the two other members of the SCM family, i.e. SCML2 and SCMH1, which are located on chromosome X and chromosome 1 respectively. The sequence alignment is showed in Additional file 2. SCML1 and SCML2 are only 15-kb apart on chromosome X (Figure 3A), and the intron and exon structure at the C terminus are highly conserved between them but with totally different N terminus. The SAM domain (sterile alpha motif) is highly conserved among all the three genes (Figure 3B). Strikingly, when comparing the amino acid sequences of the SAM domain among different primate species, SCML1 have much more between-specie amino acid changes than those of SCML2 and SCMH1. In the M0 analysis, the ω values for SCMH1 and SCML2 are 0.179 and 0.325 respectively, which are much smaller than that of SCML1 (ω = 1.169). This is an average over all sites in the protein and all lineages in the tree, therefore, suggesting a dominant role of purifying selection in the evolution of SCML2 and SCMH1. With the use of free ratio model, we calculated the dN/dS values of each primate lineages for SCML2 and SCMH1, and all the lineages show strong functional constraint (purifying selection) or neutral evolution (dN/ds ≤ 1) (Figure 2B, 2C). The expression pattern of the three genes in human is different from each other though all of them are highly expressed in testis [see Additional file 3] implying functional divergence after gene duplication.
Figure 3.
Gene structure and sequence comparison among SCML1, SCML2 and SCMH1. A) SCML1 and SCML2 are both located on the X-chromosome and the transcription directions are indicated by the arrows. B) The protein sequence alignment among the three genes. The SAM domains are highlighted. Sterile alpha motif (SAM) domains are known to exhibit diverse protein-protein interaction modes[57]
Expression analysis of SCML1
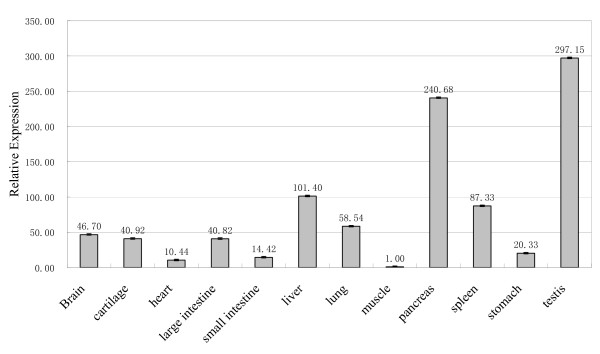
Having shown that the evolution of SCML1 in primates is consistent with positive selection, by dissecting its expression pattern, we attempt to understand the driving force and the functional consequence of selection. We first detect the expression pattern of SCML1 by testing 12 different tissue types in 1–2 year old macaques and the result is shown in Figure 4. Testis and pancreas have the highest expression when compared with the other tissues. In human, according to the micro-array expression data, SCML1 is also preferentially expressed in testis [39]. The abundant expression of SCML1 in testis confirms its involvement in male reproduction. However, in humans [see Additional file 3], liver, fetal liver, and pituitary all show higher expression than pancreas, which is different from the expression pattern of rhesus macaque, implicating functional modifications of SCML1 during primate evolution. This is consistent with the proposed positive selection on SCML1 in primates.
Figure 4.
The relative expression levels of SCML1 in different tissues. A 1–2 year old male rhesus monkey was sampled and tested using real time quantitative PCR. The pancreas and testis showed the highest expression of SCML1. The numbers above the columns are the relative levels of expression.
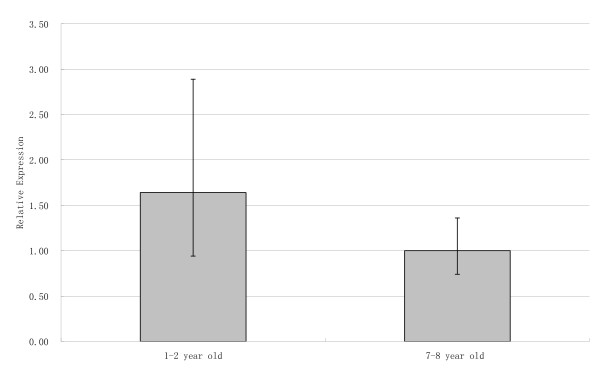
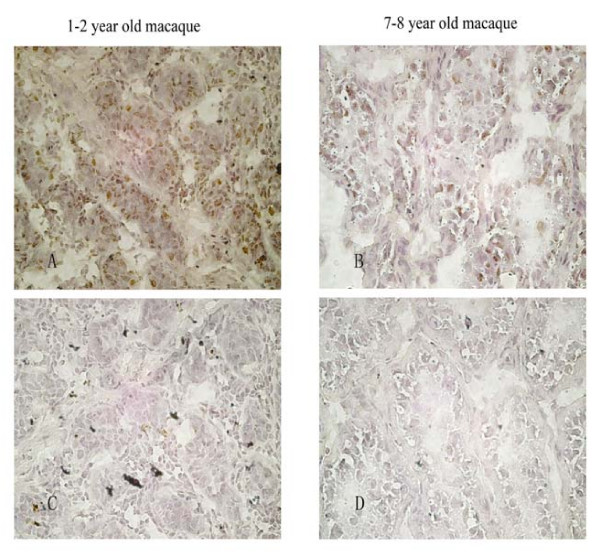
We then detect the potential expression change of SCML1 during male sexual maturation and a significant change was detected in testis. We compared two age groups, i.e. 1–2 year old monkeys (sexually immature) and 7–8 year old monkeys (sexually matured). The result indicates that both age groups have abundant expression of SCML1, and there is a significant reduction (about 60%) in the adult group (p = 0.015, two-tailed t test) (Figure 5). We next carried out immunohistochemical analysis of the two age groups (Figure 6). The result shows that SCML1 is preferentially expressed in the germ stem cells of testis (spermatogonial stem cells in 1–2 year old monkeys, and spermatogonia in 7–8 year old monkeys) [40-48], and again the 1–2 year old group has higher expression than the adult group. The higher expression in the 1–2 year old monkeys is probably due to the relative abundance of stem cells at the sexually immature stage. The preferential expression in the stem cells of testis suggests that SCML1 is likely involved in spermatogenesis during sexual maturation in rhesus macaque.
Figure 5.
The relative expression levels of SCML1 in testis during male sexual maturation, which was tested using real time quantitative PCR. Two age groups were tested including the 1–2 year old macaque group and the 7–8 year old macaque group. Ten individuals were sampled for each group.
Figure 6.
The in-situ immunohistochemical analysis of SCML1 during testis development. Both the 1–2 year old (A, C) and 7–8 year old macaques (B, D) were tested (40×). The expression of SCML1 is mostly located in spermatogonial stem cells (1–2 year old), and spermatogonia (7–8 year old). C and D are the controls without adding SCML1 mouse antibodies.
Discussion
We demonstrate that SCML1 evolves rapidly in primates, which was caused by Darwinian positive selection. Genes expressed exclusively or preferentially in testis are likely involved in male reproduction and have been shown to evolve rapidly under positive selection in previous studies ([1-3,49-55]. Our observation of rapid evolution in SCML1 provides another example of male reproductive gene under Darwinian positive selection in primates.
Darwinian positive selection may lead to functional changes of the target genes during evolution. The SAM domain located in the C terminal (Figure 1) is the only known functional domain of SCML1[56]. Among primates, the amino acid sequences of the SAM domains are relatively conserved across species and there is one site under positive selection (Figure 1). The SAM domain is known to exhibit diverse protein-protein interaction modes, and is involved in developmental regulation [57]. Through functional domain prediction [56-63], besides of the SAM domain, we identified a total of six fragments[64,65] (amino acid position 1–9, 13–23, 72–79, 88–114, 125–157 and 224–239) in SCML1 containing potential functional domains. For example, the fragment 1–9 is a signal peptide. The positively selected sites using 95% cutoff are listed in Table 1, and most of them are also located in the potential functional domains other than the structural domains (4/1 and 5/2 for model M2a and model M8 respectively). This distribution bias of the positively selected sites indicates that Darwinian positive selection on SCML1 targets the putative functional domains, which is consistent with the proposed functional modification of SCML1 during primate evolution.
Sexual selection is the favored explanation for the observed adaptive evolution of male reproductive genes [35,66]. The immunohistochemical and RT-PCR data suggests that SCML1 is important for the development and normal function of testis in primates. Therefore, it is reasonable to propose that the adaptive evolution of SCML1 in primates is likely due to sexual selection[4]. Sperm competition, one of the major mechanisms for sexual selection has been used to define the driving force of selection in promiscuous species, e.g. chimpanzee and human, which seems to explain the observed adaptive evolution of SCML1 since both chimpanzee and human are among the rapidly evolving lineages (ω > 1, Figure 2A). However, gibbon is a monogamous species with a high ω value, and the highly promiscuous rhesus monkey does not show accelerated evolution (ω = 0.09). Therefore, the branch-specific rapid evolution of SCML1 in primates does not provide consistent support for the sexual selection hypothesis. Other evolutionary mechanisms need to be tested, e.g. speciation [67-70].
The origin of SCML1 probably occurred at the early stage of mammalian radiation about 100 million years ago because we do not identify SCML1 in non-mammalian species, neither in mouse and rat, but in dog, cow and primates. The comparison of evolutionary and expression patterns among the three genes of the same SCML family suggests that the rapid evolution of SCML1 likely led to function modification of testis development and spermatogenesis in primates [71-73].
Conclusion
The adaptive evolution of SCML1 in primates provides a new case in understanding the evolutionary process of genes involved in primate male reproduction.
Abbreviations
Hum: human – Homo sapiens; Chi: chimpanzee – Pan troglodytes; Gor: gorilla – Gorilla gorilla; Ora: orangutan – Pongo pygmaeus; HLG: white-browed gibbon – Bunopithecus hoolock; WCG: white-cheeked gibbon – Nomascus leucogenys; Mac: rhesus monkeys – Macaca mulatta; SCM: Yunnan snub-nosed monkey – Rhinopithecus bieti; Mar: common marmoset – Callithrix jacchus.
Authors' contributions
BS and HW conceived the project. HW executed the sequencing, data analysis, immunohistochemical analysis, real-time quantitative RT-PCR, data mining and statistical analysis. BS supervised the project execution. BS and HW wrote the manuscript.
Supplementary Material
The PCR primer sequences for SCML1.
The protein sequence alignment of SCML2 and SCMH1.
The expression patterns of SCML1, SCML2 and SCMH1 in normal human tissues.
Contributor Information
Hai-hui Wu, Email: haihuiwu2002@gmail.com.
Bing Su, Email: sub@mail.kiz.ac.cn.
Acknowledgements
We thank Drs. Qi XB, Wang YQ and Han L for their help in data analysis. We are also thankful to the technical help of Zhang H and Yu YC. This study was supported by grants from the National 973 project of China (2007CB947701and 2007CB815705), the Chinese Academy of Sciences (KSCX1-YW-R-34), the National Natural Science Foundation of China (30525028, 30630013 and 30623007), and the Natural Science Foundation of Yunnan Province of China.
References
- Clark NL, Aagaard JE, Swanson WJ. Evolution of reproductive proteins from animals and plants. Reproduction. 2006;131(1):11–22. doi: 10.1530/rep.1.00357. [DOI] [PubMed] [Google Scholar]
- Swanson WJ, Vacquier VD. The rapid evolution of reproductive proteins. Nat Rev Genet. 2002;3(2):137–144. doi: 10.1038/nrg733. [DOI] [PubMed] [Google Scholar]
- Panhuis TM, Clark NL, Swanson WJ. Rapid evolution of reproductive proteins in abalone and Drosophila. Philos Trans R Soc Lond B Biol Sci. 2006;361(1466):261–268. doi: 10.1098/rstb.2005.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen-Seaman MI, Li WH. Evolution of the hominoid semenogelin genes, the major proteins of ejaculated semen. Journal of molecular evolution. 2003;57(3):261–270. doi: 10.1007/s00239-003-2474-x. [DOI] [PubMed] [Google Scholar]
- Vosse E van de, Walpole SM, Nicolaou A, Bent P van der, Cahn A, Vaudin M, Ross MT, Durham J, Pavitt R, Wilkinson J. et al. Characterization of SCML1, a new gene in Xp22, with homology to developmental polycomb genes. Genomics. 1998;49(1):96–102. doi: 10.1006/geno.1998.5224. [DOI] [PubMed] [Google Scholar]
- Duncan IM. Polycomblike: a gene that appears to be required for the normal expression of the bithorax and antennapedia gene complexes of Drosophila melanogaster. Genetics. 1982;102(1):49–70. doi: 10.1093/genetics/102.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke A, DeCamillis M, Zink D, Cheng N, Brock HW, Paro R. Polycomb and polyhomeotic are constituents of a multimeric protein complex in chromatin of Drosophila melanogaster. The EMBO journal. 1992;11(8):2941–2950. doi: 10.1002/j.1460-2075.1992.tb05364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin EC, Adler PN. The Polycomb group gene Posterior Sex Combs encodes a chromosomal protein. Development. 1993;117(2):641–655. doi: 10.1242/dev.117.2.641. [DOI] [PubMed] [Google Scholar]
- Jones RS, Gelbart WM. The Drosophila Polycomb-group gene Enhancer of zeste contains a region with sequence similarity to trithorax. Mol Cell Biol. 1993;13(10):6357–6366. doi: 10.1128/mcb.13.10.6357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonie A, D'Andrea R, Paro R, Saint R. Molecular characterisation of the Polycomblike gene of Drosophila melanogaster, a trans-acting negative regulator of homeotic gene expression. Development. 1994;120(9):2629–2636. doi: 10.1242/dev.120.9.2629. [DOI] [PubMed] [Google Scholar]
- Schumacher A, Magnuson T. Murine Polycomb- and trithorax-group genes regulate homeotic pathways and beyond. Trends Genet. 1997;13(5):167–170. doi: 10.1016/S0168-9525(97)01133-5. [DOI] [PubMed] [Google Scholar]
- Gould A. Functions of mammalian Polycomb group and trithorax group related genes. Curr Opin Genet Dev. 1997;7(4):488–494. doi: 10.1016/S0959-437X(97)80075-5. [DOI] [PubMed] [Google Scholar]
- Takihara Y, Tomotsune D, Shirai M, Katoh-Fukui Y, Nishii K, Motaleb MA, Nomura M, Tsuchiya R, Fujita Y, Shibata Y. et al. Targeted disruption of the mouse homologue of the Drosophila polyhomeotic gene leads to altered anteroposterior patterning and neural crest defects. Development. 1997;124(19):3673–3682. doi: 10.1242/dev.124.19.3673. [DOI] [PubMed] [Google Scholar]
- Montini E, Buchner G, Spalluto C, Andolfi G, Caruso A, den Dunnen JT, Trump D, Rocchi M, Ballabio A, Franco B. Identification of SCML2, a second human gene homologous to the Drosophila sex comb on midleg (Scm): A new gene cluster on Xp22. Genomics. 1999;58(1):65–72. doi: 10.1006/geno.1999.5755. [DOI] [PubMed] [Google Scholar]
- Berger J, Kurahashi H, Takihara Y, Shimada K, Brock HW, Randazzo F. The human homolog of Sex comb on midleg (SCMH1) maps to chromosome 1p34. Gene. 1999;237(1):185–191. doi: 10.1016/S0378-1119(99)00285-1. [DOI] [PubMed] [Google Scholar]
- Francis NJ, Saurin AJ, Shao Z, Kingston RE. Reconstitution of a functional core polycomb repressive complex. Mol Cell. 2001;8(3):545–556. doi: 10.1016/S1097-2765(01)00316-1. [DOI] [PubMed] [Google Scholar]
- Shao Z, Raible F, Mollaaghababa R, Guyon JR, Wu CT, Bender W, Kingston RE. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell. 1999;98(1):37–46. doi: 10.1016/S0092-8674(00)80604-2. [DOI] [PubMed] [Google Scholar]
- King IF, Francis NJ, Kingston RE. Native and recombinant polycomb group complexes establish a selective block to template accessibility to repress transcription in vitro. Mol Cell Biol. 2002;22(22):7919–7928. doi: 10.1128/MCB.22.22.7919-7928.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomotsune D, Takihara Y, Berger J, Duhl D, Joo S, Kyba M, Shirai M, Ohta H, Matsuda Y, Honda BM. et al. A novel member of murine Polycomb-group proteins, Sex comb on midleg homolog protein, is highly conserved, and interacts with RAE28/mph1 in vitro. Differentiation. 1999;65(4):229–239. doi: 10.1046/j.1432-0436.1999.6540229.x. [DOI] [PubMed] [Google Scholar]
- Sathyamurthy A, Allen MD, Murzin AG, Bycroft M. Crystal structure of the malignant brain tumor (MBT) repeats in Sex Comb on Midleg-like 2 (SCML2) J Biol Chem. 2003;278(47):46968–46973. doi: 10.1074/jbc.M306469200. [DOI] [PubMed] [Google Scholar]
- Grubach L, Juhl-Christensen C, Rethmeier A, Olesen LH, Aggerholm A, Hokland P, Ostergaard M. Gene expression profiling of Polycomb, Hox and Meis genes in patients with acute myeloid leukaemia. Eur J Haematol. 2008. [DOI] [PubMed]
- Qi X-B, Yang S, Zheng H-K, Wang Y-Q, Liao C-H, Liu Y, Chen X-H, Shi H, Yu X-J, Lin AA. et al. Detecting positive Darwinian selection in brain-expressed genes during human evolution. Chinese Science Bulletin. 2007;52(3):324–335. doi: 10.1007/s11434-007-0062-y. [DOI] [Google Scholar]
- Goodman M, Porter CA, Czelusniak J, Page SL, Schneider H, Shoshani J, Gunnell G, Groves CP. Toward a phylogenetic classification of Primates based on DNA evidence complemented by fossil evidence. Mol Phylogenet Evol. 1998;9(3):585–598. doi: 10.1006/mpev.1998.0495. [DOI] [PubMed] [Google Scholar]
- The ensembl website. http://www.ensembl.org
- Kumar S, Tamura K, Nei M. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Briefings in bioinformatics. 2004;5(2):150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular biology and evolution. 2007;24(8):1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Page SL, Goodman M. Catarrhine phylogeny: noncoding DNA evidence for a diphyletic origin of the mangabeys and for a human-chimpanzee clade. Mol Phylogenet Evol. 2001;18(1):14–25. doi: 10.1006/mpev.2000.0895. [DOI] [PubMed] [Google Scholar]
- Yang Z, Swanson WJ. Codon-substitution models to detect adaptive evolution that account for heterogeneous selective pressures among site classes. Mol Biol Evol. 2002;19(1):49–57. doi: 10.1093/oxfordjournals.molbev.a003981. [DOI] [PubMed] [Google Scholar]
- Nielsen R, Yang Z. Likelihood models for detecting positively selected amino acid sites and applications to the HIV-1 envelope gene. Genetics. 1998;148(3):929–936. doi: 10.1093/genetics/148.3.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. Complexity of the simplest phylogenetic estimation problem. Proceedings. 2000;267(1439):109–116. doi: 10.1098/rspb.2000.0974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Goldman N, Friday A. Comparison of models for nucleotide substitution used in maximum-likelihood phylogenetic estimation. Mol Biol Evol. 1994;11(2):316–324. doi: 10.1093/oxfordjournals.molbev.a040112. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R. Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol Biol Evol. 2002;19(6):908–917. doi: 10.1093/oxfordjournals.molbev.a004148. [DOI] [PubMed] [Google Scholar]
- Wong WS, Yang Z, Goldman N, Nielsen R. Accuracy and power of statistical methods for detecting adaptive evolution in protein coding sequences and for identifying positively selected sites. Genetics. 2004;168(2):1041–1051. doi: 10.1534/genetics.104.031153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Primates' sequencing database. http://genome.wustl.edu/genome_group.cgi?GROUP=1
- Wyckoff GJ, Wang W, Wu CI. Rapid evolution of male reproductive genes in the descent of man. Nature. 2000;403(6767):304–309. doi: 10.1038/35002070. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol Biol Evol. 2000;17(1):32–43. doi: 10.1093/oxfordjournals.molbev.a026236. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R, Goldman N, Pedersen AM. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics. 2000;155(1):431–449. doi: 10.1093/genetics/155.1.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Wong WS, Nielsen R. Bayes empirical bayes inference of amino acid sites under positive selection. Molecular biology and evolution. 2005;22(4):1107–1118. doi: 10.1093/molbev/msi097. [DOI] [PubMed] [Google Scholar]
- GNF SymAtlas database. http://symatlas.gnf.org/SymAtlas/
- Clermont Y, Leblond CP. Differentiation and renewal of spermatogonia in the monkey, Macacus rhesus. Am J Anat. 1959;104:237–273. doi: 10.1002/aja.1001040204. [DOI] [PubMed] [Google Scholar]
- Clermont Y. Two classes of spermatogonial stem cells in the monkey (Cercopithecus aethiops) Am J Anat. 1969;126(1):57–71. doi: 10.1002/aja.1001260106. [DOI] [PubMed] [Google Scholar]
- Clermont Y. Kinetics of spermatogenesis in mammals: seminiferous epithelium cycle and spermatogonial renewal. Physiol Rev. 1972;52(1):198–236. doi: 10.1152/physrev.1972.52.1.198. [DOI] [PubMed] [Google Scholar]
- Clermont Y, Antar M. Duration of the cycle of the seminiferous epithelium and the spermatogonial renewal in the monkey Macaca arctoides. Am J Anat. 1973;136(2):153–165. doi: 10.1002/aja.1001360204. [DOI] [PubMed] [Google Scholar]
- de Rooij DG, van Alphen MM, Kant HJ van de. Duration of the cycle of the seminiferous epithelium and its stages in the rhesus monkey (Macaca mulatta) Biol Reprod. 1986;35(3):587–591. doi: 10.1095/biolreprod35.3.587. [DOI] [PubMed] [Google Scholar]
- De Rooij DG, Van Dissel-Emiliani FM, Van Pelt AM. Regulation of spermatogonial proliferation. Ann N Y Acad Sci. 1989;564:140–153. doi: 10.1111/j.1749-6632.1989.tb25894.x. [DOI] [PubMed] [Google Scholar]
- Ehmcke J, Schlatt S. A revised model for spermatogonial expansion in man: lessons from non-human primates. Reproduction. 2006;132(5):673–680. doi: 10.1530/rep.1.01081. [DOI] [PubMed] [Google Scholar]
- Ehmcke J, Hubner K, Scholer HR, Schlatt S. Spermatogonia: origin, physiology and prospects for conservation and manipulation of the male germ line. Reprod Fertil Dev. 2006;18(1–2):7–12. doi: 10.1071/RD05119. [DOI] [PubMed] [Google Scholar]
- Ehmcke J, Wistuba J, Schlatt S. Spermatogonial stem cells: questions, models and perspectives. Hum Reprod Update. 2006;12(3):275–282. doi: 10.1093/humupd/dmk001. [DOI] [PubMed] [Google Scholar]
- Gavrilets S. Rapid evolution of reproductive barriers driven by sexual conflict. Nature. 2000;403(6772):886–889. doi: 10.1038/35002564. [DOI] [PubMed] [Google Scholar]
- Grus WE, Zhang J. Rapid turnover and species-specificity of vomeronasal pheromone receptor genes in mice and rats. Gene. 2004;340(2):303–312. doi: 10.1016/j.gene.2004.07.037. [DOI] [PubMed] [Google Scholar]
- Podlaha O, Webb DM, Zhang J. Accelerated evolution and loss of a domain of the sperm-egg-binding protein SED1 in ancestral primates. Molecular biology and evolution. 2006;23(10):1828–1831. doi: 10.1093/molbev/msl066. [DOI] [PubMed] [Google Scholar]
- Rooney AP, Zhang J. Rapid evolution of a primate sperm protein: relaxation of functional constraint or positive Darwinian selection? Mol Biol Evol. 1999;16(5):706–710. doi: 10.1093/oxfordjournals.molbev.a026153. [DOI] [PubMed] [Google Scholar]
- Rooney AP, Zhang J, Nei M. An unusual form of purifying selection in a sperm protein. Mol Biol Evol. 2000;17(2):278–283. doi: 10.1093/oxfordjournals.molbev.a026307. [DOI] [PubMed] [Google Scholar]
- Zhang J, Rosenberg HF. Diversifying selection of the tumor-growth promoter angiogenin in primate evolution. Mol Biol Evol. 2002;19(4):438–445. doi: 10.1093/oxfordjournals.molbev.a004099. [DOI] [PubMed] [Google Scholar]
- Wang X, Zhang J. Rapid evolution of mammalian X-linked testis-expressed homeobox genes. Genetics. 2004;167(2):879–888. doi: 10.1534/genetics.103.025072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: identification of signaling domains. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(11):5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CA, Bowie JU. SAM domains: uniform structure, diversity of function. Trends Biochem Sci. 2003;28(12):625–628. doi: 10.1016/j.tibs.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Peterson AJ, Kyba M, Bornemann D, Morgan K, Brock HW, Simon J. A domain shared by the Polycomb group proteins Scm and ph mediates heterotypic and homotypic interactions. Mol Cell Biol. 1997;17(11):6683–6692. doi: 10.1128/mcb.17.11.6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TM. SAM breaks its stereotype. Nat Struct Biol. 2003;10(9):677–679. doi: 10.1038/nsb0903-677. [DOI] [PubMed] [Google Scholar]
- Tsutsui K, Asai Y, Fujimoto A, Yamamoto M, Kubo M, Hatta N. A novel p63 sterile alpha motif (SAM) domain mutation in a Japanese patient with ankyloblepharon, ectodermal defects and cleft lip and palate (AEC) syndrome without ankyloblepharon. Br J Dermatol. 2003;149(2):395–399. doi: 10.1046/j.1365-2133.2003.05423.x. [DOI] [PubMed] [Google Scholar]
- Zhang H, Azevedo RB, Lints R, Doyle C, Teng Y, Haber D, Emmons SW. Global regulation of Hox gene expression in C. elegans by a SAM domain protein. Dev Cell. 2003;4(6):903–915. doi: 10.1016/S1534-5807(03)00136-9. [DOI] [PubMed] [Google Scholar]
- Ponting CP. SAM: a novel motif in yeast sterile and Drosophila polyhomeotic proteins. Protein Sci. 1995;4(9):1928–1930. doi: 10.1002/pro.5560040927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J, Ponting CP, Hofmann K, Bork P. SAM as a protein interaction domain involved in developmental regulation. Protein Sci. 1997;6(1):249–253. doi: 10.1002/pro.5560060128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The DisEMBL on line. http://biodatabase.org/index.php/DisEMBL
- The Smart site on line. http://smart.embl-heidelberg.de/
- Dorus S, Evans PD, Wyckoff GJ, Choi SS, Lahn BT. Rate of molecular evolution of the seminal protein gene SEMG2 correlates with levels of female promiscuity. Nat Genet. 2004;36(12):1326–1329. doi: 10.1038/ng1471. [DOI] [PubMed] [Google Scholar]
- Gavrilets S. Waiting time to parapatric speciation. Proceedings. 2000;267(1461):2483–2492. doi: 10.1098/rspb.2000.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrilets S, Acton R, Gravner J. Dynamics of speciation and diversification in a metapopulation. Evolution. 2000;54(5):1493–1501. doi: 10.1111/j.0014-3820.2000.tb00695.x. [DOI] [PubMed] [Google Scholar]
- Gavrilets S, Li H, Vose MD. Patterns of parapatric speciation. Evolution. 2000;54(4):1126–1134. doi: 10.1111/j.0014-3820.2000.tb00548.x. [DOI] [PubMed] [Google Scholar]
- Nei M, Zhang J. Molecular origin of species. Science. 1998;282(5393):1428–1429. doi: 10.1126/science.282.5393.1428. [DOI] [PubMed] [Google Scholar]
- Prince VE, Pickett FB. Splitting pairs: the diverging fates of duplicated genes. Nat Rev Genet. 2002;3(11):827–837. doi: 10.1038/nrg928. [DOI] [PubMed] [Google Scholar]
- Scannell DR, Byrne KP, Gordon JL, Wong S, Wolfe KH. Multiple rounds of speciation associated with reciprocal gene loss in polyploid yeasts. Nature. 2006;440(7082):341–345. doi: 10.1038/nature04562. [DOI] [PubMed] [Google Scholar]
- van Hoof A. Conserved functions of yeast genes support the duplication, degeneration and complementation model for gene duplication. Genetics. 2005;171(4):1455–1461. doi: 10.1534/genetics.105.044057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The GEO database. http://www.ncbi.nlm.nih.gov/geo
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The PCR primer sequences for SCML1.
The protein sequence alignment of SCML2 and SCMH1.
The expression patterns of SCML1, SCML2 and SCMH1 in normal human tissues.