Abstract
Preimplantation mouse embryos express both classical (class Ia) and nonclassical (class Ib) MHC class I proteins, and yet are not rejected by the maternal immune system. Although the function of the embryonic MHC class Ia proteins is unknown, one MHC class Ib protein, Qa-2, the product of the preimplantation embryo development (Ped) gene, actually enhances reproductive success. Similar in structure to MHC class Ia proteins, Qa-2 protein is a trimer of the alpha (heavy) chain, β2 microglobulin and a bound peptide. Studies on the folding, assembly and trafficking of MHC class Ia molecules to the cell surface have revealed this process to be dependent on multiple protein chaperone molecules, but information on the role of chaperone molecules in Qa-2 expression is incomplete. Here, we report the detection of mRNA for four chaperone molecules (TAP1, TAP2, calnexin and tapasin) in preimplantation embryos. We then focused on the role of the MHC-dedicated chaperone, tapasin, on Qa-2 protein expression. First, we demonstrated that tapasin protein is expressed by preimplantation embryos. Then, we used tapasin knockout mice to evaluate the role of tapasin in Qa-2 protein expression on both T cells and preimplantation embryos. We report here that optimal cell surface expression of Qa-2 is dependent on tapasin in both T cells and preimplantation embryos. Identification of the molecules involved in regulation of MHC class I protein expression in early embryos is an important first step in gaining insight into mechanisms of escape of embryos from destruction by the maternal immune system.
Keywords: Qa-2, MHC, TAP, calnexin, tapasin, preimplantation embryo
1. Introduction
The major histocompatibility complex (MHC) is a region of the vertebrate genome that encodes three types of proteins, class I, class II and class III, that are involved in immune surveillance and response. Class I proteins, the subject of this paper, may be further divided into classical MHC class Ia proteins and nonclassical MHC class Ib proteins. Classical MHC class Ia proteins present endogenous peptides to the T cell receptor (TCR), signaling the presence of foreign cytosolic proteins to cytotoxic T cells (CTL). The role of nonclassical MHC class Ib proteins in the immune response is less clear, and includes functions unrelated to the immune system (reviewed in Margulies et al., 2003).
One MHC class Ib protein of particular interest to reproductive immunology is the nonpolymorphic murine class Ib protein Qa-2, which is encoded by the preimplantation embryo development (Ped) gene in the Q region of the mouse MHC (reviewed in Warner et al., 2001, 2004; Warner, 2007). Qa-2 positive mouse strains exhibit faster embryo cleavage during preimplantation development than Qa-2 negative mouse strains, and have a greater chance of survival to birth. Qa-2 positive mice also have higher birth weight, higher weaning weight and lower blood pressure as adults compared to their Qa-2 negative counterparts (Warner et al., 1991; Watkins et al., 2006).
Aside from a defined function in reproduction, Qa-2 has a function also in the immune system. Qa-2 is expressed by most somatic cells and in several immune-privileged locations, such as the anterior chamber of the eye and the testis (Niederkorn et al., 1999; Ungchusri et al., 2001). Qa-2 has been shown to play a role in CTL-mediated tumor rejection and may also serve as an inhibitory ligand for an unknown natural killer (NK) cell receptor (Chiang et al., 2002, 2004).
Similar to classical MHC class Ia molecules, stable cell surface expression of Qa-2 requires the proper folding and assembly of a trimer consisting of the Qa-2 alpha (heavy) chain (encoded in the MHC), β2 microglobulin (β2m) (encoded outside the MHC) and a short (approximately 9 amino acid) peptide (Rotzschke et al., 1993; He et al., 2001). MHC class Ia proteins utilize a highly orchestrated set of protein chaperones to mediate assembly and trafficking of the trimer to the cell surface. In the current model of the mechanism of MHC class Ia expression, the newly translated MHC alpha chain enters the endoplasmic reticulum (ER). In the ER, this heavy chain first interacts with the lectin chaperone calnexin, which facilitates its folding into a native conformation. After dissociation from calnexin, the heavy chain is able to bind to β2m and is brought into proximity of the peptide loading complex, consisting of calreticulin, ERp57, tapasin and the transporter associated with antigen processing (TAP) (reviewed in Cresswell et al., 2005). Calreticulin is the soluble homolog of calnexin, which further aids the proper folding of the MHC class Ia heavy chain. ERp57, a thiol reductase, is covalently linked to the chaperone tapasin. Tapasin (TAP-associated glycoprotein) is an MHC-dedicated chaperone which facilitates the loading of high affinity peptides into the MHC peptide-binding groove and stabilizes the interaction of the MHC molecule with TAP (Sadasivan et al., 1996; Grandea et al., 2000, 2001; Elliott et al., 2005). However, tapasin dependence is allele-specific, so some MHC class I proteins can bind optimal peptides in the absence of tapasin (Williams et al., 2002; Park et al., 2003b). TAP is an essential component of this loading complex, consisting of two subunits (TAP1 and TAP2) which join to form a transmembrane heterodimer in the ER. The TAP complex transports peptides (derived from ubiquitinated proteins degraded in the cytosol by proteasomes) across the ER membrane into the lumen, making the peptides available for loading onto MHC class Ia molecules (Spies et al., 1992; Cresswell et al., 2005). In normally functioning cells, these chaperones all function to create a stable trimeric complex of MHC class Ia heavy chain, β2m and bound peptide. At this point, the complete MHC class Ia molecule can be released from the ER via the Golgi apparatus to reach the surface of the cell.
Much less is known about which chaperone molecules are involved in the processing of MHC class Ib proteins in general and, specifically, Qa-2. Our laboratory has shown previously that Qa-2 is TAP1-dependent for optimal cell surface expression in embryos and lymphocytes (Ke et al., 2000). Tabaczewski and Stroynowski (1994) have shown that the TAP2-negative murine cell line, RMA-S, has severely diminished Qa-2 expression, further implicating a role for the TAP complex in Qa-2 trafficking.
We have analyzed mRNA expression patterns of four key MHC class I chaperones, TAP1, TAP2, calnexin and tapasin, in preimplantation embryos to determine if these could potentially facilitate MHC protein assembly and trafficking. We focused then on whether or not the MHC class I-dedicated chaperone, tapasin, is required for cell surface expression of Qa-2 in both T cells and preimplantation embryos.
2. Materials and Methods
2.1. Mice and embryo collection
C57BL/6, CBA/Ca and TAP1 knockout mice were purchased from the Jackson Laboratory (Bar Harbor, ME) and bred in our animal facilities. Tapasin knockout mice were a kind gift from Dr. Luc Van Kaer at Vanderbilt University. TAP1 knockout (TAP-/-) and tapasin knockout (tpn-/-) mice were both on a C57BL/6 genetic background and housed in facilities for immunocompromised mice. All mouse strains were housed in an Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC) approved facility in a day-night controlled room (14hr day/10hr night; lights on at 4am EST) and given food and water ad libitum. The protocols followed the NIH guidelines and were approved by Northeastern University’s IACUC.
To produce embryos for our experiments on mRNA levels, C57BL/6 female mice were superovulated with 5IU of equine chorionic gonadotropin (eCG; Sigma, St. Louis, MO) followed by 10IU of human chorionic gonadotropin (hCG; Sigma) 48 hours later. Female mice were then mated with individual C57BL/6 male mice overnight. Plug-positive female mice were sacrificed by cervical dislocation and embryos used for RNA isolation were collected at 17, 41, 53, 65, 77 and 96 hours post-hCG injection, corresponding to the 1-cell, 2-cell, 4-cell, 8-cell, morula and blastocyst stages of development. One cell embryos were torn from the swollen ampulla and treated with 0.5mg/ml hyaluronidase (Sigma) to remove cumulus cells. Two to 8-cell embryos were flushed from the oviduct with KSOMAA (Millipore, Billerica, MA), while morula and blastocysts were flushed from the uteri with the same medium. All embryos were washed twice in KSOMAA and a final wash was performed in 1xPBS. Pools of embryos (320 1-cell, 160 2-cell, 80 4-cell, 40 8-cell, 20 morula and 10 blastocyst) were collected in a minimal volume of PBS and stored with 1U of RNase inhibitor (Applied Biosystems; Foster City, CA) and 2μl of lysis buffer at -80°C until use.
Embryos used for detection of tapasin protein and Qa-2 protein were collected at 39 hours post-hCG injection (for 2-4 cell embryos) or 63 hours post-hCG (for 5-8 cell embryos).
2.2. RNA isolation and reverse transcription
RNA was isolated from pools of C57BL/6 embryos using Stratagene’s Absolutely RNA Nanoprep Kit, designed for RNA isolation from fewer than 1,000 cells (Stratagene, La Jolla, CA). During the cell lysis step, 1μl (106 copies) of pAW109 RNA (Applied Biosystems) was added to serve as an exogenous control. The RNA was treated with DNase during the isolation procedure. RNA was eluted with 10μl elution buffer and used directly for reverse transcription. RNA was isolated from T lymphocytes (T cells) with an Absolutely RNA Miniprepe Kit for use as a positive control. T cells were purified as described later in section 2.6.
Complementary DNA (cDNA) was synthesized by priming with random hexamers. The reaction mixture contained 1x PCR buffer, 8mM MgCl2, 1.6mM dNTPs, 40U RNase inhibitor, 100U MuLV reverse transcriptase, 2.5μM random hexamers and 6μl RNA in a total volume of 40μl (all reagents from Applied Biosystems). The reverse transcription reaction was carried out in a Perkin-Elmer DNA Thermal Cycler 480 (Foster City, CA) under the following conditions: 37°C for 190 minutes, 99°C for 5 minutes and 4°C soak.
2.3. Quantitative real-time RT-PCR
Primers and thermocyling protocols were optimized for detection of TAP1, TAP2, calnexin, tapasin and HPRT mRNAs, and pAW109 synthetic RNA. The sequence of primers, their source, fluorescence detection method and thermocycling conditions are presented in Table I. All PCR reactions utilized SYBR green fluorescent dye detection, except tapasin detection which utilized a Taqman probe and FAM detection. The SYBR green PCR reactions contained 1xSYBR Green PCR master mix, various concentrations of each primer, nuclease-free water and 2μl of cDNA. The TAP1 primers have been previously described (Pearce et al., 1993), and the TAP2, calnexin and HPRT primer sequences were obtained from the PrimerBank database (Wang et al., 2003). We designed the tapasin primers and probe using Applied Biosystem’s ‘Assay by Design’ program. Tapasin PCR reactions contained 1xTaqman master mix, 1xassay mix containing primer and probes and 2μl of cDNA. Thermocycling and fluorescence detection were carried out using an ABI Prism 7000 (Applied Biosystems). During optimization, amplicon size was confirmed via agarose gel electrophoresis.
Table I.
Real-time PCR primers and reaction conditions
| Primer Name | Sequence 5’-3’ | Primer ID | Thermocycling protocol | Size Location PCR Efficiency (E)d |
|---|---|---|---|---|
|
TAP1 Fora
TAP1 Rev |
GGCAACCAGCTACGGGTC
CCGGTAGCACCCTCCTCT |
N/Ac | 50°C 2 min, 95°C 10 min, 40 cycles of 95°C 15 sec, 66°C 1 min | 186 bp
Exons 10-11 E=2.06 |
|
TAP2 Forb
TAP2 Rev |
AACACTACTGATGAAGCGGTTG
CGAATAGCGAGGGATTAACGTCT |
PrimerBank ID 7549791a3 | 50°C 2 min, 95°C 10 min, 40 cycles of 95°C 15 sec, 60°C 30 sec, 72°C 30 sec; 72°C 10 min | 118 bp
Exons 2-3 E=2.01 |
|
Calnexin Forb
Calnexin Forb |
ATTTTGCTGACTCCTTTGACAGA
ATTTTGCTGACTCCTTTGACAGA |
PrimerBank ID 6671664a2 | 50°C 2 min, 95°C 10 min, 40 cycles of 95°C 15 sec, 60°C 30 sec, 72°C 30 sec; 72°C 10 min | 154 bp
Exons 3-5 E=1.94 |
|
Tapasin For
Tapasin Rev |
CCAGATCTTGACCCAAAGCTATACT
GCGGCCAGGAGCATTC Probe: CCGGGTCATCCACCTTG |
Applied Biosystems: Assays By Design 4337222-4337224 | 50°C 2 min, 95°C 10 min, 40 cycles of 95°C 15 sec, 60°C 1 min | 61 bp
Exons 2-3 E=2.01 |
|
HPRT Forb
HPRT Rev |
GTTAAGCAGTACAGCCCCAAA
AGGGCATATCCAACAACAAAC |
PrimerBank ID 7305155a2 | 50°C 2 min, 95°C 10 min, 40 cycles of 95°C 15 sec, 60°C 30 sec, 72°C 30 sec; 72°C 10 min | 131 bp
Exons 6-8 E= N/A |
|
pAW109 For
pAW109 Rev |
GTCTCTGAATCAGAAATCCTTCTATC
CATGTCAAATTTCACTGCTTCATCC |
Applied Biosystems DM151/DM152 | 50°C 2 min, 95 C 10 min, 40 cycles of 95C 15 sec, 62C 1 min | 301 bp
E=2.05 |
TAP1 primers were from Pearce et al. (1993);
TAP2, calnexin and HPRT primers were from the PrimerBank website (http://pga.mgh.harvard.edu/primerbank/) (Wang et al., 2003); All reactions utilized SYBR Green detection, except for tapasin which utilized a Taqman probe and FAM dye detection.
N/A = not applicable;
E= PCR efficiency.
Two pools of embryos (from C57BL/6 mice) per stage of preimplantation development were analyzed for expression of mRNA from the genes described in Table 1. Results were analyzed via the Pfaffl equation (Pfaffl, 2001) which computes the relative expression of each gene at the various embryo stages, normalized to the expression of a reference RNA. The Pfaffl equation, shown below, considers the efficiency (E) of each PCR reaction in the calculation.
The PCR efficiencies were determined via a standard curve (repeated in triplicate) of serially diluted mouse T cell cDNA, and the average efficiency (E) is shown in Table 1. In this case, pAW109 was used as the reference RNA against each target chaperone mRNA. Relative mRNA expression at each stage of preimplantation development (experimental) was compared to the two-cell embryo stage (control). A two-tailed, type 2 (equal variance), non-paired t-test was used for statistical analyses.
2.4. Conventional RT-PCR
In order to verify the presence of full-length tapasin mRNA transcripts, conventional RT-PCR was performed. Previously described primers (Seliger et al., 2001) were first used to amplify an 848bp region spanning tapasin exons 3-6 (forward 5’ GAGCCTGTCGTCATCACCAT; reverse 5’ AGCACCTTGAGGAGTCCGAG). A second semi-nested reaction was then used to detect a 465bp product [semi-nested reverse 5’ CACAACGGGTGCTGGTGTTAG (Chiang et al., 2003)]. In both primary and semi-nested PCR reactions, the reaction mixtures contained 1x Platinum PCR Supermix (Invitrogen), 10μM forward and reverse primers and 2μl cDNA from preimplantation embryos (collected and processed as described in previous sections) or 2μl of the primary PCR reaction.
2.5. Immunofluorescence microscopy
Early preimplantation embryos from C57BL/6 mice (2-4 cell embryos) were analyzed for presence of tapasin protein via immunofluorescence microscopy. Embryos were washed twice with PBS plus 1% BSA and 0.1% sodium azide (PBSAZ). Samples were then fixed in 3.7% formaldehyde for 30 minutes and permeabilized with 0.5% Triton X-100 for 20 minutes. All incubations and washes were performed in PBSAZ with 0.1% Triton X-100. Embryos were incubated with either 2μg/ml goat polyclonal anti-tapasin IgG (sc-; Santa Cruz Biotechnology, Santa Cruz, CA) or 2μg/ml normal goat IgG (control antibody; Santa Cruz Biotechnology) for 35 minutes. This was followed by incubation with the labeled secondary antibody, Alexa-Fluor-488 donkey anti-goat IgG (Santa Cruz Biotechnology), and further washing.
Qa-2 cell surface protein expression was analyzed by immunofluorescence microscopy in 5-8 cell embryos from two mouse strains: C57BL/6 (Qa-2 positive control) and tapasin knockout mice. Embryos were incubated with biotinylated anti-Qa-2 antibody (clone 1-9-9 at 10μg/ml; SouthernBiotech, Birmingham, AL) for 30-40 minutes. After 5 washes (10 minute soaks for all washes) in PBSAZ, embryos were incubated with 100nM streptavidin-conjugated quantum dots (Qdot605; Invitrogen, Carlsbad, CA). Embryos were counterstained with Hoechst (Invitrogen) during the final 5 washes and then fixed in formaldehyde as described above.
After staining, all embryos were placed in microdrops of 1xPBSAZ on a glass-bottom imaging dish (Mat-Tek Corp., Ashland, MA) and imaged using the DIC/brightfield and epifluorescence modalities on the Keck 3D Fusion Microscope located at Northeastern University. For more information see the Keck Microscope website http://www.keck3dfm.neu.edu/.
2.6. Flow cytometry
Flow cytometry was used to analyze Qa-2 (MHC class Ib) and H-2Kb (MHC class Ia) protein surface expression on T cells from C57BL/6, CBA/Ca, TAP1 knockout and tapasin knockout mice. Splenic lymphocytes were isolated via density centrifugation over Histopaque (Sigma) and then enriched for CD3+ T cells via a negative selection column (R & D Systems, Minneapolis, MN). T cells were first incubated with either 5μg/ml mouse-anti-Qa-2 antibody (clone 1-9-9, eBioscience) or mouse-anti H-2Kb antibody (eBioscience) for 30-40 minutes, followed by incubation with FITC-conjugated goat anti-mouse IgG (MP Biomedicals, Solon, OH). Washes and incubations were all performed with 1xPBSAZ. Data were collected using a FACScan flow cytometer from Becton Dickinson (Franklin Lakes, NJ) and analyzed using CellQuest software.
3. Results
3.1. Chaperone molecule mRNAs are expressed in preimplantation embryos
Our laboratory has shown previously that Qa-2 mRNA is expressed at all stages of preimplantation embryo development and that Qa-2 protein can be detected on preimplantation embryos by using a variety of techniques, including ELISA and Immuno-PCR (Warner et al., 1987; McElhinny et al., 1998). This implies that the chaperone molecules necessary for Qa-2 surface expression must be expressed in preimplantation embryos. We undertook therefore the analysis of mRNA levels for four chaperone molecules, TAP1, TAP2, calnexin and tapasin, in preimplantation embryos. Real-time PCR analysis was performed on two pools of C57BL/6 embryos representing each stage of preimplantation development. Each pool of embryos contained approximately 320 cells (320 1-cell, 160 2-cell, 80 4-cell, 40 8-cell, 20 morula, 10 blastocysts). The results represent the relative expression of mRNA per embryo, as normalized against exogenously added pAW109 synthetic RNA. The MHC class I chaperone molecules analyzed (TAP1, TAP2, calnexin and tapasin) exhibited different patterns of relative mRNA expression (Figure 1). As expected, the housekeeping gene HPRT and the exogenous control pAW109 (not shown) were expressed in all samples. The PCR amplicons were confirmed to be the correct molecular weight by agarose gel electrophoresis (Figure 2a). One feature common to all the mRNA levels analyzed was the high level of expression of mRNA at the 1-cell (zygote) stage, presumably due to maternal message stores. The results were consistent among all the samples, except for TAP2 mRNA, which was not detected in one of two morula samples and in neither of the blastocyst stage samples (Figure 2b).
Figure 1.
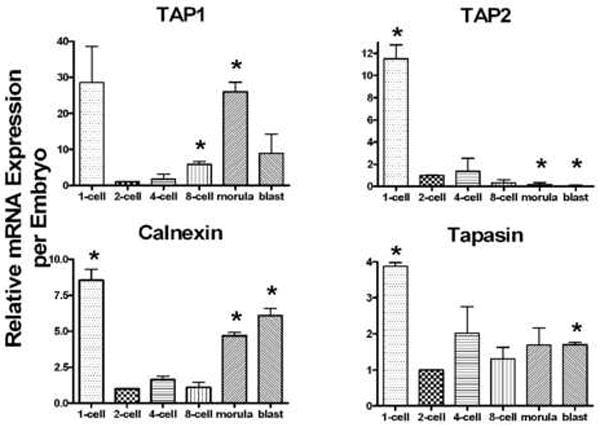
Relative mRNA expression of MHC class I chaperone molecules in preimplantation embryos. Expression was compared to the 2-cell stage, and P≤ 0.05 was considered significant (*). Due to the high standard deviation, TAP1 mRNA expression at the 1-cell stage was not significantly different from the 2-cell level (P=0.11).
Figure 2.
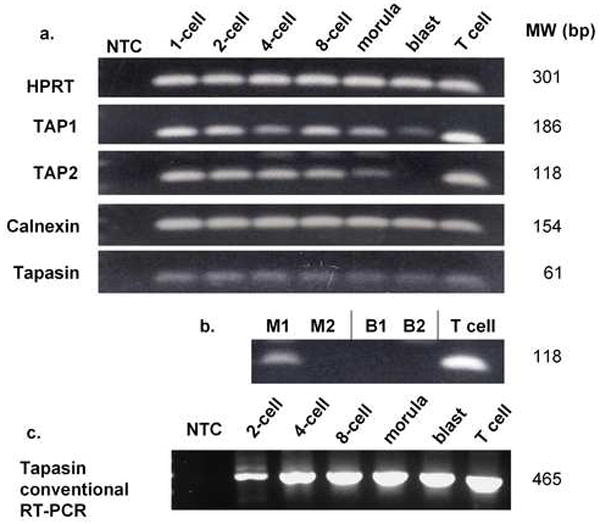
Agarose gel electrophoresis confirmation of real-time and conventional PCR results. a. Chaperone molecule mRNA in preimplantation embryos from one out of two samples analyzed via real-time RT-PCR. Both samples showed the same results except for TAP2. b. TAP2 mRNA from both embryo samples. TAP2 mRNA was not detected in one out of two morula samples and in neither blastocyst sample. M1,2= morula samples 1 and 2. B1,2= blastocyst samples 1 and 2. c. Conventional semi-nested RT-PCR detection of a long tapasin transcript in preimplantation embryos.
3.2 Full- length tapasin mRNA is expressed in preimplantation embryos
Since tapasin is an MHC class I-dedicated chaperone, and has not been examined previously in embryos, we chose this chaperone molecule for further study. The tapasin primers used for quantitative RT-PCR only spanned a short sequence (61bp in exons 2-3). Therefore, we utilized conventional RT-PCR and previously published primer sequences to confirm that longer mRNA sequences were present in preimplantation embryos. In the first round of RT-PCR, a faint 848bp product was detected in all stages tested (2-cells to blastocyst) spanning exons 3-6 (data not shown). A semi-nested RT-PCR was then utilized to confirm the presence and specificity of the tapasin transcript. In the semi-nested PCR, a 465bp product was present in all preimplantation embryo stages (Figure 2c).
3.3 Tapasin protein is detectable in wild-type (C57BL/6) embryos
Using immunofluorescence microscopy, we detected tapasin protein in the cytosol of the blastomeres of two and four-cell embryos (Figure 3). The positive fluorescence signal detected with specific antibody was significantly above the background level of fluorescence seen when using a nonspecific control antibody. In three separate experiments, a total of 16 embryos were stained with anti-tapasin antibody and a total of 15 embryos were stained with a control antibody. All samples gave the same results. A representative sample of embryos is shown in Figure 3.
Figure 3.
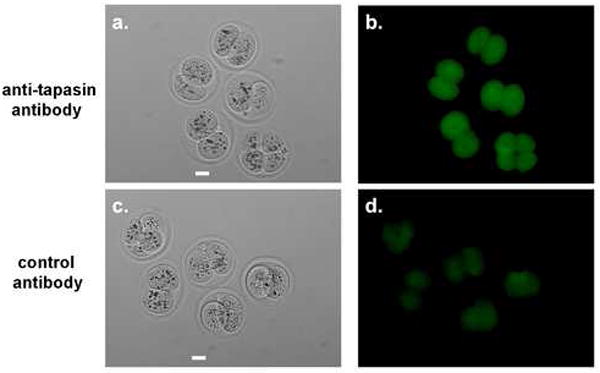
Tapasin protein detection in early preimplantion stage mouse embryos. Embryos were incubated with either anti-tapasin specific primary antibody (a,b) or control IgG primary antibody (c,d), followed by detection with AlexaFluor-488 conjugated secondary antibody. Panels a and c are brightfield images; panels b and d are Alexa-Fluor-488 epifluorescence images. Scale bar = 25μm
3.4. Qa-2 is dependent on tapasin for optimal cell surface expression in T cells
In order to test the dependence of Qa-2 protein expression on tapasin, we used tapasin knockout mice. We first analyzed tapasin dependence of Qa-2 in T cells, which are known to express high levels of Qa-2 protein, as a model system.
The surface expression of Qa-2 on T cells was analyzed in tapasin knockout mice and compared to wild-type Qa-2 positive (C57BL/6), Qa-2 null (CBA/Ca) and TAP1 knockout mice. The expression of H-2Kb on T cells from these same mice was analyzed also as a control, since this protein is known to be tapasin-dependent (Garbi et al., 2000; Grandea et al., 2000). Tapasin knockout mice showed significantly decreased Qa-2 cell surface expression in T cells compared to wild-type mice (Figure 4a). These data clearly show that Qa-2 is dependent on tapasin for optimal cell surface expression. Interestingly, Qa-2 surface expression in tapasin-/- and TAP1-/- T cells is similarly suppressed (Figure 4a). However, tapasin-/- T cells exhibited a broader range of Qa-2 expression than the TAP1-/- T cells.
Figure 4.
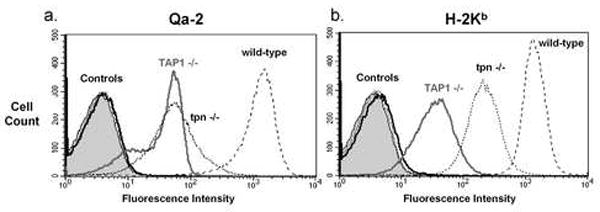
Qa-2 expression on T cells from tapasin knockout mice. a. Surface expression of Qa-2 on T cells from C57BL/6 (wild-type), TAP1 knockout (TAP1-/-) and tapasin knockout (tpn-/-) mice. Controls were T cells without primary antibody and T cells from CBA/Ca (Qa-2 and H-2Kb negative) mice. b. Surface expression of H-2Kb on T cells from C57BL/6 (wild-type), TAP1 knockout (TAP1-/-) and tapasin knockout (tpn-/-) mice. Controls were T cells without primary antibody and T cells from CBA/Ca (Qa-2 and H-2Kb negative) mice.
The cell surface expression of the MHC class Ia protein H-2Kb on T cells was markedly different from expression of Qa-2 in these same mouse strains. Like Qa-2, T cells from tapasin knockout mice had decreased cell surface expression of H-2Kb compared to wild-type mice. However, TAP1-/- T cells had significantly lower cell surface expression of H-2Kb compared to tapasin-/- T cells. To quantify this difference, we analyzed the percent reduction of expression of Qa-2 versus H-2Kb in both TAP1-/-and tapasin -/- T cells. We measured the median fluorescence of the knockout strains as a percentage of wild-type median fluorescence. Five separate mice from each mouse strain were analyzed in four separate experiments. Qa-2 surface expression was found to be 2-3.5% of wild-type expression in both TAP-/- and tapasin-/- T cells (no significant difference). In contrast, H-2Kb surface expression was significantly (P<0.05) more affected by loss of TAP1 (1.5% of wild-type) than loss of tapasin (11% of wild-type).
3.5. Qa-2 is dependent on tapasin for cell surface expression in preimplantation embryos
After tapasin dependence of Qa-2 was established in T cells, we examined the role of tapasin in Qa-2 expression in preimplantation embryos, where Qa-2 contributes to embryo health and viability. We were able to develop a sensitive imaging technique utilizing quantum dots to allow Qa-2 protein to be detected by immunofluorescence microscopy on the embryo surface. Note that previous detection of Qa-2 on the embryo cell surface required the use of ELISA or Immuno-PCR because conventional immunofluorescence was not sensitive enough to detect Qa-2 on the embryo cell surface (Warner et al., 1987; McElhinny et al., 1998). We used this new method to determine whether Qa-2 cell surface expression is affected by the absence of tapasin in tapasin-/-embryos. First, we stained Qa-2 positive embryos (C57BL/6, n=34) as a positive control (Figure 5a,b). Qa-2 protein was detected as punctate cell surface staining on most, but not all, of the Qa-2 positive C57BL/6 embryos tested (Qa-2 detected on 28/34 embryos; Figure 5 a,b). We do not understand why Qa-2 was not detected on all C57BL/6 embryos tested, although it seems likely that the low level of Qa-2 expression on preimplantation embryos may be near the limit of detection for our immunofluorescence staining protocol. Next, we stained tapasin -/- embryos (n=26) and showed that all lacked detectable Qa-2 protein surface expression (Figure 5 c,d). Therefore, our results show unequivocally that Qa-2 expression is dependent on tapasin for optimal cell surface expression in preimplantation embryos.
Figure 5.
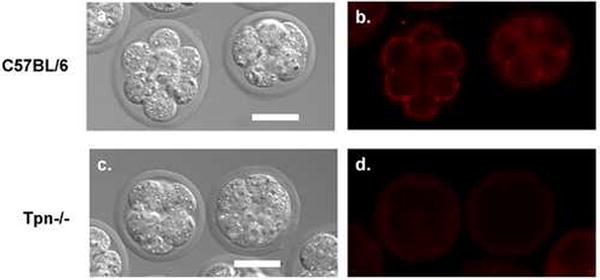
Analysis of Qa-2 cell surface protein expression on control and tapasin knockout embryos. Qa-2 was detected via immunofluorescence on Qa-2 positive (control) C57BL/6 embryos (a,b). In contrast, Qa-2 was not detected on tapasin knockout embryos (c,d). Panels a and c are differential interference contrast (DIC) images; panels b and d are Qdot605 epifluorescence images. Scale bar = 25μm
4. Discussion
We have shown that mRNA for four chaperone molecules involved in MHC class I folding, assembly and trafficking, TAP1, TAP2, calnexin and tapasin, are expressed by preimplantation mouse embryos. MHC class Ia and class Ib proteins are expressed at low levels in preimplantation embryos, with increasing Qa-2 cell surface expression in later-stage preimplantation embryos (Warner et al., 1987, 1993). Therefore, it seemed likely that the chaperone molecules necessary for optimal MHC class I protein expression would be present in preimplantation embryos and that is, indeed, what we determined.
We report here that TAP1 mRNA was detectable at all stages of preimplantation embryo development, while TAP2 mRNA was detectable from the 1-cell to the 8-cell stage (Figure 1). TAP1 and TAP2 proteins form a transmembrane heterodimer that functions to translocate short peptides from the cytosol to the lumen of the endoplasmic reticulum for loading into the peptide-binding groove of MHC class I molecules. In a previous study, Cooper et al. (1998) were unable to detect mRNA for TAP1 until the blastocyst stage and did not detect mRNA for TAP2 at any stage of preimplantation development, leading them to conclude that MHC class I molecules probably do not display peptide antigens at this stage of development. In contrast, our results support the view that MHC class I proteins probably do present peptide antigens on the cellular surface of preimplantation embryos because mRNA for both TAP1 and TAP2 was detected. The detection of TAP2 mRNA in our study is most likely due to increased sensitivity of our mRNA isolation and detection methods. However, the use of a different mouse strain in the Cooper et al. (1998) study may have resulted in differences between our two studies.
We report here on calnexin mRNA in preimplantation mouse embryos. Calnexin protein expression has been previously detected in human oocytes, zygotes and four-cell embryos (Balakier et al., 2002), as well as in mouse oocytes (Calvert et al., 2003). Although calnexin protein expression has not yet been measured in mouse embryos, it seems likely that it is present based on the human embryo data. Calnexin is important in MHC class Ia protein folding, but is also known to be involved in the folding of other ER glycoproteins (reviewed in Williams, 2006). Due to its unique glycosylphosphatidylinositol (GPI) anchor, Qa-2 lacks the proposed MHC class I calnexin-binding site and does not interact with calnexin in vitro (Margolese et al., 1993). However, unlike Qa-2 protein, MHC class Ia proteins do have a calnexin binding site. Therefore, although it seems unlikely that there is a role for calnexin in Qa-2 protein folding, it is likely that calnexin is involved in MHC class Ia folding in embryos.
Tapasin, like TAP1/TAP2, is dedicated to MHC class I protein assembly. We report here the presence of tapasin in preimplantation embryos at both the mRNA and protein levels (Figures 1 and 3). This detection of tapasin in embryos establishes a likely role for tapasin in Qa-2 peptide loading. Since the expression of some MHC class I molecules is tapasin-independent, we used tapasin knockout mice to decipher whether or not Qa-2 protein expression is tapasin-dependent. We used first T cells as a model system because they are easy to harvest, available in large quantities and express high levels of Qa-2 protein. Analyses performed using flow cytometry allowed a quantitative assessment of Qa-2 cell surface expression on T cells from tapasin knockout mice and a comparison of expression of Qa-2 on tapasin knockout with wild-type mice (Figure 4). We found that Qa-2 is dependent on tapasin for optimal cell surface expression on T cells.
After completing analysis of tapasin dependence of Qa-2 in T cells, we then moved on to preimplantation embryos. First, a new, highly sensitive immunofluorescence technique was developed to enable Qa-2 to be imaged on the embryo cell surface. Using this technique, it was shown that Qa-2 expression is dependent on tapasin for cell surface expression on preimplantation embryos. Unlike positive control (C57BL/6) embryos, tapasin knockout embryos lacked detectable Qa-2 protein on their cell surface (Figure 5). Therefore, we have shown that expression of Qa-2 on the surface of preimplantation embryos is dependent on tapasin.
Our data on T cells (Figure 4) show that the dependence of Qa-2 expression on tapasin is significantly different from its MHC class Ia relative, H-2Kb. Our data on H-2Kb expression in T cells agrees with previous work by several other groups, in which TAP-/- mice were shown to exhibit lower H-2Kb expression in splenic lymphocytes than tapasin -/- mice (Garbi et al., 2000; Grandea et al., 2000). The difference in H-2Kb surface expression in T cells from these two different knockout strains is believed to be related to the different functions of TAP and tapasin. In TAP1 knockout mice, peptides for MHC class I presentation are unable to be actively transported into the endoplasmic reticulum via the TAP complex and are therefore not available for binding to the MHC molecule, preventing its expression as a stable trimer on the cell surface. In tapasin knockout mice, appropriate peptides enter the ER, but the various functions of tapasin (peptide-optimization, facilitation of peptide loading and retention of suboptimal MHC) are not available (reviewed in Elliott et al., 2005). Thus, for H-2Kb cell surface expression, the functions of TAP are more critical than those of tapasin.
The difference in tapasin dependence between Qa-2 and H-2Kb may be due to the differences in peptide-binding groove structure and peptide specificity between these two proteins. Although Qa-2 is able to display a wide range of both self and nonself peptides, its peptide specificity differs from H-2Kb (Joyce et al., 1994). Peptides capable of binding to Qa-2 contain two dominant residues, a histidine at position 7 (P7) and a hydrophobic residue (leucine, isoleucine or phenylalanine) at position 9 (P9) (Tabaczewski et al., 1997). There are also several auxiliary binding positions, which can vary from peptide to peptide. In contrast, classical MHC class I molecules tend to bind peptides with anchor residues closer to the N-terminus (P5 and P8 for H-2Kb) (Margulies et al., 2003). The crystal structure of Qa-2 reveals a peptide-binding groove and peptide fit distinct from that of H-2Kb and other classical MHC class Ia molecules (He et al., 2001). The Qa-2 peptide binding groove is shallow and the peptide ‘bulges’ away from the groove in a central location (P2-P7), unlike the more deeply imbedded peptides of MHC class Ia molecules (He et al., 2001). These differences in peptide motifs and in the structures of the peptide-binding grooves between Qa-2 and H-2Kb may significantly influence the degree of tapasin dependence of these two molecules, rendering Qa-2 more dependent on tapasin for binding its unique peptides.
Although it is interesting to compare the tapasin dependence of the two mouse MHC class I proteins, Qa-2 and H-2Kb, it is especially interesting to compare the tapasin-dependence of Qa-2 and its human homolog, HLA-G. It has been previously shown that HLA-G exhibits tapasin dependence (Park et al., 2003a). Several groups have shown that a single amino acid residue in human MHC class I molecules determines tapasin-dependence, with some HLA proteins and alleles exhibiting tapasin dependence and others exhibiting intermediate dependence or tapasin-independence (Williams et al., 2002; Park et al., 2003b). However, the correlation of particular amino acids with tapasin dependence has not been evaluated for murine class I MHC proteins. Here, we have shown that Qa-2, like its human homolog HLA-G, exhibits strong tapasin dependence.
It should be noted that Qa-2 and HLA-G are homologs, not orthologs. Orthologous genes evolve from a single ancestral gene during speciation. DNA sequencing analysis has shown that approximately 80% of mouse genes have a strict human ortholog (Waterston et al., 2002). MHC class I genes evolved after the divergence of mouse. and human and are therefore not orthologs (reviewed in Kumanovics et al., 2003). Interestingly, of the 20% non-orthologous genes between mouse and human, many are involved in reproduction and immunity (Waterston et al., 2002). The basis for the classification of Qa-2 and HLA-G as homologs is their shared sequence and structural similarities (Shiroishi et al., 2006) as well as their functional homology (Comiskey et al., 2007). It is not possible to test for tapasin dependence of HLA-G expression on human embryos due to the scarcity of human embryos available for research and the prohibition of federal funding for human embryo research. However, due to the homology between Qa-2 and HLA-G, it seems likely that HLA-G expression will be found to be tapasin dependent in human embryos.
Returning to the enigma that embryos are not rejected by the maternal immune system even though they express MHC class I proteins, it has been more than 50 years since Medawar (1953) first highlighted the allograft nature of the fetus during pregnancy and the need for regulation of the immune system to achieve a successful pregnancy. In fact, reduced MHC expression has been shown recently to reduce reproductive fitness in a β2-microglobulin deficient mouse model (Cooper et al., 2007). Although some progress has been made towards understanding the mechanism by which this immunoregulation occurs, our knowledge is far from complete. Here, we describe the detection of chaperone molecules necessary for MHC class I expression in preimplantation embryos. We focused on one particular MHC class I molecule, Qa-2 protein, because as the product of the mouse Ped gene it is known to influence reproductive success. We have shown that Qa-2 is dependent on the MHC-dedicated chaperone, tapasin, for optimal cell surface expression in embryos. Based on the data reported here and in other published data (Cresswell et al., 2005), we have diagramed a summary of the likely pathway by which MHC class I molecules are folded, assembled and transported to the cell surface (Figure 6). The analysis of MHC class I chaperone molecules, presented here, represents an important initial step in understanding the regulation of MHC class I protein expression on early embryos. This should facilitate the eventual complete understanding of how embryos that express MHC class I molecules escape immunological surveillance.
Figure 6.
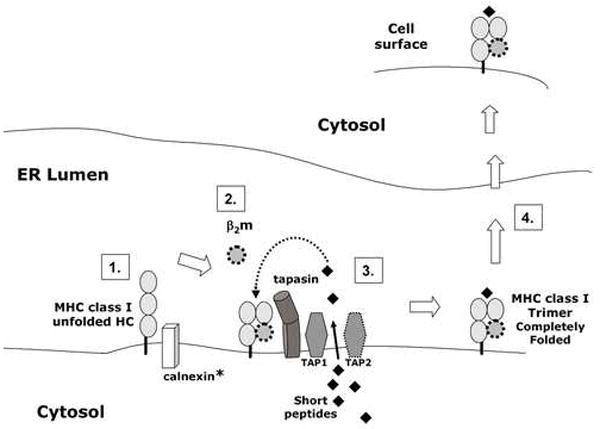
Diagram of likely pathway for MHC class Ia and class Ib cell surface expression. The illustration is based on results presented in this paper and a recent review of MHC class I antigen presentation (Cresswell et al., 2005). 1. In the endoplasmic reticulum (ER), the MHC class Ia heavy chain (HC) interacts with calnexin to facilitate folding into its native conformation. *However, Qa-2 (MHC class Ib) lacks the calnexin binding site and is likely to fold in the absence of calnexin. 2. Properly folded MHC class I heavy chain noncovently binds to β2 microglobulin (β2m). 3. The TAP complex (TAP1/TAP2), tapasin, MHC heavy chain and β2m are part of the peptide loading complex. The TAP complex actively transports short peptides, produced by proteosome-mediated degradation of proteins, into the ER lumen from the cytosol. Tapasin facilitates the loading of high affinity peptide into the MHC class I peptide binding groove. 4. After the MHC class I trimer (heavy chain, β2m and peptide) is assembled, the MHC class I protein is transported to the embryo cell surface via the Golgi apparatus. ER=endoplasmic reticulum; HC= heavy chain; β2m= β2 microglobulin; TAP = transporter associated with antigen processing.
Acknowledgments
This research was supported by NIH grant HD39215 and the Bernard M. Gordon Center for Subsurface Sensing and Imaging Systems at Northeastern University (NSF EEC-9986821). We thank Michele Mammolenti and Robert Crooker for expert animal care, Dr. Gary Laevsky and William Warger for help with microscopy, and Judith Newmark for critical review of the manuscript. We thank also Dr. Luc Van Kaer at Vanderbilt University for the kind gift of the tapasin knockout mice.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Balakier H, Dziak E, Sojecki A, Librach C, Michalak M, Opas M. Calcium-binding proteins and calcium-release channels in human maturing oocytes, pronuclear zygotes and early preimplantation embryos. Hum Reprod. 2002;17:2938–2947. doi: 10.1093/humrep/17.11.2938. [DOI] [PubMed] [Google Scholar]
- Calvert ME, Digilio LC, Herr JC, Coonrod SA. Oolemmal proteomics -identification of highly abundant heat shock proteins and molecular chaperones in the mature mouse egg and their localization on the plasma membrane. Reprod Biol Endocrinol. 2003;1:27. doi: 10.1186/1477-7827-1-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang EY, Henson M, Stroynowski I. The nonclassical major histocompatibility complex molecule Qa-2 protects tumor cells from NK cell- and lymphokine-activated killer cell-mediated cytolysis. J Immunol. 2002;168:2200–2211. doi: 10.4049/jimmunol.168.5.2200. [DOI] [PubMed] [Google Scholar]
- Chiang EY, Henson M, Stroynowski I. Correction of defects responsible for impaired Qa-2 class Ib MHC expression on melanoma cells protects mice from tumor growth. J Immunol. 2003;170:4515–4523. doi: 10.4049/jimmunol.170.9.4515. [DOI] [PubMed] [Google Scholar]
- Chiang EY, Stroynowski I. A nonclassical MHC class I molecule restricts CTL-mediated rejection of a syngeneic melanoma tumor. J Immunol. 2004;173:4394–4401. doi: 10.4049/jimmunol.173.7.4394. [DOI] [PubMed] [Google Scholar]
- Comiskey M, Domino KE, Warner CM. HLA-G is found in lipid rafts and can act as a signaling molecule. Hum Immunol. 2007;68:1–11. doi: 10.1016/j.humimm.2006.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JC, Dealtry GB, Ahmed MA, Arck PC, Klapp BF, Blois SM, Fernandez N. An impaired breeding phenotype in mice with a genetic deletion of beta-2 microglobulin and diminished MHC class I expression: role in reproductive fitness. Biol Reprod. 2007;77:274–279. doi: 10.1095/biolreprod.106.057125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JC, Fernandez N, Joly E, Dealtry GB. Regulation of major histocompatibility complex and TAP gene products in preimplantation mouse stage embryos. Am J Reprod Immunol. 1998;40:165–171. doi: 10.1111/j.1600-0897.1998.tb00408.x. [DOI] [PubMed] [Google Scholar]
- Cresswell P, Ackerman AL, Giodini A, Peaper DR, Wearsch PA. Mechanisms of MHC class I-restricted antigen processing and cross-presentation. Immunol Rev. 2005;207:145–157. doi: 10.1111/j.0105-2896.2005.00316.x. [DOI] [PubMed] [Google Scholar]
- Elliott T, Williams A. The optimization of peptide cargo bound to MHC class I molecules by the peptide-loading complex. Immunol Rev. 2005;207:89–99. doi: 10.1111/j.0105-2896.2005.00311.x. [DOI] [PubMed] [Google Scholar]
- Garbi N, Tan P, Diehl AD, Chambers BJ, Ljunggren HG, Momburg F, Hammerling GJ. Impaired immune responses and altered peptide repertoire in tapasin-deficient mice. Nat Immunol. 2000;1:234–238. doi: 10.1038/79775. [DOI] [PubMed] [Google Scholar]
- Grandea AG, 3rd, Golovina TN, Hamilton SE, et al. Impaired assembly yet normal trafficking of MHC class I molecules in Tapasin mutant mice. Immunity. 2000;13:213–222. doi: 10.1016/s1074-7613(00)00021-2. [DOI] [PubMed] [Google Scholar]
- Grandea AG, 3rd, Van Kaer L. Tapasin: an ER chaperone that controls MHC class I assembly with peptide. Trends Immunol. 2001;22:194–199. doi: 10.1016/s1471-4906(01)01861-0. [DOI] [PubMed] [Google Scholar]
- He X, Tabaczewski P, Ho J, Stroynowski I, Garcia KC. Promiscuous antigen presentation by the nonclassical MHC Ib Qa-2 is enabled by a shallow, hydrophobic groove and self-stabilized peptide conformation. Structure (Camb) 2001;9:1213–1224. doi: 10.1016/s0969-2126(01)00689-x. [DOI] [PubMed] [Google Scholar]
- Joyce S, Tabaczewski P, Angeletti RH, Nathenson SG, Stroynowski I. A nonpolymorphic major histocompatibility complex class Ib molecule binds a large array of diverse self-peptides. J Exp Med. 1994;179:579–588. doi: 10.1084/jem.179.2.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke X, Warner CM. Regulation of Ped gene expression by TAP protein. J Reprod Immunol. 2000;46:1–15. doi: 10.1016/s0165-0378(99)00053-4. [DOI] [PubMed] [Google Scholar]
- Kumanovics A, Takada T, Lindahl KF. Genomic organization of the mammalian MHC. Annu Rev Immunol. 2003;21:629–657. doi: 10.1146/annurev.immunol.21.090501.080116. [DOI] [PubMed] [Google Scholar]
- Margolese L, Waneck GL, Suzuki CK, Degen E, Flavell RA, Williams DB. Identification of the region on the class I histocompatibility molecule that interacts with the molecular chaperone, p88 (calnexin, IP90) J Biol Chem. 1993;268:17959–17966. [PubMed] [Google Scholar]
- Margulies DH, McCluskey J. The major histocompatibility complex and its encoded proteins. In: Paul W, editor. Fundamental Immunology. Lippincott, Williams and Wilkins; Philadelphia, PA: 2003. pp. 571–605. [Google Scholar]
- McElhinny AS, Kadow N, Warner CM. The expression pattern of the Qa-2 antigen in mouse preimplantation embryos and its correlation with the Ped gene phenotype. Mol Hum Reprod. 1998;4:966–971. doi: 10.1093/molehr/4.10.966. [DOI] [PubMed] [Google Scholar]
- Medawar P. Some immunological and endocrinological problems raised by the evolution of viviparity in vertebrates. Symp Soc Exp Biol. 1953;7:320–338. [Google Scholar]
- Niederkorn JY, Chiang EY, Ungchusri T, Stroynowski I. Expression of a nonclassical MHC class Ib molecule in the eye. Transplantation. 1999;68:1790–1799. doi: 10.1097/00007890-199912150-00025. [DOI] [PubMed] [Google Scholar]
- Park B, Ahn K. An essential function of tapasin in quality control of HLA-G molecules. J Biol Chem. 2003a;278:14337–14345. doi: 10.1074/jbc.M212882200. [DOI] [PubMed] [Google Scholar]
- Park B, Lee S, Kim E, Ahn K. A single polymorphic residue within the peptide-binding cleft of MHC class I molecules determines spectrum of tapasin dependence. J Immunol. 2003b;170:961–968. doi: 10.4049/jimmunol.170.2.961. [DOI] [PubMed] [Google Scholar]
- Pearce RB, Trigler L, Svaasand EK, Peterson CM. Polymorphism in the mouse Tap-1 gene. Association with abnormal CD8+ T cell development in the nonobese nondiabetic mouse. J Immunol. 1993;151:5338–5347. [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotzschke O, Falk K, Stevanovic S, Grahovac B, Soloski MJ, Jung G, Rammensee HG. Qa-2 molecules are peptide receptors of higher stringency than ordinary class I molecules. Nature. 1993;361:642–644. doi: 10.1038/361642a0. [DOI] [PubMed] [Google Scholar]
- Sadasivan B, Lehner PJ, Ortmann B, Spies T, Cresswell P. Roles for calreticulin and a novel glycoprotein, tapasin, in the interaction of MHC class I molecules with TAP. Immunity. 1996;5:103–114. doi: 10.1016/s1074-7613(00)80487-2. [DOI] [PubMed] [Google Scholar]
- Seliger B, Wollscheid U, Momburg F, Blankenstein T, Huber C. Characterization of the major histocompatibility complex class I deficiencies in B16 melanoma cells. Cancer Res. 2001;61:1095–1099. [PubMed] [Google Scholar]
- Shiroishi M, Kuroki K, Rasubala L, et al. Structural basis for recognition of the nonclassical MHC molecule HLA-G by the leukocyte Ig-like receptor B2 (LILRB2/LIR2/ILT4/CD85d) Proc Natl Acad Sci USA. 2006;103:16412–16417. doi: 10.1073/pnas.0605228103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spies T, Cerundolo V, Colonna M, Cresswell P, Townsend A, DeMars R. Presentation of viral antigen by MHC class I molecules is dependent on a putative peptide transporter heterodimer. Nature. 1992;355:644–646. doi: 10.1038/355644a0. [DOI] [PubMed] [Google Scholar]
- Tabaczewski P, Chiang E, Henson M, Stroynowski I. Alternative peptide binding motifs of Qa-2 class Ib molecules define rules for binding of self and nonself peptides. J Immunol. 1997;159:2771–2781. [PubMed] [Google Scholar]
- Tabaczewski P, Stroynowski I. Expression of secreted and glycosylphosphatidylinositol-bound Qa-2 molecules is dependent on functional TAP-2 peptide transporter. J Immunol. 1994;152:5268–5274. [PubMed] [Google Scholar]
- Ungchusri T, Chiang EY, Brown G, Chen M, Tabaczewski P, Timares L, Stroynowski I. Widespread expression of the nonclassical class I Qa-2 antigens in hemopoietic and nonhemopoietic cells. Immunogenetics. 2001;53:455–467. doi: 10.1007/s002510100347. [DOI] [PubMed] [Google Scholar]
- Wang X, Seed B. A PCR primer bank for quantitative gene expression analysis. Nucleic Acids Res. 2003;31:e154. doi: 10.1093/nar/gng154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner CM. Immunological aspects of embryo development. In: Cohen J, Elder K, editors. Human Embryo Evaluation and Selection. Parthenon Publishing Group; United Kingdom: 2007. In press. [Google Scholar]
- Warner CM, Brenner CA. Genetic regulation of preimplantation embryo survival. Curr Top Dev Biol. 2001;52:151–192. doi: 10.1016/s0070-2153(01)52011-6. [DOI] [PubMed] [Google Scholar]
- Warner CM, Brownell MS, Rothschild MF. Analysis of litter size and weight in mice differing in Ped gene phenotype and the Q region of the H-2 complex. J Reprod Immunol. 1991;19:303–313. doi: 10.1016/0165-0378(91)90042-o. [DOI] [PubMed] [Google Scholar]
- Warner CM, Gollnick SO. Expression of H-2K major histocompatibility antigens on preimplantation mouse embryos. Biol Reprod. 1993;48:1082–1087. doi: 10.1095/biolreprod48.5.1082. [DOI] [PubMed] [Google Scholar]
- Warner CM, Gollnick SO, Flaherty L, Goldbard SB. Analysis of Qa-2 antigen expression by preimplantation mouse embryos: possible relationship to the preimplantation-embryo-development (Ped) gene product. Biol Reprod. 1987;36:611–616. doi: 10.1095/biolreprod36.3.611. [DOI] [PubMed] [Google Scholar]
- Warner CM, Newmark JA, Comiskey M, et al. Genetics and imaging to assess oocyte and preimplantation embryo health. Reprod Fertil Dev. 2004;16:729–741. doi: 10.1071/rd04088. [DOI] [PubMed] [Google Scholar]
- Waterston RH, Lindblad-Toh K, Birney E, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Watkins A, Wilkins A, Osmond C, Warner CM, Comiskey M, Hanson M, Fleming TP. The influence of mouse Ped gene expression on postnatal development. J Physiol. 2006;571:211–220. doi: 10.1113/jphysiol.2005.099192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams AP, Peh CA, Purcell AW, McCluskey J, Elliott T. Optimization of the MHC class I peptide cargo is dependent on tapasin. Immunity. 2002;16:509–520. doi: 10.1016/s1074-7613(02)00304-7. [DOI] [PubMed] [Google Scholar]
- Williams DB. Beyond lectins: the calnexin/calreticulin chaperone system of the endoplasmic reticulum. J Cell Sci. 2006;119:615–623. doi: 10.1242/jcs.02856. [DOI] [PubMed] [Google Scholar]


