Abstract
cDNAs of Anopheles gambiae Defensin 2 (AgDef2), Defensin 3 (AgDef3) and Defensin 4 (AgDef4), identified in the genome sequence, have been characterized and their expression profiles investigated. In contrast to both typical defensins and insect antimicrobial peptides generally, the newly identified defensins were not upregulated with acute-phase kinetics following immune challenge in insects or cell culture. However, mRNA abundance of AgDef2, AgDef3 and AgDef4 increased significantly during the larval stages. Promoter analysis of all three genes failed to identify putative immune response elements previously identified in other mosquito defensin genes. As previous studies failed to identify these larval-specific defensins, it seems likely that further antimicrobial peptide genes with nontypical expression profiles will be identified as more genome sequences become available.
Keywords: defensin genes, mosquito, Anopheles gambiae, larval-specific, antimicrobial peptide
Introduction
Mosquitoes are among the most important vectors of human disease with Anopheles gambiae being the principal vector of human malaria in sub-Saharan Africa. In response to infection, mosquitoes mount an effective immune response (Lehane et al., 2004). A primary defence mechanism, forming part of the innate immune response, is the transcription of antimicrobial peptide (AMP) genes. Insect AMPs can be divided into four major families based on sequence similarity, namely the cecropins, cysteine-rich antimicrobial peptides (which include the defensins), glycine-rich polypeptides and proline-rich peptides (Meister et al., 1997). Insect defensins form a key component of innate immunity and have been found in every species investigated (reviewed in Bulet & Stocklin, 2005). They are small, cationic peptides, typically 33-46 amino acids, characterized by six highly conserved cysteine residues that form three intramolecular disulphide bridges. This results in their characteristic 3D structure consisting of an N-terminal loop, an α-helix and two antiparallel β sheets (Cornet et al., 1995), which is distinct from the structure of mammalian defensins.
An insect defensin (DmDef) was first fully characterized in Drosophila melanogaster (Dimarcq et al., 1994). Subsequently, Defensin 1 (AgDef1) was isolated from An. gambiae (Richman et al., 1996; Eggleston et al., 2000) and this led to a comprehensive range of studies on its expression profiles and activity against both bacteria and malaria parasites (Dimopoulos et al., 1997, 1998; Richman et al., 1997; Vizioli et al., 2001a; Blandin et al., 2002; Meredith et al., 2006). Three additional putative defensin genes were predicted following analysis of the An. gambiae genome (Christophides et al., 2002). Other AMPs isolated from An. gambiae include cecropins 1 to 3 (Vizioli et al., 2000; Zheng & Zheng, 2002) with a fourth cecropin plus an attacin identified in the genome sequence (Christophides et al., 2002) and Gambicin (Vizioli et al., 2001b), a novel cysteine-rich peptide. Defensins, cecropins and gambicin have been isolated from a number of other mosquitoes, including the characterisation of defensins A (AaDefA) and C (AaDefC) (Chalk et al., 1995; Lowenberger et al., 1995; Cho et al., 1996) and cecropins A, B and C (Lowenberger et al., 1999a; Sun & Fallon, 2002) from Aedes mosquitoes. Additional putative AMP genes have also been identified following analysis of the newly released Aedes aegypti genome sequence (Waterhouse et al., 2007). These include two more defensins (AaDefD and AaDefE), an attacin, additional cecropins as well as putative diptericin and holotricin genes.
In D. melanogaster, the ‘model organism’ for Diptera, seven distinct AMPs are induced following immune challenge. Involvement of the Rel signal transduction pathways Toll and Imd has been demonstrated for all D. melanogaster AMPs (reviewed in Engstrom, 1999) and NF-κB binding sites, which bind Rel-type transcription factors, are conserved in their proximal regulatory regions. Publication of the An. gambiae genome sequence enabled many components of both the Toll and Imd signal transduction pathways to be identified (Christophides et al., 2002). More recently, Luna et al. (2006) have demonstrated the involvement of both pathways in the expression of An. gambiae Cecropin 1, AgDef1 and Gambicin. Following pathogen challenge, insect AMPs are typically upregulated with acute-phase kinetics, such that mosquito peptides and cDNAs are usually isolated from immune challenged insects or cell lines. In addition, cis-regulatory elements resembling those involved in the mammalian acute-phase response are commonly identified within insect AMP proximal promoter regions and we have demonstrated the requirement for NF-κB binding sites in the immune stimulation of both AgDef1 and AaDefA (Meredith et al., 2006).
In common with other insect AMPs, mosquito defensin transcripts have been observed at a low level in unchallenged, naïve populations (Dimarcq et al., 1994; Dimopoulos et al., 1997; Lowenberger et al., 1999b) and basal levels of transcription are evident in reporter assays (Meredith et al., 2006). However, to date, all well-characterized insect AMPs have been shown to be upregulated following immune stimulation, as would be anticipated from their profiles of cis-regulatory elements associated with the Rel signal transduction pathway. Alongside this immune stimulation, there is also evidence for AMP upregulation in the absence of external immune challenge during metamorphosis, because DmDef, AgDef1 and AaDefC are all up-regulated during the pupal stage (Dimarcq et al., 1994; Richman et al., 1996; Lowenberger et al., 1999c).
Here we describe the characterization and expression profiles of three newly identified defensin genes, first predicted in the An. gambiae genome sequence. In common with our previous characterisation of AgDef1, we have named these new genes AgDef2, AgDef3 and AgDef4. Although transcripts for all three peptides indicate basal levels of expression during all life stages, we were unable to demonstrate immune stimulation consistent with an acute-phase response. This was despite using highly sensitive reporter assays and real-time quantitative PCR. Instead, we find that high and consistent levels of upregulation for all three genes are confined to the larval stages.
Results
Identification and characterisation of AgDef2, AgDef3 and AgDef4 cDNAs
The An. gambiae PEST strain genomic sequence identified three putative defensin genes (Christophides et al., 2002), in addition to the previously characterized AgDef1 (Eggleston et al., 2000; Meredith et al., 2006). The four An. gambiae defensin genes are dispersed, with AgDef1 located at 3L (42A), AgDef2 at 2R (19D), AgDef3 at 2L (27A) and AgDef4 at 2L (22B).
Full-length cDNA sequences were identified by RACE using infected blood-fed mosquito cDNA for AgDef3 and cDNA from unchallenged mid-stage larvae for AgDef2 and AgDef4. 5′ and 3′ RACE products for all three genes were cloned and sequenced, together with the internal PCR product for AgDef3. This enabled us to piece together the entire cDNA sequences of the An. gambiae Keele strain defensin genes. These sequence data have been submitted to the DDBJ/EMBL/GenBank databases under accession numbers AY973195 (AgDef2), AY907825 (AgDef3) and AY973196 (AgDef4). The cDNA for AgDef2 was 292 bp with 5′ and 3′ UTRs of 35 and 14 bp, respectively. For AgDef3, a nested 5′ RACE reaction generated two different sized products. Of 96 clones screened, 85 had smaller inserts corresponding to a transcription start site 63 bp upstream of the putative initiating AUG. This is subsequently referred to as the major transcription start site (TSS). The remaining 11 clones had larger inserts representing an additional minor TSS, 97 bp upstream of the translation start site. Transcription from the AgDef3 major TSS resulted in a cDNA of 362 bp, with a 63 bp 5′ UTR and a 95 bp 3′ UTR. For AgDef4 the cDNA was 748 bp with 5′ and 3′ UTRs of 113 and 349 bp, respectively (Fig. 1). The Keele strain AgDef2 and AgDef3 cDNA sequences were co-linear with the published PEST strain genomic sequence. Conversely, alignment of the AgDef4 cDNA sequence with the published genomic sequence identified an intron of 103 bp after nucleotide 211 of the cDNA (Fig. 1) which is bounded by highly conserved splice donor and acceptor sites. Amino acid alignments of the Keele and PEST strain inferred defensin peptide sequences identified a single coding change, from threonine to alanine at the C terminus of AgDef3. There were no nucleotide substitutions between the Keele and PEST AgDef2 sequences, six silent substitutions between the AgDef3 sequences (two in the coding region and four in the 3′ UTR) and 10 differences between the AgDef4 sequences (two substitutions in the 5′ UTR and eight changes or deletions in the Keele strain 3′ UTR). Putative polyadenylation signals (AATAAA) were identified at nucleotides 267-272, 340-345 and 533-538 for AgDef2, 3 and 4, respectively. For AgDef2, which has a very short 3′ UTR, this putative signal is just within the coding region.
Figure 1.
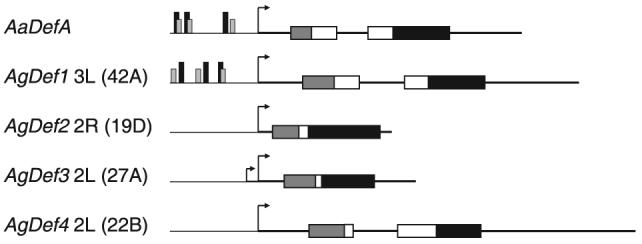
A schematic of mosquito defensin genes. Anopheles gambiae AgDef2, 3 and 4 are aligned with the previously characterized AgDef1 and Aedes aegypti AaDefA (Cho et al., 1996; Meredith et al., 2006). Chromosome number and location are indicated for the An. gambiae genes. The transcription start site (TSS, arrow) separates promoter region (fine line) from transcribed region (bold line). Functional transcription factor binding sites within 200 bp of the TSS (vertical bars) as identified in (Meredith et al., 2006) are κB (black) and C/EBP (grey). No such homologous regions have been located in AgDef2, 3 or 4. Translated regions are indicated by horizontal boxes and show signal sequence (grey), pro-piece (which is separated by an intron in AaDefA, AgDef1 and AgDef4; white) and mature defensin (black).
Phylogenetic analysis has placed DmDef and AgDef1 in Clade I, together with the Aedes defensins, whereas the AgDef2, AgDef3 and AgDef4 peptides are assigned to Clade IV alongside other highly divergent defensins (Christophides et al., 2002). The inferred preprodefensin sequences for the three newly identified An. gambiae defensins were aligned with DmDef, AgDef1 and the two characterized Ae. aegypti defensins (Fig. 2). Amino acid sequences for the new An. gambiae defensins indicate preprodefensin-like open reading frames of 80 amino acids for AgDef2, 67 amino acids for AgDef3 and 94 amino acids for AgDef4, compared to 102, 98, 99 and 92 amino acids for AgDef1, AaDefA, AaDefC and DmDef, respectively (Fig. 2). The deduced amino-terminal regions for AgDef2, AgDef3 and AgDef4 gave good signal sequence predictions with Signal P (Nielsen et al., 1997), but with different predicted cleavage sites between VSG and TT for AgDef2, GEA and QL for AgDef3 and TFA and NP for AgDef4. The signal sequence (‘pre’ region) precedes a ‘prosegment’, which is cleaved to convert the peptide to an active mature defensin. This ‘pro’ region shows considerable length and sequence variation. For AgDef4 the ‘pre’ and ‘pro’ regions, of 27 and 35 amino acids respectively, are of similar length to those of AgDef1 (Fig. 2). Conversely, AgDef2 and AgDef3 have signal sequences of 20 and 22 amino acids respectively but very short ‘pro’ regions of just six and four amino acids respectively. The mature defensins for AgDef2, 3 and 4 are not preceded by a putative lysine-arginine proteolytic cleavage site, which is conserved in other insect defensins including AgDef1 (Dimarcq et al., 1994). There are however other exceptions, with Tenebrio molitor Tenecin1 and Apis mellifera Defensin predicted to cleave at lysine-valine (Casteels-Josson et al., 1994; Moon et al., 1994) and Stomoxys calcitrans Defensin 1 predicted to cleave at valine-alanine, (Lehane et al., 1997).
Figure 2.
Alignment of preprodefensin sequences. Deduced amino acid sequences for Anopheles gambiae AgDef1, AgDef2, AgDef3 and AgDef4 are aligned with Drosophila melanogaster DmDef (Dimarcq et al., 1994) and Aedes aegypti AaDefA and AaDefC (Chalk et al., 1995; Lowenberger et al., 1995; Cho et al., 1996) using Clustal with the PAM250 residue weight table. The top panel shows PrePro sequences with the putative signal peptide underlined. The lower panel aligns mature peptide sequences with the conserved cysteine residues boxed.
The predicted mature peptides for AgDef2, 3 and 4 retain six characteristic conserved cysteine residues (Fig. 2) but vary in length. The classic defensins from D. melanogaster (DmDef), Ae. aegypti (AaDefA and C) and An. gambiae (AgDef1) are all 40 amino acids in length with 24 out of the 40 amino acids being completely conserved. The mature defensins expressed from AgDef2, 3 and 4 are 54, 40 and 31 amino acids, respectively, and, apart from the six conserved cysteine residues, only two glycines are conserved among all three (Fig. 2) and only one of these is also conserved in AgDef1, AaDefA, AaDefC and DmDef.
Transcription profiles of AgDef2, AgDef3 and AgDef4
Real-time PCR, quantified relative to 18S rRNA, was employed to investigate mRNA abundance levels of AgDef2, 3 and 4. As AgDef1, in common with other defensins, is upregulated following immune challenge, total RNA extracted from infected and non-infected mosquitoes was used in initial experiments. AgDef3 mRNA was detectable at very low levels in control adult mosquitoes; however we were unable to show an increase in abundance following a blood meal, an infected blood meal, sterile injection or bacterial injection (data not shown). By including RNA samples from different life stages in the study, we identified a significant increase in mRNA abundance for AgDef2, 3 and 4 from the larval stages (Fig. 3). Expression in embryos, pupae and adults was extremely low, with no significant difference in mRNA abundance at these life stages (P > 0.05). For all three genes, mRNA abundance in larvae was significantly increased compared to that in embryos. AgDef3 and AgDef4 mRNAs were significantly increased (P < 0.05) during all larval instars and AgDef2 was significantly up-regulated (P < 0.05) in the later larval instars 2/3 and 4. Abundance of mRNA peaked during larval instars 2/3 for AgDef2 and AgDef3 (Fig. 3A and B) but the peak was earlier (larval instar 1) for AgDef4 (Fig. 3C). To investigate whether larval AgDef3 transcriptional activity could be immune stimulated, fourth instar larvae were challenged with bacteria and left to recover for 12 h, a time at which AgDef1 had previously been shown to be upregulated by bacterial challenge (Richman et al., 1996). No further increase in mRNA abundance was detected in fourth instar larvae after sterile or bacterial injection or following bacterial infection by feeding.
Figure 3.
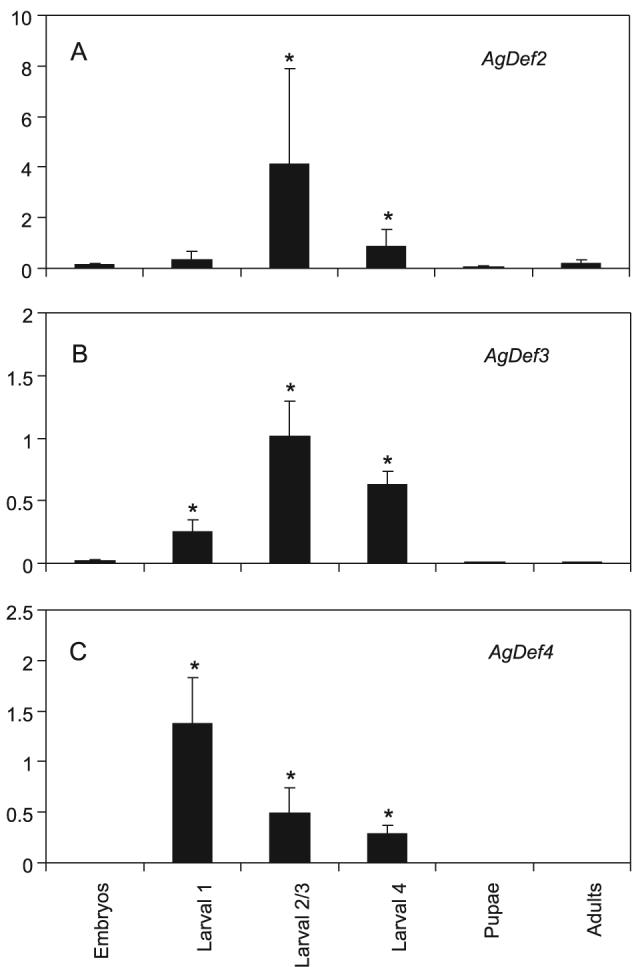
Transcription profiles for Anopheles gambiae defensins in unchallenged life stages (A, AgDef2; B, AgDef3 and C, AgDef4). The graphs show relative mRNA abundance levels for AgDef2, 3 and 4 (y-axis) following quantitative real-time PCR on cDNA from different life stages (x-axis). Bars represent means with standard errors from triplicate reactions performed on three separate occasions. Statistical analysis was on data normalized to fourth larval instar, with significant differences to mRNA abundance in embryos indicated by asterisks (P < 0.05 for all comparisons).
Expression from the AgDef2, AgDef3 and AgDef4 promoters
Using PCR amplification we cloned approximately 1.4 kb of promoter sequence, including the entire 5′ UTR, for AgDef2, 3 and 4. Nucleotides from −1337 to +35 (AgDef2), −1321 to +63 (AgDef3) and −1228 to +113 (AgDef4) were cloned into the luciferase reporter plasmid pGL3-Basic. Constructs were verified by sequence analysis and data submitted to the DDBJ/EMBL/GenBank databases under accession numbers DQ137803 (AgDef2), AY907824 (AgDef3) and DQ137804 (AgDef4).
Inspection of the genomic sequences surrounding the transcription start sites (TSS) identified putative arthropod initiator sequences (consensus TCAGT, Cherbas & Cherbas, 1993) either one base pair downstream of the TSS (AgDef2, 4 and AgDef3 minor TSS) or overlapping the start site (AgDef3 major TSS). In addition, putative TATA boxes were identified 21 and 31 nucleotides upstream of the transcription start site for AgDef2 and 4, respectively, but not for AgDef3. We were unable to identify downstream promoter elements (consensus GWCG, Willy et al., 2000) in close proximity to the TSS for any of the genes. TFSearch software (http://www.rwcp.or.jp/papia/) was used to identify putative transcription factor binding sites within these promoter regions. A number of putative NF-κB binding sites, with at least 80% homology to the insect consensus (GGGRNTYYYY, Kappler et al., 1993) were identified in the AgDef2 and AgDef3 promoter sequences. However, none were within 200 base pairs of the TSS (AgDef2 −378 to −369, −417 to −408, −867 to −876 and −1090 to −1081; AgDef3 −816 to −807 and −933 to −924), as observed previously for the classical mosquito defensins AgDef1 and AaDefA (Fig. 1 and Meredith et al., 2006). Also within the promoters of these classical defensins, we had previously identified C/EBP binding sites (consensus TKNNGYAAK, Ryden & Beemon, 1989) which overlap, or are very closely associated with the NF-κB binding sites and which are also required for transcriptional regulation (Meredith et al., 2006). As a result of their plasticity, a large number of putative C/EBP binding sequences were identified in the AgDef2, 3 and 4 promoters, but none were associated with the putative NF-κB binding sites closest to the TSS in AgDef2, and other putative binding sites for NF-κB were located a considerable distance from the TSS. We additionally identified a number of putative binding sites for GATA factors (consensus WGATAR, Evans et al., 1990) within the AgDef2, 3 and 4 promoters, but none were within 12 bp of putative NF-κB binding sites as reported for other insect immune genes (Kadalayil et al., 1997). Dorsal binding sites (consensus NGRGAAAANCN, Thisse et al., 1991) also bind Rel-like transcription factors and D. melanogaster Dorsal has been implicated in the expression of both Defensin (Han & Ip, 1999) and Drosomycin in the larval fat body (Manfruelli et al., 1999). We identified a number of putative Dorsal binding sites in all three promoters (AgDef2: −584 to −574, −681 to −691; AgDef3: −157 to −147, −260 to −250 and AgDef4: −353 to −363 and −707 to −716). We also identified putative Dorsal binding sites in the AgDef1 promoter region (−18 to −108 and −193 to −183), but their involvement in AgDef1 expression has not been investigated. In conclusion, we were unable to identify putative transcription factor binding sites indicative of conventional immune regulation of classical insect defensins.
Following transfection into An. gambiae Sua 4.0 cells, basal luciferase activity was detected from all three promoter constructs, although levels varied among promoters. Basal activities compared to the promoterless pGL3 control were threefold higher for AgDef2, ninefold higher for AgDef3 and 37-fold higher for AgDef4 (Fig. 4). For comparison, basal activity of the classical defensin AgDef1 was 25-fold higher than the promoterless control. We had previously shown that immune stimulation with lipopolysaccharide (LPS) and Micrococcus luteus, but not laminarin, significantly upregulated the activity of AgDef1 (Meredith et al., 2006 and Fig. 4). Following immune stimulation of the new defensin reporter constructs, only that for AgDef3 treated with M. luteus showed a modest but significant upregulation (P = 0.0062, Fig. 4). This contrasts with no detectable immune stimulation of AgDef3 by M. luteus in whole adults or larvae using real-time PCR and most probably reflects the extreme sensitivity of the cell-based dual-luciferase reporter assay. Although the activity of AgDef2 was significantly altered following challenge with both M. luteus and laminarin but not Escherichia coli, this reflected a modest downregulation (0.83 and 0.87 times basal activity, respectively) and is probably also a reflection of the sensitivity of the dual-luciferase assay. The activity of the AgDef4 promoter was not significantly altered by immune challenge.
Figure 4.
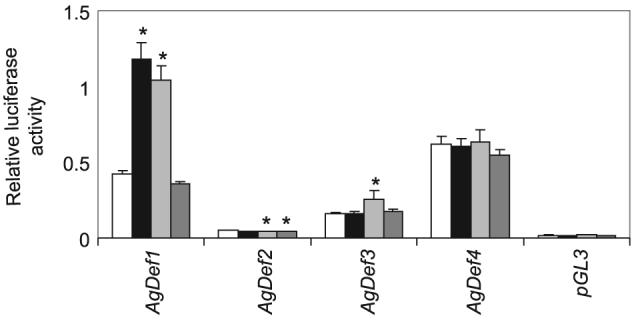
Normalized luciferase activities of Anopheles gambiae defensin promoters following transfection of luciferase reporter constructs into Sua 4.0 cells. Relative light units (y-axis) are shown for reporter constructs in untreated cells (open bars) or immune stimulated cells: 100 μg/ml LPS (black bars); 2 × 108 bacterial/ml – Micrococcus luteus (light grey bars); 20 μg/ml laminarin (dark grey bars). The means and standard errors of eight replicates, performed on two separate occasions, are shown. Significant differences between luciferase activities in untreated and immune stimulated cells are indicated by asterisks (P < 0.05).
We note that the response to septic injury of the Drosophila Diptericin promoter increases during the third larval instar and is linked to an increase in transcriptional activity by the steroid ecdysone in the salivary gland (Meister & Richards, 1996). We identified a putative ecdysone response element in the AgDef3 promoter at nucleotides −517 to −529 (consensus RGKTCANTGAMCY, Cherbas et al., 1991). Functionality of this putative element was tested by the addition of 20-hydroxyecdysone to cells transfected with the AgDef3 reporter construct, but no increase in luciferase activity was observed (data not shown).
Discussion
We have described the gene structure and expression profiles of three putative defensin genes, AgDef2, AgDef3 and AgDef4, previously identified in the An. gambiae genome sequence (Christophides et al., 2002). We have demonstrated low level constitutive expression for all three genes, at all life stages and shown significant increases in mRNA abundance during the larval stages. We believe this to be the first report of AMP genes whose upregulation is exclusively linked to the larval stages. We were unable to increase mRNA abundance of AgDef3 following infection of adults with parasites or bacteria, or challenge of larvae with bacteria. By means of a very sensitive luciferase reporter assay we attempted immune upregulation of all three promoters using a range of stimuli. However, we were only able to identify modest changes in activity that were not detectable in intact insects. This lack of an acute-phase response is consistent with the apparent absence of proximal NF-κB motifs in the promoter regions of all three genes. Defensin, or defensin-like genes, from both St. calcitrans and Spodoptera frugiperda are also reported to lack functional NF-κB like motifs and show atypical up-regulation (Munks et al., 2001; Volkoff et al., 2003).
For An. gambiae, mRNA abundance of all three newly characterized defensins was significantly upregulated during larval stages. Levels of AgDef2 and AgDef3 mRNA peaked during the second and third instars and AgDef4 mRNA was most abundant during the first larval instar. Expression of these genes might be a response to bacterially infected water or could represent developmental regulation during larval stages to pre-empt bacterial infection. The former seems unlikely as we were unable to stimulate an acute-phase response either in cell culture or of AgDef3 in intact insects. We can not, however, rule out the possibility of post-transcriptional immune regulation, particularly because there is evidence for translational control of other defensins expressed by Ae. aegypti (Bartholomay et al., 2004) and St. calcitrans (Hamilton et al., 2002).
Developmental regulation of other insect AMP genes has been reported (although always in addition to immune stimulation), including dual roles in immunity and development postulated for sapecins, which are expressed at both embryonic and early pupal stages in the flesh fly Sarcophaga peregrina (Matsuyama & Natori, 1988; Natori et al., 1999). Upregulation in non-induced early pupae has also been reported for Dipteran defensins (Dimarcq et al., 1994; Lowenberger et al., 1999c). Lowenberger et al. suggested that the observed expression of Ae. aegypti Defensin C in callow pupae could protect against bacteria released from the larval gut during histolysis or by the entry of pathogens through small tears in the soft cuticle prior to sclerotization. An. gambiae defensins expressed during larval stages could similarly offer protection during histolysis or following larval moults. These defensins could be transcribed prior to a known bacterial challenge as was postulated for St. calcitrans midgut defensins (Munks et al., 2001). More recently, AaDefA has been shown to co-localize with phenoloxidase, suggesting a potential role for defensin in the phenoloxidase-based melanization response (Hillyer & Christensen, 2005). Such a role would be consistent with defensin expression following larval moults or in callow pupae.
Whereas the previously characterized Dipteran defensins are in Clades I and III, Christophides et al. (2002) grouped the three new An. gambiae defensins in Clade IV alongside other highly divergent defensins. This group also includes Heliothis virescens Heliomicin and Sa. peregrina Sapecin B. Heliomicin is exclusively antifungal, with similarities to D. melanogaster Drosomycin (Lamberty et al., 1999), whereas Sapecin B is antibacterial (Yamada & Natori, 1993). Yamada and Natori reported significant similarity between Sapecin B and the scorpion venom toxin charybdotoxin. Like charybdotoxin, Sapecin B was found to be a potent inhibitor of calcium-activated potassium channels (Shimoda et al., 1994) and was detected in the brain of naïve larvae (Lee et al., 1995). AgDef4 is truncated in the loop region between the first and second cysteines, with four amino acids compared to 12 in the other An. gambiae defensins. In this respect AgDef4 closely resembles both Sapecin B with six amino acids and charybdotoxin with five amino acids. The AgDef4 mature peptide shares 33% identity with Sapecin B compared to 37% identity with AgDef3. It remains to be established whether AgDef4 has evolved a role in regulation of potassium channels. The mature AgDef2 is considerably larger than classical defensins, at 54 amino acids, because of a carboxy-terminal extension also identified in bee defensins from A. mellifera and Bombus pascuorum (Casteels-Josson et al., 1994; Klaudiny et al., 2005). The carboxy-terminal extension in bee defensins is reported to adopt an alpha-helical structure, stabilized by amidation, although its function is unknown. The recent analysis of immune-related genes in Ae. aegypti identified two further defensins, AaDefD and AaDefE (Waterhouse et al., 2007). Phylogenetic analysis has placed AaDefD in an orthologous group with AaDefA, AaDefC, AgDef1 and DmDef. Although separate, AaDefE is more closely related to these classical defensins than either AgDef2, AgDef3 or AgDef4. Thus, if these An. gambiae defensins have acquired novel functions, the same may not be true of the newly identified Ae. aegypti defensins. It is interesting to note that analysis of the Ae. aegypti genome sequence also identified a total of 10 putative cecropin genes, compared to four each in An. gambiae and D. melanogaster, in addition to putative diptericin and holotricin genes, all indicative of a large degree of diversity among mosquito AMPs.
An exhaustive study on the evolution of mosquito defensins in particular, and insect defensins in general, has recently been published (Dassanayake et al., 2007). This study identified 65 defensin sequences based on their similarity to AgDef1. The resulting phylogenetic tree also positions AgDef3 and AgDef4 close to Lepidoptera defensins, including Heliomicin, and in the same domain as scorpion venom toxin, although AgDef2 is placed in a different domain. Five of the defensins included in the study, including AgDef3, are predicted to have lost their cationic nature, important for host-membrane interaction, and may therefore possess novel functions.
In mammals, defensin peptides tend to be either constitutively expressed on mucosal surfaces or induced with acute-phase kinetics (reviewed in Kaiser & Diamond, 2000) and AgDef2, 3 or 4 could fall into the former category during larval stages. Developmental expression of mammalian defensins is also well-documented with sheep β-defensin 2 (SBD-2) having significantly greater tissue distribution in foetal and neonatal lambs than adult sheep (Meyerholz et al., 2004). In common with insect Sapecin, mammalian β-defensins have been shown to participate in cellular differentiation and growth in vitro (Frye et al., 2001; McDermott et al., 2001) and it is proposed that mammalian β-defensins are regulated by cellular proliferation and differentiation in addition to microbial pathogens and inflammatory cytokines. Similar expression stimuli could be present during the rapidly growing larval stages of insects such as An. gambiae.
Multiple cecropin AMP genes have been characterized from An. gambiae, Ae. aegypti and D. melanogaster (Kylsten et al., 1990; Sun & Fallon, 2002; Zheng & Zheng, 2002). These genes are clustered within a single locus and would be expected to be under the transcriptional control of the same enhancer and repressor elements. D. melanogaster has a single defensin gene, but other insects present multiple defensin genes. The defensin genes previously characterized from Ae. aegypti are also tightly clustered (Lowenberger et al., 1999c) yet maintain different expression profiles in response to infection (Lowenberger et al., 1999b). Such clustering of related genes is indicative of an evolutionary origin through gene duplication. It was therefore surprising that the four defensin genes in An. gambiae are physically dispersed, which would suggest an independent origin. This is supported by the differences in expression profile among the four genes. Several studies on immune stimulated An. gambiae insects or cell lines failed to isolate transcripts or peptides for AgDef2, 3 or 4, presumably because of their larval specificity. We anticipate that the diversity apparent in An. gambiae defensins might be more widely seen in other insects or vertebrates as more genomes are sequenced or existing genomes are analysed in greater depth. Additional functions have been reported for other insect defensins, which is surprising considering their size. These include a growth factor-like activity for Sapecin in embryonic and pupal stages (Komano et al., 1991) and potent inhibition of calcium-activated potassium channels for Sapecin B (Shimoda et al., 1994). It remains to be seen whether the diversity apparent among the four defensins of An. gambiae reflects additional or alternative functions and why this mosquito species expresses three distinct defensins during the larval stages.
Experimental procedures
Treatment and collection of insect samples
Anopheles gambiae Keele strain mosquitoes (Hurd et al., 2005) were maintained at 26 °C ± 1 °C and 80% humidity in a 12-h light : 12-h dark photoperiod. Stock larvae were reared under standardized conditions (Jahan & Hurd, 1997) and adults were fed 10% glucose ad libitum. For RNA extraction, mosquitoes were collected on ice and frozen immediately at −70 °C. Adult females were collected 3 days post-emergence or 24 h post-bloodmeal, which was given at day 6. Plasmodium yoelii nigeriensis infected mice provided infected bloodmeals. Bacterial challenge was with a mixture of E. coli K12 RM148 and M. luteus. CO2-anaesthetized adults were sham or bacterially injected with 1 μl of lauria broth or a bacterial suspension (OD600 = 2.0) respectively, into the thorax. Insects were left to recover for 18 h before storing at −70 °C. Challenged fourth instar larvae were either left in bacterially infected water for 1 h or pricked with a sharp needle dipped in bacterial slurry. Both groups were allowed to recover for 12 h before freezing. Adult and larval samples were taken at a time when AgDef1 expression was detected (Richman et al., 1996).
5′ and 3′ RACE analysis
5′ and 3′ RACE reactions, performed on total RNA extracted from unchallenged mid-stage larvae for AgDef2 and AgDef4 or P. y. nigeriensis infected adults for AgDef3, were used to amplify cDNA sequences. All reactions used 5 μg total RNA in the GeneRacer protocol (Invitrogen Ltd, Paisley, UK), together with GeneRacer primers and gene-specific primers, following the manufacturer’s instructions. 5′ RACE reactions used gene-specific primers Def2intR (5′-GTTGCAGTAGCCTCCCGTTTTG-3′) for AgDef2, nested primers Def3R2 (5′-GTTGGCCCATTTCGGTCCTTCGTT-3′) followed by Def3R1 (5′-AAGCAGGGCCACCAACAGGAAGGAT-3′) for AgDef3 and Def45race (5′-GTTGCAGCACGCCGGGAAGTTGTTTTG-3′)) for AgDef4. 3′ RACE reactions used primers Def2intF (5′-ACTTATATGTGCAATCGCCGTGTC-3′) for AgDef2, Def3F2 (5′-AAATTCGCCGTAGTATCCTTCCTGTTG-3′) followed by Def3F1 (5′-GTCTGGACGTGGCGCTGGATCTTGTAAC-3′) for AgDef3 and Def4intF (5′-GATTGGTGCCTGGTGCTTTAGTGG-3′) for AgDef4. AgDef2 and AgDef4 5′ and 3′ RACE products overlapped, generating complete cDNA sequences. For AgDef3 the central 272 bp of the cDNA, to overlap 5′ and 3′ RACE sequences, was amplified with Def3F3 (5′-CGGACAGTCAATTACGCAGAAA-3′) and Def3R3 (5′-CTCACTACCAGCTCCTCCACCAT-3′). RT-PCR and cloning reagents were all from Invitrogen. Primers were synthesized by Proligo (Paris, France) and reactions used Platinum Taq DNA polymerase High Fidelity (Invitrogen). The resulting PCR products were TOPO cloned (Invitrogen) and sequenced (Lark Technologies, Takeley, UK) prior to analysis.
Quantitative real-time PCR
cDNA was generated from total RNA as described above, with the modification that 5 μg RNA was used in reverse transcription with 250 ng random primers (Invitrogen). A dilution of this cDNA (1 in 4 or 1 in 8) was used for each quantitative PCR reaction. Reactions were performed with an ABI Prism 7000 Sequence Detection System (Applied Biosystems, Warrington, UK) with plates, TaqMan Universal PCR Mastermix and assays from the same company. Assays by Design (Applied Biosystems) included forward and reverse primers with FAM-labelled probe. Assay Def2292Q-DF2E used primers 5′-GGGAACGACAGTTACATTACAATCC-3′ and 5′-GCTCGGCATGTGCATAGC-3′ with probe 5′-CTGTTGCAGTAGCCTCC-3′, AgDef3-sNN-df3b used primers 5′-TCTTCGCGGACAGTCAATTACG-3′ and 5′-ATACTACGGCGAATTTCATCTTGGA-3′ with probe 5′-CTGCTCTGCGATTTC-3′ and assay Def4398Q-INTR used primers 5′-ACCGAGATGACGTTCGCTAATC-3′ and 5′-ATCTGGAAGGGCTGGATGTG-3′ with probe 5′-CACTTTCTCCCAACTCGC-3′. Relative quantification was to the eukaryotic 18S rRNA endogenous control (Applied Biosystems part number 4319413E) and used the Standard Curve Method (separate tubes) detailed in Applied Biosystems User Bulletin #2. For statistical analysis, data normalized relative to fourth instar larval expression were compared by one-way anova using log-transformed data from triplicate reactions performed on two or three independent samples.
Construction of reporter plasmids
Genomic DNA was extracted from 18 mosquitoes using the GeneElute Mammalian Genomic Miniprep Kit (Sigma-Aldrich, Poole, UK) following the manufacturer’s instructions, except that insects were crushed in lysis solution T using a Pellet Pestle (Anachem Ltd, Luton, UK). Promoter regions from −1337 to +35 (AgDef2), −1321 to +63 (AgDef3) and −1228 to +113 (AgDef4), to include the 5′ UTR, were PCR amplified from genomic DNA using Platinum Taq DNA polymerase High Fidelity (Invitrogen). Forward and reverse primers (Proligo) were Def2promF (5′-CGTACGCGTGCTTGTTGCTGATACTGCGG-3′) with Def2promR (5′-GAGATCTAATGATGATACAAAGACGAGGAA-3′), Def3promF (5′-GGTACCACGCGTACAAACCGGCAGCGATGA-3′) with Def3promR (5′-CTCGAGATCTTGGATGCTGCTCTGCGATT-3′) or Def4promF (5′-TCGCTAGCATGCGGGACAAGACGATAATGA-3′) with Def4promR (5′-TAAGCTTGGTTGTGCAAAATTTCACCAAAA-3′). Primers introduced 5′ Mlu1 and 3′ BglII restriction sites for AgDef2 and AgDef3 or 5′ Nhe1 and 3′ HindIII restriction sites for AgDef4. PCR products, cloned into pCR2.1-TOPO (Invitrogen), were sequenced prior to transfer into the luciferase expression vector pGL3-Basic (Promega, Southampton, UK) using the introduced restriction sites. Construction of the comparable AgDef1 reporter plasmid, p5AgDef1, has been described previously (Meredith et al., 2006). The Actin 5C-Renilla transfection control plasmid contains the Drosophila Actin 5C promoter (Pinkerton et al., 2000) in pRL-null (Promega).
Cell culture and transfection
Anopheles gambiae Sua 4.0 cells (Catteruccia et al., 2000) were maintained at 27 °C in Schneider’s medium supplemented with 10% foetal bovine serum, 50 U/ml penicillin and 50 μg/ml streptomycin (Sigma-Aldrich). Transfection was mediated by FuGENE 6 (Roche Diagnostic Ltd, Lewes, UK) following the manufacturer’s instructions. Briefly, 5 × 105 cells were plated in 500 μl medium in individual wells of 24-well plates and grown to 60-70% confluence at 27 °C with 5% CO2. Cells were transfected using 6 μl FuGENE per four wells together with 4 μg defensin promoter reporter plasmid and 10 ng Actin 5C-Renilla transfection control plasmid in a total volume of 120 μl serum free medium. Immune challenge, 24 h post transfection, was 100 μg/ml LPS (Escherichia coli serotype 026:B6, 15 × 106 EU/mg), 20 μg/ml laminarin (all from Sigma-Aldrich) or 102 heat killed bacteria (E. coli K12 RM148 or M. luteus) per cell (Dimopoulos et al., 1997). 20-hydroxyecdysone (Sigma-Aldrich) in ethanol was added to a final concentration of 1 μM (Muller et al., 1999).
Dual-luciferase reporter assays
Firefly and Renilla luciferase activities were measured using the Dual-Glo system (Promega). Briefly, cells were lysed in 24-well plates with 75 μl PBS and 75 μl luciferase reagent and transferred to a white 96-well plate to measure firefly luciferase activity. Renilla activity was measured following the addition of 75 μl Stop & Glo reagent. Following normalization to Renilla activity, luciferase data from two experiments, with four replicates in each, were pooled for analysis. Log-transformed data, checked for normal distribution, were analysed by anova (General Linear Model) with Tukey’s pairwise comparisons.
Acknowledgements
We would like to thank Derric Nimmo for bacterial injection of mosquitoes and Hans-Michael Muller for providing the Sua 4.0 cell line. We are grateful for financial support from the Medical Research Council (G0000707 and G0001004 to Eggleston and Lehane) and the Wellcome Trust (059349 to Eggleston and ME02579 to Eggleston and Hurd).
References
- Bartholomay LC, Fuchs JF, Cheng LL, Beck ET, Vizioli J, Lowenberger C, et al. Reassessing the role of defensin in the innate immune response of the mosquito, Aedes aegypti. Insect Mol Biol. 2004;13:125–132. doi: 10.1111/j.0962-1075.2004.00467.x. [DOI] [PubMed] [Google Scholar]
- Blandin S, Moita LF, Kocher T, Wilm M, Kafatos FC, Levashina EA. Reverse genetics in the mosquito Anopheles gambiae: targeted disruption of the Defensin gene. EMBO Rep. 2002;3:852–856. doi: 10.1093/embo-reports/kvf180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulet P, Stocklin R. Insect antimicrobial peptides: structures, properties and gene regulation. Protein Pept Lett. 2005;12:3–11. doi: 10.2174/0929866053406011. [DOI] [PubMed] [Google Scholar]
- Casteels-Josson K, Zhang W, Capaci T, Casteels P, Tempst P. Acute transcriptional response of the honeybee peptide-antibiotics gene repertoire and required post-translational conversion of the precursor structures. J Biol Chem. 1994;269:28569–28575. [PubMed] [Google Scholar]
- Catteruccia F, Nolan T, Blass C, Muller HM, Crisanti A, Kafatos FC, et al. Toward Anopheles transformation: minos element activity in anopheline cells and embryos. Proc Natl Acad Sci USA. 2000;97:2157–2162. doi: 10.1073/pnas.040568397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalk R, Albuquerque CM, Ham PJ, Townson H. Full sequence and characterization of two insect defensins: immune peptides from the mosquito Aedes aegypti. Proc R Soc Lond B Biol Sci. 1995;261:217–221. doi: 10.1098/rspb.1995.0139. [DOI] [PubMed] [Google Scholar]
- Cherbas L, Cherbas P. The arthropod initiator: the capsite consensus plays an important role in transcription. Insect Biochem Mol Biol. 1993;23:81–90. doi: 10.1016/0965-1748(93)90085-7. [DOI] [PubMed] [Google Scholar]
- Cherbas L, Lee K, Cherbas P. Identification of ecdysone response elements by analysis of the Drosophila Eip28/29 gene. Genes Dev. 1991;5:120–131. doi: 10.1101/gad.5.1.120. [DOI] [PubMed] [Google Scholar]
- Cho WL, Fu YC, Chen CC, Ho CM. Cloning and characterization of cDNAs encoding the antibacterial peptide, defensin A, from the mosquito, Aedes aegypti. Insect Biochem Mol Biol. 1996;26:395–402. doi: 10.1016/0965-1748(95)00108-5. [DOI] [PubMed] [Google Scholar]
- Christophides GK, Zdobnov E, Barillas-Mury C, Birney E, Blandin S, Blass C, et al. Immunity-related genes and gene families in Anopheles gambiae. Science. 2002;298:159–165. doi: 10.1126/science.1077136. [DOI] [PubMed] [Google Scholar]
- Cornet B, Bonmatin JM, Hetru C, Hoffmann JA, Ptak M, Vovelle F. Refined three-dimensional solution structure of insect defensin A. Structure. 1995;3:435–448. doi: 10.1016/s0969-2126(01)00177-0. [DOI] [PubMed] [Google Scholar]
- Dassanayake RS, Silva Gunawardene YI, Tobe SS. Evolutionary selective trends of insect/mosquito antimicrobial defensin peptides containing cysteine-stabilized alpha/beta motifs. Peptides. 2007;28:62–75. doi: 10.1016/j.peptides.2006.09.022. [DOI] [PubMed] [Google Scholar]
- Dimarcq JL, Hoffmann D, Meister M, Bulet P, Lanot R, Reichhart JM, et al. Characterization and transcriptional profiles of a Drosophila gene encoding an insect defensin. A study in insect immunity. Eur J Biochem. 1994;221:201–209. doi: 10.1111/j.1432-1033.1994.tb18730.x. [DOI] [PubMed] [Google Scholar]
- Dimopoulos G, Richman A, Muller HM, Kafatos FC. Molecular immune responses of the mosquito Anopheles gambiae to bacteria and malaria parasites. Proc Natl Acad Sci USA. 1997;94:11508–11513. doi: 10.1073/pnas.94.21.11508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimopoulos G, Seeley D, Wolf A, Kafatos FC. Malaria infection of the mosquito Anopheles gambiae activates immune-responsive genes during critical transition stages of the parasite life cycle. Embo J. 1998;17:6115–6123. doi: 10.1093/emboj/17.21.6115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggleston P, Lu W, Zhao Y. Genomic organization and immune regulation of the defensin gene from the mosquito, Anopheles gambiae. Insect Mol Biol. 2000;9:481–490. doi: 10.1046/j.1365-2583.2000.00212.x. [DOI] [PubMed] [Google Scholar]
- Engstrom Y. Induction and regulation of antimicrobial peptides in Drosophila. Dev Comp Immunol. 1999;23:345–358. doi: 10.1016/s0145-305x(99)00016-6. [DOI] [PubMed] [Google Scholar]
- Evans T, Felsenfeld G, Reitman M. Control of globin gene transcription. Annu Rev Cell Biol. 1990;6:95–124. doi: 10.1146/annurev.cb.06.110190.000523. [DOI] [PubMed] [Google Scholar]
- Frye M, Bargon J, Gropp R. Expression of human beta-defensin-1 promotes differentiation of keratinocytes. J Mol Med. 2001;79:275–282. doi: 10.1007/s001090100200. [DOI] [PubMed] [Google Scholar]
- Hamilton JV, Munks RJ, Lehane SM, Lehane MJ. Association of midgut defensin with a novel serine protease in the blood-sucking fly Stomoxys calcitrans. Insect Mol Biol. 2002;11:197–205. doi: 10.1046/j.1365-2583.2002.00325.x. [DOI] [PubMed] [Google Scholar]
- Han ZS, Ip YT. Interaction and specificity of Rel-related proteins in regulating Drosophila immunity gene expression. J Biol Chem. 1999;274:21355–21361. doi: 10.1074/jbc.274.30.21355. [DOI] [PubMed] [Google Scholar]
- Hillyer JF, Christensen BM. Mosquito phenoloxidase and defensin colocalize in melanization innate immune responses. J Histochem Cytochem. 2005;53:689–698. doi: 10.1369/jhc.4A6564.2005. [DOI] [PubMed] [Google Scholar]
- Hurd H, Taylor PJ, Adams D, Underhill A, Eggleston P. Evaluating the costs of mosquito resistance to malaria parasites. Evolution Int J Org Evolution. 2005;59:2560–2572. [PMC free article] [PubMed] [Google Scholar]
- Jahan N, Hurd H. The effects of infection with Plasmodium yoelii nigeriensis on the reproductive fitness of Anopheles stephensi. Ann Trop Med Parasitol. 1997;91:365–369. doi: 10.1080/00034989760987. [DOI] [PubMed] [Google Scholar]
- Kadalayil L, Petersen UM, Engstrom Y. Adjacent GATA and kappa B-like motifs regulate the expression of a Drosophila immune gene. Nucleic Acids Res. 1997;25:1233–1239. doi: 10.1093/nar/25.6.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser V, Diamond G. Expression of mammalian defensin genes. J Leukoc Biol. 2000;68:779–784. [PubMed] [Google Scholar]
- Kappler C, Meister M, Lagueux M, Gateff E, Hoffmann JA, Reichhart JM. Insect immunity. Two 17 bp repeats nesting a kappa B-related sequence confer inducibility to the diptericin gene and bind a polypeptide in bacteria-challenged Drosophila. EMBO J. 1993;12:1561–1568. doi: 10.1002/j.1460-2075.1993.tb05800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaudiny J, Albert S, Bachanova K, Kopernicky J, Simuth J. Two structurally different defensin genes, one of them encoding a novel defensin isoform, are expressed in honeybee Apis mellifera. Insect Biochem Mol Biol. 2005;35:11–22. doi: 10.1016/j.ibmb.2004.09.007. [DOI] [PubMed] [Google Scholar]
- Komano H, Homma K, Natori S. Involvement of sapecin in embryonic cell proliferation of Sarcophaga peregrina (flesh fly) FEBS Lett. 1991;289:167–170. doi: 10.1016/0014-5793(91)81061-c. [DOI] [PubMed] [Google Scholar]
- Kylsten P, Samakovlis C, Hultmark D. The cecropin locus in Drosophila; a compact gene cluster involved in the response to infection. EMBO J. 1990;9:217–224. doi: 10.1002/j.1460-2075.1990.tb08098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamberty M, Ades S, Uttenweiler-Joseph S, Brookhart G, Bushey D, Hoffmann JA, et al. Insect immunity. Isolation from the lepidopteran Heliothis virescens of a novel insect defensin with potent antifungal activity. J Biol Chem. 1999;274:9320–9326. doi: 10.1074/jbc.274.14.9320. [DOI] [PubMed] [Google Scholar]
- Lee SR, Kurata S, Natori S. Molecular cloning of cDNA for sapecin B, an antibacterial protein of Sarcophaga, and its detection in larval brain. FEBS Lett. 1995;368:485–487. doi: 10.1016/0014-5793(95)00717-n. [DOI] [PubMed] [Google Scholar]
- Lehane MJ, Aksoy S, Levashina E. Immune responses and parasite transmission in blood-feeding insects. Trends Parasitol. 2004;20:433–439. doi: 10.1016/j.pt.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Lehane MJ, Wu D, Lehane SM. Midgut-specific immune molecules are produced by the blood-sucking insect Stomoxys calcitrans. Proc Natl Acad Sci USA. 1997;94:11502–11507. doi: 10.1073/pnas.94.21.11502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowenberger C, Bulet P, Charlet M, Hetru C, Hodgeman B, Christensen BM, et al. Insect immunity: isolation of three novel inducible antibacterial defensins from the vector mosquito, Aedes aegypti. Insect Biochem Mol Biol. 1995;25:867–873. doi: 10.1016/0965-1748(95)00043-u. [DOI] [PubMed] [Google Scholar]
- Lowenberger C, Charlet M, Vizioli J, Kamal S, Richman A, Christensen BM, et al. Antimicrobial activity spectrum, cDNA cloning, and mRNA expression of a newly isolated member of the cecropin family from the mosquito vector Aedes aegypti. J Biol Chem. 1999a;274:20092–20097. doi: 10.1074/jbc.274.29.20092. [DOI] [PubMed] [Google Scholar]
- Lowenberger CA, Kamal S, Chiles J, Paskewitz S, Bulet P, Hoffmann JA, et al. Mosquito-Plasmodium interactions in response to immune activation of the vector. Exp Parasitol. 1999b;91:59–69. doi: 10.1006/expr.1999.4350. [DOI] [PubMed] [Google Scholar]
- Lowenberger CA, Smartt CT, Bulet P, Ferdig MT, Severson DW, Hoffmann JA, et al. Insect immunity: molecular cloning, expression, and characterization of cDNAs and genomic DNA encoding three isoforms of insect defensin in Aedes aegypti. Insect Mol Biol. 1999c;8:107–118. doi: 10.1046/j.1365-2583.1999.810107.x. [DOI] [PubMed] [Google Scholar]
- Luna C, Hoa NT, Lin H, Zhang L, Nguyen HL, Kanzok SM, et al. Expression of immune responsive genes in cell lines from two different Anopheline species. Insect Mol Biol. 2006;15:721–729. doi: 10.1111/j.1365-2583.2006.00661.x. [DOI] [PubMed] [Google Scholar]
- Manfruelli P, Reichhart JM, Steward R, Hoffmann JA, Lemaitre B. A mosaic analysis in Drosophila fat body cells of the control of antimicrobial peptide genes by the Rel proteins Dorsal and DIF. Embo J. 1999;18:3380–3391. doi: 10.1093/emboj/18.12.3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama K, Natori S. Purification of three antibacterial proteins from the culture medium of NIH-Sape-4, an embryonic cell line of Sarcophaga peregrina. J Biol Chem. 1988;263:17112–17116. [PubMed] [Google Scholar]
- McDermott AM, Redfern RL, Zhang B. Human beta-defensin 2 is up-regulated during re-epithelialization of the cornea. Curr Eye Res. 2001;22:64–67. doi: 10.1076/ceyr.22.1.64.6978. [DOI] [PubMed] [Google Scholar]
- Meister M, Richards G. Ecdysone and insect immunity: the maturation of the inducibility of the diptericin gene in Drosophila larvae. Insect Biochem Mol Biol. 1996;26:155–160. doi: 10.1016/0965-1748(95)00076-3. [DOI] [PubMed] [Google Scholar]
- Meister M, Lemaitre B, Hoffmann JA. Antimicrobial peptide defense in Drosophila. Bioessays. 1997;19:1019–1026. doi: 10.1002/bies.950191112. [DOI] [PubMed] [Google Scholar]
- Meredith JM, Munks RJ, Grail W, Hurd H, Eggleston P, Lehane MJ. A novel association between clustered NF-kappaB and C/EBP binding sites is required for immune regulation of mosquito Defensin genes. Insect Mol Biol. 2006;15:393–401. doi: 10.1111/j.1365-2583.2006.00635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerholz DK, Gallup JM, Grubor BM, Evans RB, Tack BF, McCray PB, Jr, et al. Developmental expression and distribution of sheep beta-defensin-2. Dev Comp Immunol. 2004;28:171–178. doi: 10.1016/s0145-305x(03)00105-8. [DOI] [PubMed] [Google Scholar]
- Moon HJ, Lee SY, Kurata S, Natori S, Lee BL. Purification and molecular cloning of cDNA for an inducible antibacterial protein from larvae of the coleopteran, Tenebrio molitor. J Biochem (Tokyo) 1994;116:53–58. doi: 10.1093/oxfordjournals.jbchem.a124502. [DOI] [PubMed] [Google Scholar]
- Muller HM, Dimopoulos G, Blass C, Kafatos FC. A hemocyte-like cell line established from the malaria vector Anopheles gambiae expresses six prophenoloxidase genes. J Biol Chem. 1999;274:11727–11735. doi: 10.1074/jbc.274.17.11727. [DOI] [PubMed] [Google Scholar]
- Munks RJ, Hamilton JV, Lehane SM, Lehane MJ. Regulation of midgut defensin production in the blood-sucking insect Stomoxys calcitrans. Insect Mol Biol. 2001;10:561–571. doi: 10.1046/j.0962-1075.2001.00296.x. [DOI] [PubMed] [Google Scholar]
- Natori S, Shiraishi H, Hori S, Kobayashi A. The roles of Sarcophaga defense molecules in immunity and metamorphosis. Dev Comp Immunol. 1999;23:317–328. doi: 10.1016/s0145-305x(99)00014-2. [DOI] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- Pinkerton AC, Michel K, O’Brochta DA, Atkinson PW. Green fluorescent protein as a genetic marker in transgenic Aedes aegypti. Insect Mol Biol. 2000;9:1–10. doi: 10.1046/j.1365-2583.2000.00133.x. [DOI] [PubMed] [Google Scholar]
- Richman AM, Bulet P, Hetru C, Barillas-Mury C, Hoffmann JA, Kafalos FC. Inducible immune factors of the vector mosquito Anopheles gambiae: biochemical purification of a defensin antibacterial peptide and molecular cloning of preprodefensin cDNA. Insect Mol Biol. 1996;5:203–210. doi: 10.1111/j.1365-2583.1996.tb00055.x. [DOI] [PubMed] [Google Scholar]
- Richman AM, Dimopoulos G, Seeley D, Kafatos FC. Plasmodium activates the innate immune response of Anopheles gambiae mosquitoes. Embo J. 1997;16:6114–6119. doi: 10.1093/emboj/16.20.6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryden TA, Beemon K. Avian retroviral long terminal repeats bind CCAAT/enhancer-binding protein. Mol Cell Biol. 1989;9:1155–1164. doi: 10.1128/mcb.9.3.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoda M, Takagi H, Kurata S, Yoshioka T, Natori S. Inhibition of the Ca(2+)-activated K(+)-channel by sapecin B, an insect antibacterial protein. FEBS Lett. 1994;339:59–62. doi: 10.1016/0014-5793(94)80384-6. [DOI] [PubMed] [Google Scholar]
- Sun D, Fallon AM. Characterization of genomic DNA encoding cecropins from an Aedes albopictus mosquito cell line. Insect Mol Biol. 2002;11:21–30. doi: 10.1046/j.0962-1075.2001.00305.x. [DOI] [PubMed] [Google Scholar]
- Thisse C, Perrin-Schmitt F, Stoetzel C, Thisse B. Sequence-specific transactivation of the Drosophila twist gene by the dorsal gene product. Cell. 1991;65:1191–1201. doi: 10.1016/0092-8674(91)90014-p. [DOI] [PubMed] [Google Scholar]
- Vizioli J, Bulet P, Charlet M, Lowenberger C, Blass C, Muller HM, et al. Cloning and analysis of a cecropin gene from the malaria vector mosquito, Anopheles gambiae. Insect Mol Biol. 2000;9:75–84. doi: 10.1046/j.1365-2583.2000.00164.x. [DOI] [PubMed] [Google Scholar]
- Vizioli J, Richman AM, Uttenweiler-Joseph S, Blass C, Bulet P. The defensin peptide of the malaria vector mosquito Anopheles gambiae: antimicrobial activities and expression in adult mosquitoes. Insect Biochem Mol Biol. 2001a;31:241–248. doi: 10.1016/s0965-1748(00)00143-0. [DOI] [PubMed] [Google Scholar]
- Vizioli J, Bulet P, Hoffmann JA, Kafatos FC, Muller HM, Dimopoulos G. Gambicin: a novel immune responsive antimicrobial peptide from the malaria vector Anopheles gambiae. Proc Natl Acad Sci USA. 2001b;98:12630–12635. doi: 10.1073/pnas.221466798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkoff AN, Rocher J, d’Alencon E, Bouton M, Landais I, Quesada-Moraga E, et al. Characterization and transcriptional profiles of three Spodoptera frugiperda genes encoding cysteine-rich peptides. A new class of defensin-like genes from lepidopteran insects? Gene. 2003;319:43–53. doi: 10.1016/s0378-1119(03)00789-3. [DOI] [PubMed] [Google Scholar]
- Waterhouse RM, Kriventseva EV, Meister S, Xi Z, Alvarez KS, Bartholomay LC, et al. Evolutionary dynamics of immune-related genes and pathways in disease-vector mosquitoes. Science. 2007;316:1738–1743. doi: 10.1126/science.1139862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willy PJ, Kobayashi R, Kadonaga JT. A basal transcription factor that activates or represses transcription. Science. 2000;290:982–985. doi: 10.1126/science.290.5493.982. [DOI] [PubMed] [Google Scholar]
- Yamada K, Natori S. Purification, sequence and antibacterial activity of two novel sapecin homologues from Sarcophaga embryonic cells: similarity of sapecin B to charybdotoxin. Biochem J. 1993;291(Pt 1):275–279. doi: 10.1042/bj2910275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng XL, Zheng AL. Genomic organization and regulation of three cecropin genes in Anopheles gambiae. Insect Mol Biol. 2002;11:517–525. doi: 10.1046/j.1365-2583.2002.00360.x. [DOI] [PubMed] [Google Scholar]



