Abstract
Changes in nuclear size and shape during the cell cycle or during development require coordinated nuclear membrane remodeling, but the underlying molecular events are largely unknown. We have shown previously that the activity of the conserved phosphatidate phosphatase Pah1p/Smp2p regulates nuclear structure in yeast by controlling phospholipid synthesis and membrane biogenesis at the nuclear envelope. Two screens for novel regulators of phosphatidate led to the identification of DGK1. We show that Dgk1p is a unique diacylglycerol kinase that uses CTP, instead of ATP, to generate phosphatidate. DGK1 counteracts the activity of PAH1 at the nuclear envelope by controlling phosphatidate levels. Overexpression of DGK1 causes the appearance of phosphatidate-enriched membranes around the nucleus and leads to its expansion, without proliferating the cortical endoplasmic reticulum membrane. Mutations that decrease phosphatidate levels decrease nuclear membrane growth in pah1Δ cells. We propose that phosphatidate metabolism is a critical factor determining nuclear structure by regulating nuclear membrane biogenesis.
Phospholipids are the major cellular components required for the assembly of biological membranes (1). The regulated production and distribution of phospholipids during the cell cycle or during development often underlie striking changes in membrane biogenesis, which, in turn, can impact on the size, shape, or number of organelles. For example, stimulation of phospholipid biosynthesis accompanies the expansion of the ER3 in professional secretory cells (2) or neurite growth during neuronal differentiation (3). Despite these interesting observations, the molecular mechanisms responsible for coupling lipid production to organelle morphology remain largely unknown.
An organelle that undergoes striking structural changes during the cell cycle is the nucleus. The nucleus is delimited by the nuclear envelope, which consists of a double lipid bilayer, the outer and inner nuclear membranes (4). The outer membrane is continuous with the ER, whereas the inner membrane faces the nucleoplasm and binds to chromatin. Nuclear membrane growth is essential for cell division. In yeast, nuclear membrane expansion allows anaphase to take place within a single nuclear compartment that partitions between mother and daughter cells (5). In metazoan cells, the nuclear membrane expands at the end of mitosis to accommodate chromatin decondensation and DNA replication (6). The source of the nuclear membrane and the mechanism by which it is added to the nuclear envelope remain unknown. Nuclear envelope remodeling is also important for cell types that undergo dramatic nuclear structure changes during their differentiation, such as mammalian blood cell types, where nuclei can be highly lobed and segmented, or spermatocytes and myocytes, where nuclei can take very elongated morphologies (7). The importance of proper nuclear envelope structure in cell physiology is underscored by the recent identification of several diseases that are associated with changes in nuclear shape and are caused by mutations in nuclear envelope proteins (8).
We have recently identified an unexpected link between phospholipid biosynthesis and nuclear structure. Mutations in Pah1p, a PA phosphatase that catalyzes the dephosphorylation of PA to DAG (9), cause proliferation of the nuclear/ER membrane and a massive expansion of the nucleus (10). Interestingly, the activity of Pah1p is regulated by Cdc28p-dependent phosphorylation during mitosis. A phosphorylation-null Pah1p mutant has higher PA phosphatase activity than the wild-type enzyme (11). Consistent with this, cells lacking Nem1p-Spo7p, the phosphatase complex that dephosphorylates Pah1p, accumulate hyperphosphorylated Pah1p and exhibit nuclear membrane proliferation like pah1Δ cells (10).
There are two key consequences of the changes in PA levels that take place in pah1 mutants. First, in yeast cells, PA regulates coordinately the transcription of many phospholipid biosynthetic enzyme genes through the PA-sensitive repressor Opi1p (12). When levels of PA are high, as in pah1Δ cells, Opi1p is excluded from the nucleus, and transcription is derepressed (13). Interestingly, Pah1p regulates the transcription of these genes through both Opi1p-dependent and Opi1p-independent mechanisms (11). Second, both PA and DAG are the precursors for the synthesis of abundant phospholipids that are used for membrane biogenesis, through the CDP-DAG and CDP-choline pathways, respectively (see Fig. 1A). Therefore, changes in PA and DAG levels in pah1 mutants impact on the cellular levels of these phospholipids (9, 14).
FIGURE 1.
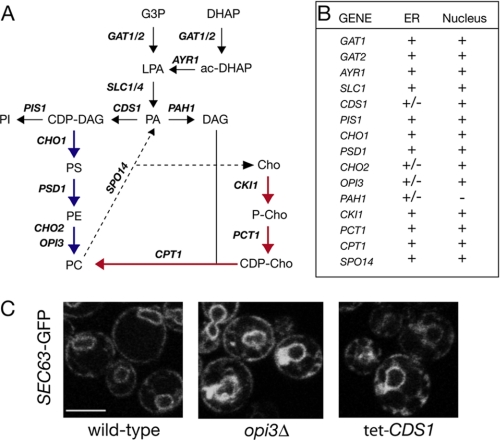
Systematic analysis of the role of phospholipid metabolism in nuclear/ER membrane structure. A, pathways of phospholipid biosynthesis and PA metabolism in budding yeast. Reactions involved in the production of cellular pools of phospholipids are shown. For simplicity, the Kennedy pathway reactions leading to PE synthesis are not shown. Blue and red arrows highlight the CDP-DAG and CDP-choline (Cho) pathways, respectively. G3P, glycerol 3-phosphate; DHAP, dihydroxyacetone phosphate. LPA, lysophosphatidic acid; PI, phosphatidylinositol; PS, phosphatidylserine. B, list of mutants tested for nuclear/ER morphology defects. Cells were transformed with a centromeric plasmid expressing SEC63-GFP and visualized by microscopy. At least two transformants per mutant were analyzed. +, no defects in nuclear or ER structure; -, severe defects; +/-, partial defects. The tet-CDS1 repressible strain was grown in the presence of 10 μg/ml doxycycline, and SEC63-GFP fluorescence was visualized after 0, 6, 12, and 24 h. The pis1 temperature-sensitive strain was grown at 30 °C and then transferred to 37 °C for 3 h. All mutants were derived from BY4742, except the tet-CDS1 and psd1Δ strains, which were derived from BY4741, and the pis1 (41) and pah1Δ (10) strains. C, representative images of wild-type, tet-CDS1 (12 h in doxycycline), and opi3Δ expressing the SEC63-GFP reporter. Scale bar = 5 μm.
How the inactivation of Pah1p leads to nuclear growth is not understood. Moreover, whether Pah1p regulates primarily ER or nuclear membrane biogenesis and whether particular lipids are associated with these changes are not known. We have shown previously that the up-regulation of phospholipid synthesis is necessary but not sufficient to drive nuclear growth in pah1Δ cells (10, 11). Therefore, there could be additional factors required for nuclear expansion. To investigate the role of phospholipid biosynthesis in nuclear membrane formation and to identify novel components involved in this process, we applied two complementary genetic approaches. First, a systematic search among mutants of known phospholipid biosynthetic enzymes revealed that only Pah1p affects nuclear structure. We therefore performed two non-biased screens looking for previously uncharacterized genes that could encode PA regulators and examined their effect on nuclear structure. Both screens identified the same gene, DGK1 (known previously as HSD1), encoding a nuclear/ER integral membrane protein. We show that Dgk1p counteracts the effects of Pah1p at the nuclear membrane by controlling PA levels. Dgk1p is a novel CTP-dependent DAG kinase that catalyzes the phosphorylation of DAG to PA. Unlike previously described DAG kinases that use ATP, Dgk1p utilizes CTP as a phosphate donor for the kinase reaction. Overexpression of DGK1 causes recruitment of PA reporter protein on the nuclear membrane and leads to its expansion. Interestingly, cortical ER membrane is not proliferating in these cells. These data reveal a specific role for PA metabolism in nuclear membrane biogenesis.
EXPERIMENTAL PROCEDURES
Yeast Strains, Media, and Growth Conditions—The yeast strains used in this study are listed in Table 1. Unless stated otherwise, the deletion mutants shown in Fig. 1 were obtained from Open Biosystems and are derived from the BY4742 strain. Knock-out of DGK1 was done by homologous recombination, transforming a dgk1::HIS3 fragment containing 5′- and 3′-flanking sequences into the wild-type (RS453) or pah1::TRP1 deletion strain to obtain the dgk1Δ or pah1Δ dgk1Δ strains, respectively. Deletion of other genes (GAT1, GAT2, PSD1, CHO2, and OPI3) into the pah1Δ strain was done by transforming the appropriate KanMX4 fragments into a pah1::TRP1 strain complemented by a YCplac33-PAH1 plasmid. The PAH1 plasmid was then shuffled out on 5-fluoroorotic acid-containing plates to yield the appropriate double deletions. Yeast cells were grown in YEPD (1% yeast extract, 2% peptone, and 2% glucose) or synthetic medium containing 2% glucose and lacking the appropriate amino acids for plasmid selection. GAL1/10-dependent overexpression was performed by changing the carbon source of early log phase cells from 2% raffinose to 2% galactose. Cells were grown at 30 °C unless mentioned otherwise. To assay growth of yeast cells in medium lacking inositol, synthetic medium was prepared using yeast nitrogen base lacking inositol (Bio 101, Inc.).
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Ref./source |
|---|---|---|
| RS453 | MATaade2-1 his3-11,15 ura3-52 leu2-3,112 trp1-1 | Ref. 43 |
| SS1026 | RS453 pah1::TRP1 | Ref. 10 |
| SS1144 | RS453 dgk1::HIS3 | This study |
| SS1147 | RS453 pah1::TRP1 dgk1::HIS3 | This study |
| SS1246 | RS453 pah1::TRP1 gat1::KanMX4 | This study |
| SS1247 | RS453 pah1::TRP1 gat2::KanMX4 | This study |
| SS1282 | RS453 pah1::TRP1 psd1::KanMX4 + YCplac33-URA3-PAH1 | This study |
| SS1301 | RS453 pah1::TRP1 cho2::KanMX4 + YCplac33-URA3-PAH1 | This study |
| SS1290 | RS453 pah1::TRP1 opi3::KanMX4 + YCplac33-URA3-PAH1 | This study |
| SS1372 | RS453 mCherry-PUS1::URA3 | This study |
Plasmids and Construction of Fusion Genes—The plasmids used in this study are listed in Table 2. The high copy library used for the suppressor screens was from American Type Culture Collection (catalog no. 37323). The construction of GAT2(G253L) and DGK1(D177A) was done by PCR-mediated mutagenesis. The wild-type DGK1 and DGK1(D177A) genes were overexpressed using the GAL1/10 promoter. The RTN2-GFP fusion was constructed by inserting the GFP fragment into the C terminus of RTN2. The fusion gene was cloned into a centromeric URA3 vector (YCplac33), and its expression was driven by the RTN2 promoter. The HEH2-GFP fusion was constructed by inserting the GFP fragment into the C terminus of HEH2. The fusion gene was cloned into a centromeric LEU2 vector (YCplac111), and its expression was driven by the NOP1 promoter. The mCherry-PUS1 fusion was constructed by introducing the mCherry fragment into the N terminus of PUS1. The fusion gene was cloned into a YIplac211 vector, and its expression was driven by the NOP1 promoter. The PtA-DGK1 and GFP-DGK1 fusions were constructed by inserting the protein A or GFP fragment, respectively, into the N terminus of DGK1; their expression was driven by the NOP1 or DGK1 promoter, respectively. The DGK1-PtA fusion was constructed by inserting the protein A fragment into the C terminus of DGK1, and its expression was driven by inducible GAL1/10 promoter.
TABLE 2.
Plasmids used in this study
| Plasmid | Description | Ref./source |
|---|---|---|
| YCplac111-SEC63-GFP | SEC63-GFP under control of SEC63 promoter into CEN/LEU2 vector | Ref. 10 |
| YCplac33-SEC63-GFP | SEC63-GFP under control of SEC63 promoter into CEN/URA3 vector | This study |
| YEplac195-PAH1-7P | PAH1-7P under control of GAL1/10 promoter into 2μ/URA3 vector | This study |
| YCplac33-GAL1/10-NEM1 | NEM1 under control of GAL1/10 promoter into CEN/URA3 vector | This study |
| pRS313-GAL1/10-SPO7 | SPO7 under control of GAL1/10 promoter into CEN/HIS3 vector | This study |
| YCplac111-GAL1/10-DGK1 | DGK1 under control of GAL1/10 promoter into CEN/LEU2 vector | This study |
| YEplac181-GAL1/10-DGK1 | DGK1 under control of GAL1/10 promoter into 2μ/LEU2 vector | This study |
| pRS313-RFP-PUS1 | PUS1 under control of NOP1 promoter into CEN/HIS3 vector | This study |
| YCplac111-GAL1/10-DGK1(D177A) | DGK1(D177A) under control of GAL1/10 promoter into CEN/LEU2 vector | This study |
| YIplac211-mCherry-PUS1 | PUS1 under control of NOP1 promoter into integrative/URA3 vector | This study |
| YCplac33-RTN2-GFP | RTN2-GFP under control of RTN2 promoter into CEN/URA3 vector | This study |
| YCplac111-HEH2-GFP | HEH2-GFP under control of NOP1 promoter into CEN/LEU2 vector | This study |
| YCplac33-PAH1 | PAH1 under control of PAH1 promoter into CEN/URA3 vector | Ref. 10 |
| pRS426-G20 | GFP-SPO20-(51-91) under control of TEF2 promoter into 2μ/URA3 vector | Ref. 26 |
| pRS424-GAL1pr-DGK | Bacterial DGK under control of GAL1 promoter into 2μ/TRP1 vector | Ref. 26 |
| YCplac111-GAT2 | GAT2 under control of GAT2 promoter into CEN/LEU2 vector | This study |
| YCplac111-GAT2(G253L) | GAT2(G253L) under control of GAT2 promoter into CEN/LEU2 vector | This study |
High Copy Number Suppressor Screens—For the PAH1-7P suppressor screen, the wild-type RS453 strain overexpressing PAH1-7P from the inducible GAL1/10 promoter in a high copy plasmid (YEplac195) was transformed with a high copy yeast genomic library (15). About 5000 transformants were screened for their ability to suppress the toxicity of the overexpressed PAH1-7P mutant on galactose plates lacking inositol (11). Library plasmids were recovered from two suppressor colonies and then retransformed into the original screening strain to verify the suppression. Sequencing of the two library plasmids followed by subcloning of various open reading frames present in the genomic fragment showed that, in both cases, the DGK1 gene was responsible for the suppression of the inositol auxotrophy of the PAH1-7P mutant.
For the NEM1-SPO7 suppressor screen, the wild-type RS453 strain overexpressing NEM1 and SPO7 from the GAL1/10 promoter in the YCplac33 and pRS313 plasmids, respectively, was transformed with the same high copy genomic library as described above. About 6000 transformants were screened for their ability to suppress the toxicity of the NEM1-SPO7 overexpression (10). Four suppressors were isolated that were found, when analyzed as described above, to contain overlapping inserts of the DGK1 gene. Further subcloning demonstrated that DGK1 was responsible for the suppression of the NEM1-SPO7 toxicity.
RNA Extractions and Quantitative PCR—Total RNA was isolated as described previously (11). Quantitative real-time PCR was performed on total RNA using an ABI 7900HT system and a SYBR Green Mastermix (Applied Biosystems) as described previously (11).
Affinity Purification of PtA-Dgk1p—The dgk1Δ strain carrying a pRS315-PtA-DGK1 fusion was grown to A600 = 1, washed once with water, and spheroplasted as described previously (16). Spheroplasts were first lysed in 150 mm KCl, 20 mm Tris-HCl (pH 8.0), and 5 mm MgCl2 supplemented with a mixture of protease inhibitors (Complete EDTA-free, Roche Applied Science) and 4-(2-aminoethyl)benzenesulfonyl fluoride. The extract was centrifuged for 15 min at 15,000 rpm (Sorvall SS34 rotor), and the pellet was resuspended in the same lysis buffer containing 1% Triton X-100. The extract was centrifuged for 30 min as described above, and the supernatant was first pre-cleared by incubation with Sepharose Fast-Flow beads (GE Healthcare) and then loaded onto a column packed with IgG-Sepharose beads (GE Healthcare). Affinity purifications of PtA-Dgk1p followed by tobacco etch virus protease-dependent elution of untagged Dgk1p were performed as described (16).
Preparation of Labeled CTP—[γ-32P]CTP was synthesized enzymatically from CDP and [γ-32P]ATP with nucleoside-diphosphate kinase as described previously (17).
DAG Kinase Assay—DAG kinase activity was measured for 20 min by following the phosphorylation of 0.1 mm DAG with 0.1 mm [γ-32P]CTP (50,000 cpm/nmol) in the presence of 50 mm Tris-HCl (pH 7.5), 1 mm Triton X-100, 5 mm CaCl2, 10 mm β-mercaptoethanol, and enzyme protein in a total volume of 0.1 mm at 30 °C. The reaction was terminated by the addition of 0.5 ml of 0.1 n HCl in methanol. The radioactive chloroform-soluble product PA was separated from the radioactive water-soluble substrate CTP by a chloroform/methanol/water phase partition (18). The radioactive chloroform phase was subjected to liquid scintillation counting. Alternatively, the product was subjected to thin layer chromatography using a solvent system of chloroform/methanol/water (65:25:4, v/v) and visualized by phosphorimaging. The DAG kinase assay was conducted in triplicate, and the average S.D. of the assay was ±5%. The enzyme reaction was linear with time and protein concentration.
Labeling and Analysis of Lipids—Steady-state labeling of phospholipids and neutral lipids with 32Pi and [2-14C]acetate, respectively, was performed as described previously (9). Phospholipids were separated by two-dimensional thin layer chromatography on silica gel plates using chloroform/methanol/ammonium hydroxide/water (45:25:2:3, v/v) and chloroform/methanol/glacial acetic acid/water (32:4:5:1, v/v) as the solvent systems for the first and second dimensions, respectively. Neutral lipids were separated by one-dimensional thin layer chromatography on silica gel plates using a solvent system of hexane/diethyl ether/glacial acetic acid (40:10:1, v/v). The identity of the labeled lipids on thin layer chromatography plates was confirmed by comparison with standards after exposure to iodine vapor. Radiolabeled lipids were visualized by phosphorimaging and subjected to ImageQuant analysis.
Microscopy—To visualize GFP, red fluorescent protein, and mCherry fusions, cells were grown in the appropriate selective medium at 30 °C to early logarithmic phase and examined with a ×63 Plan Apochromat objective on a Zeiss Axiovert 200 M inverted microscope with an LSM 510 confocal laser scanning attachment. Cells in Figs. 2D and 6C were viewed using a Zeiss Axiophot fluorescence microscope equipped with a CCD camera, and images were recorded using IP Labs software.
FIGURE 2.
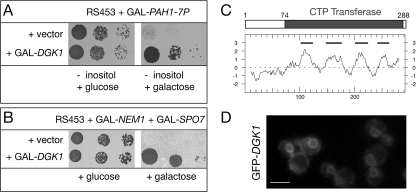
DGK1 rescues the lethality of dephosphorylated PAH1. A, identification of DGK1 as a high copy suppressor of the inositol auxotrophy of PAH1-7P. The wild-type RS453 strain carrying a high copy vector expressing the non-phosphorylated PAH1-7P mutant from a galactose-inducible promoter was transformed with the indicated plasmids. Transformants were grown on synthetic plates lacking inositol and supplemented with glucose (left panel) or galactose (right panel) at 30 °C. B, identification of DGK1 as a high copy suppressor of the lethality of NEM1-SPO7 overexpression. RS453 cells carrying plasmids coexpressing NEM1 and SPO7 from a galactose-inducible promoter were transformed with the indicated plasmids. Transformants were grown on synthetic plates supplemented with glucose (left panel) or galactose (right panel) at 30°C. C, primary structure and hydropathy plot of Dgk1p. The gray box indicates the conserved cytidylyltransferase domain within Dgk1p. Below, the hydropathy plot of Dgk1p is given (42), using a window of 19 amino acids. The black lines indicate the positions of the predicted transmembrane domains. D, GFP-Dgk1p localizes to the nuclear membrane and ER. A GFP-DGK1 fusion under the control of the DGK1 promoter was expressed from a centromeric plasmid in a dgk1Δ strain. Cells were grown in early logarithmic phase and visualized by fluorescent microscopy. Scale bar =5 μm.
FIGURE 6.
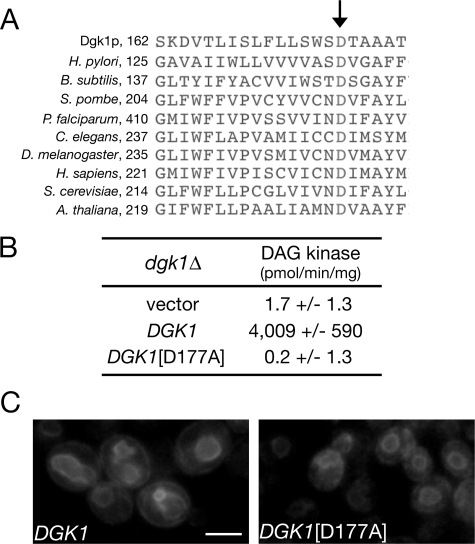
The DAG kinase activity of Dgk1p is required for nuclear membrane expansion. A, multiple sequence alignment of Dgk1p (residues 162-182) with cytidylyltransferases from the indicated species. The first residue of each sequence segment is indicated. The arrow points to the conserved aspartate residue. B, the DGK1(D177A) mutant lacks DAG kinase activity. The dgk1Δ strain was transformed with an empty centromeric plasmid or with the same plasmid carrying DGK1 or DGK1(D177A) under the control of the GAL1/10 promoter. Cell extracts from the corresponding strains were assayed for diacylglycerol kinase activity. Values and errors were calculated from three experiments. C, DGK1(D177A) does not promote nuclear membrane growth. The RS453 strain expressing a SEC63-GFP fusion was transformed with the galactose-inducible DGK1 or DGK1(D177A) construct from B, grown to early logarithmic phase in galactose-containing medium, and inspected by microscopy.
Antibodies—The antibody used to detect protein A fusions was from DAKO (catalog no. Z0113).
RESULTS
Systematic Analysis of the Function of Phospholipid Biosynthetic Enzymes in Nuclear Structure—Pah1p is a conserved Mg2+-dependent PA phosphatase required for phospholipid and triacylglycerol synthesis (9). Inactivation of Pah1p leads to changes in PA and phospholipid levels (9) and results in a massive expansion of the nuclear/ER membrane (10). To address the possible role of distinct lipid species in nuclear structure, we first asked whether inhibition of other phospholipid biosynthetic steps could have a similar effect on the nuclear/ER membrane. Using a SEC63-GFP fusion as a reporter, we analyzed nuclear/ER membrane structure in a series of mutants lacking the enzymes catalyzing key steps in PA, phosphatidylinositol, phosphatidylserine, PE, and PC biosynthesis (highlighted in Fig. 1A). Partial defects in peripheral ER structure were observed in cells lacking Cho2p and Opi3p, involved in the final steps of PC methylation (Fig. 1C). Depletion of Cds1p, the essential cytidylyltransferase that utilizes PA to synthesize phospholipids via the CDP-DAG pathway, also partially affects cortical ER structure (Fig. 1C). However, nuclear membrane proliferation and nuclear expansion, similar to those seen in pah1Δ cells, were not observed in any of the mutants tested. Therefore, among the different phospholipid biosynthetic steps, the Pah1p-mediated dephosphorylation of PA to DAG plays a unique role in nuclear structure.
Genetic Screens for Factors Regulating PA Metabolism Identify Dgk1p—We reasoned that, if PA regulates nuclear/ER membrane biogenesis, then the activity of other proteins that control PA levels might also impact on nuclear/ER structure. We therefore undertook two screens to identify novel genes encoding PA regulators. Both screens are based on our previous finding that the PA phosphatase activity of Pah1p, which controls cellular concentration of PA (9), is inhibited by phosphorylation (11). Expression of constitutively dephosphorylated Pah1p, generated by mutating its phosphorylation sites (PAH1-7P mutant), increases its PA phosphatase activity, represses transcription of phospholipid biosynthetic enzymes, and results in lethality in medium lacking inositol (11). This phenotype is consistent with a decrease in cellular PA levels. Similarly, overexpression of the Nem1p-Spo7p phosphatase complex, which dephosphorylates Pah1p, is also lethal (10). We therefore screened for high copy number suppressors that can rescue the lethality of the overexpression of PAH1-7P (screen I) or the NEM1-SPO7 phosphatase (screen II). Both screens identified independently the same gene, DGK1 (known previously as HSD1). The suppression of the PAH1-7P and NEM1-SPO7 lethality was confirmed by overexpressing DGK1 in the respective strains using the inducible GAL1/10 promoter (Fig. 2, A and B, respectively). DGK1/HSD1 was originally isolated in a screen for suppressors of a sly1 mutant involved in ER-to-Golgi trafficking (19), but its function and mechanism of suppression remained so far unknown. Its gene product, Dgk1p, is a 32.8-kDa protein that consists of a short N-terminal hydrophilic region (residues 1-73) followed by four putative transmembrane domains containing a predicted cytidylyltransferase domain (residues 74-288) (Fig. 2C). BLAST searches identified putative Dgk1p orthologs in several other yeast species, but not any apparent related proteins in higher eukaryotes.4 A GFP-Dgk1p fusion, expressed under the control of the endogenous DGK1 promoter, localized to the ER and nuclear membrane (Fig. 2D).
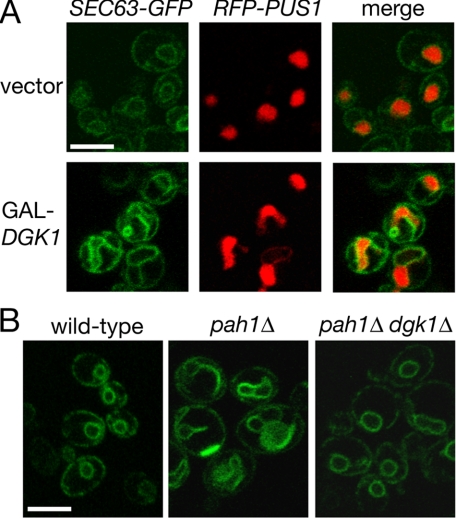
Dgk1p Counteracts the Function of Pah1p by Controlling PA Levels—Cells lacking DGK1 show no apparent growth defects or gross nuclear/ER structure abnormalities.4 Given that NEM1-SPO7 overexpression inhibits nuclear expansion during mitosis (10) and that a previous study reported the appearance of membranous tubules in DGK1-overexpressing cells (19), we first asked whether high levels of DGK1 could rescue the lethality of the NEM1-SPO7 overexpression by promoting nuclear growth. Indeed, wild-type cells overexpressing DGK1 from a galactose-inducible promoter contain enlarged and irregularly shaped nuclei and a striking nuclear/ER membrane proliferation that is reminiscent of that originally observed in cells lacking PAH1 (Fig. 3A). Because the overproduction of DGK1 phenocopies the morphological defects of pah1Δ cells, we next asked whether the complementary possibility would be true, i.e. whether lack of DGK1 would rescue the nuclear expansion that takes place in pah1Δ cells. Although pah1Δ dgk1Δ cells are still temperature-sensitive,4 they have normal nuclear morphology in that the nuclear/ER membrane labeling by SEC63-GFP looks identical to that of wild-type cells (Fig. 3B). Therefore, the function of Dgk1p in the nuclear structure appears to counteract that of Pah1p.
FIGURE 3.
DGK1 opposes the function of PAH1 at the nuclear membrane. A, overexpression of DGK1 causes nuclear/ER membrane proliferation and nuclear expansion. The RS453 strain expressing the plasmid-borne SEC63-GFP and RFP-PUS1 reporters was transformed with a centromeric plasmid expressing DGK1 under the control of a galactose-inducible promoter or an empty vector. Cells were grown in galactose-containing medium and visualized by confocal microscopy. Scale bar = 5 μm. B, deletion of DGK1 restores normal nuclear structure in pah1Δ cells. Wild-type RS453, pah1Δ, and pah1Δ dgk1Δ cells transformed with a centromeric plasmid expressing SEC63-GFP were visualized by confocal microscopy. Scale bar = 5 μm.
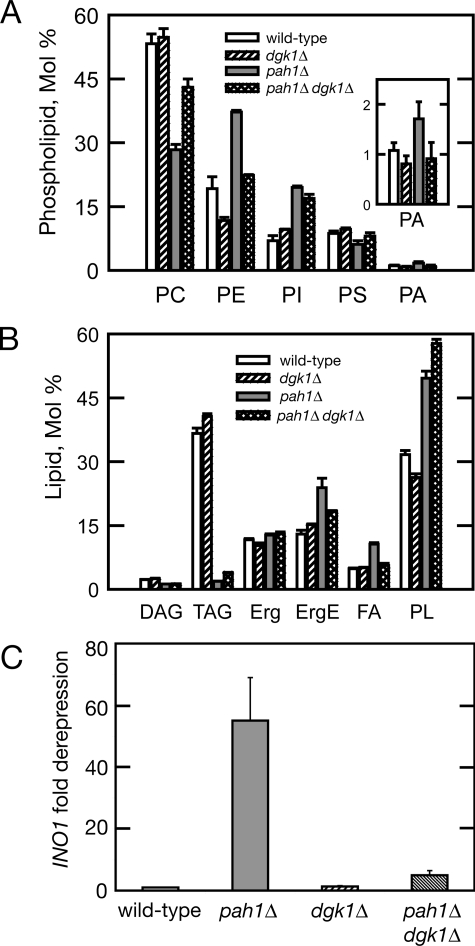
Because the accumulation of PA contributes to the transcriptional up-regulation of phospholipid biosynthetic genes and to the increased membrane biogenesis in pah1Δ cells, we next asked whether the lack of DGK1 rescues the nuclear membrane proliferation of pah1Δ cells by affecting cellular PA levels. To test this hypothesis, we compared the phospholipid composition of pah1Δ dgk1Δ cells with that of the corresponding single mutants and wild-type strain. Indeed, deletion of DGK1 returns the PA levels of the pah1Δ cells to normal (Fig. 4A). Consistent with this, mRNA levels of INO1 that respond to the intracellular PA concentration (13) are brought back to their respective wild-type values (Fig. 4C). The levels of PE in pah1Δ dgk1Δ are also brought back to normal, whereas the levels of PC are partially corrected. However, the abnormal phosphatidylinositol (Fig. 4A) and major neutral lipid triacylglycerol (Fig. 4B) levels of the pah1Δ mutant are not affected by the deletion of DGK1. Taken together, these data show that the function of DGK1 in the nuclear membrane counteracts that of PAH1 by controlling cellular PA levels.
FIGURE 4.
Deletion of DGK1 restores normal PA and INO1 transcript levels in pah1Δ cells. A, phospholipid composition of wild-type, dgk1Δ, pah1Δ, and pah1Δ dgk1Δ cells in the exponential phase of growth. The indicated yeast strains were grown to exponential phase in the presence of 32Pi (10 μCi/ml). Phospholipids were extracted and separated by two-dimensional thin layer chromatography. Images of 32P-labeled phospholipids were visualized by phosphorimaging and subjected to ImageQuant analysis. The percentages shown for the individual phospholipids were normalized to the total 32P-labeled chloroform-soluble fraction that included sphingolipids and unidentified phospholipids. Each data point represents the mean ± S.D. of three experiments. PI, phosphatidylinositol; PS, phosphatidylserine. B, neutral lipid composition of wild-type, dgk1Δ, pah1Δ, and pah1Δ dgk1Δ cells in the stationary phase of growth. The indicated yeast strains were grown to stationary phase in the presence of [2-14C]acetate (1 μCi/ml). Lipids were extracted and separated by one-dimensional thin layer chromatography. Images of 14C-labeled lipids were visualized by phosphorimaging and subjected to ImageQuant analysis. The percentages shown for the individual lipids were normalized to the total 14C-labeled chloroform-soluble fraction. Each data point represents the mean ± S.D. of three experiments. TAG, triacylglycerol; Erg, ergosterol; ErgE, ergosterol ester; FA, fatty acid; PL, phospholipid. C, INO1 mRNA levels of wild-type, pah1Δ, dgk1Δ, and pah1Δ dgk1Δ cells. The mRNA levels of INO1 were analyzed in the indicated strains grown in YEPD medium by quantitative real-time reverse transcription-PCR. Amplification of each sample was performed in duplicate and normalized to RTG2. Values and errors were calculated from three independent experiments. Values are expressed as -fold derepression over the values measured in the wild-type strain.
Dgk1p Is a Novel CTP-dependent DAG Kinase—The fact that the dgk1Δ mutation returns cellular PA levels in pah1Δ cells to normal raised the possibility that the activity of Dgk1p might be directly involved in the synthesis of PA. The cytidylyltransferase domain present in Dgk1p shows significant similarity to the PA cytidylyltransferase Cds1p (also known as CDP-DAG synthase), the enzyme that synthesizes CDP-DAG from CTP and PA. Cds1p can also catalyze the reverse reaction that leads to PA synthesis (20, 21). To investigate whether DGK1 can function as a CDP-DAG synthase, we asked whether (a) overexpression of DGK1 could rescue a conditional CDS1 mutant and (b) overexpression of CDS1 could rescue, like DGK1, the toxicity of PAH1-7P. In both cases, the results were negative.4 Taken together, these data suggest that Dgk1p does not function as a CDP-DAG synthase in vivo.
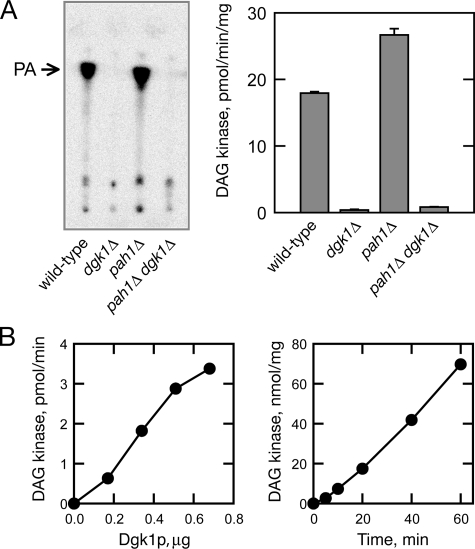
A second reaction that can directly generate PA is the phosphorylation of DAG. Although there is no homolog in Saccharomyces cerevisiae to any of the known ATP-dependent DAG kinases in bacteria or higher eukaryotes (22, 23), a previous report showed that PA is produced from membrane lipids using CTP as a phosphate donor (24). This, coupled with the fact that Dgk1p contains a CTP transferase domain, prompted us to test whether Dgk1p could be a CTP-dependent DAG kinase (25). We measured DAG kinase activity in extracts derived from wild-type, dgk1Δ, pah1Δ, and pah1Δ dgk1Δ cells using [γ-32P]CTP as a phosphate donor. As shown in Fig. 5A, the dgk1Δ mutation alone and in combination with pah1Δ resulted in a complete loss of the activity catalyzing the formation of PA from DAG and CTP. This result indicated that DGK1 encodes a DAG kinase activity. To confirm this, Dgk1p was partially purified as a PtA-Dgk1p fusion over an IgG-Sepharose column, followed by removal of the PtA tag by tobacco etch virus protease cleavage, and assayed for DAG kinase activity (Fig. 5B). The purified protein catalyzed a dose- and time-dependent phosphorylation of DAG using CTP as a phosphate donor (Fig. 5B). Taken together, these data show that Dgk1p is a novel CTP-dependent DAG kinase enzyme. The fact that Dgk1p uses CTP, rather than ATP, as a phosphate donor, explains why it does not show homology to any of the previously described DAG kinases from other species. The enzymological properties of this novel enzyme are described in the accompanying article (25).
FIGURE 5.
Dgk1p is a novel CTP-dependent DAG kinase. A, the dgk1Δ mutation abolishes diacylglycerol kinase activity. Cell extracts were prepared from wild-type, dgk1Δ, pah1Δ, and pah1Δ dgk1Δ cells at the exponential phase of growth, and 50 μg of protein from each cell extract was assayed for diacylglycerol kinase activity. Left panel, the 32P-labeled reaction product was extracted with chloroform, separated by thin layer chromatography using a solvent system of chloroform/methanol/water (65:25:4), and visualized by phosphorimaging. Right panel, the 32P-labeled reaction product was extracted with chloroform and subjected to liquid scintillation counting. Each data point represents the mean ± S.D. of triplicate enzyme determinations. B, Dgk1p exhibits DAG kinase activity. The DAG kinase activity of partially purified Dgk1p was measured with the indicated protein content (left panel) and for the indicated time intervals (right panel).
Dgk1p-derived PA Accumulates on the Nuclear Membrane—Next, we asked whether the DAG kinase activity of Dgk1p is responsible for nuclear/ER membrane growth. The CTP transferase domain of Dgk1p exhibits significant similarity to the corresponding domain of cytidylyltransferases, presumably because both need to bind to CTP for catalysis. Multiple sequence alignment of Dgk1p with cytidylyltransferases from many species identified an aspartate residue (Asp177) within its catalytic domain that is essentially conserved in all cytidylyltransferases examined (Fig. 6A). A dgk1Δ strain expressing a mutant DGK1 allele in which this residue was changed to alanine (DGK1(D177A)) did not display any CTP-dependent DAG kinase activity (Fig. 6B) and, unlike wild-type DGK1, did not trigger nuclear/ER membrane expansion in galactose medium (Fig. 6C). Therefore, the DAG kinase activity of Dgk1p is responsible for nuclear membrane expansion.
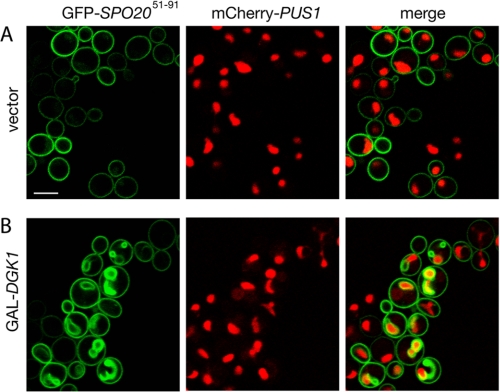
If the DAG kinase activity of Dgk1p causes nuclear expansion, then one would expect that PA levels in intracellular membranes would rise following the induction of Dgk1p. To visualize the intracellular location of PA accumulation, we followed the distribution of a GFP fusion of a short domain of the t-SNARE (target membrane soluble N-ethylmaleimide-sensitive factor attachment protein receptor) Spo20p (GFP-Spo20p-(51-91)). This GFP fusion is responsive to changes in PA levels in vivo and has been used previously as a PA sensor in both yeast (26) and mammalian (27) cells. Cells expressing GFP-Spo20p-(51-91) were transformed with either an empty vector or a GAL1/10-driven DGK1 plasmid and transferred into galactose-containing medium. Consistent with previous data (26), in control cells, GFP-Spo20p-(51-91) was found at the plasma membrane (Fig. 7A). When cells carrying GAL-DGK1 were transferred into galactose medium, the PA sensor relocalized to internal membranes (Fig. 7B). Interestingly, in the majority of these cells (79% of the cells with internal membrane staining), the PA-enriched membranes formed around the irregularly shaped nucleus and often extended into the cytoplasm (Fig. 7B). Overexpression of bacterial Dgk1p, which has been shown to relocalize to internal membranes in yeast (26), resulted in a similar relocation of the PA sensor to the nuclear membrane.4 Taken together, these results indicate that Dgk1p-produced PA accumulates on intracellular membranes around the nucleus.
FIGURE 7.
Overexpression of Dgk1p causes relocation of a PA-binding reporter at the nuclear membrane. The RS453 strain expressing the mCherry-PUS1 fusion and the GFP-SPO20-(51-91) reporter was transformed with a 2 μ vector (A) or the same vector carrying DGK1 (B). Cells were grown in raffinose, transferred into galactose-containing medium for 6 h, and viewed by confocal microscopy. Scale bar = 5 μm.
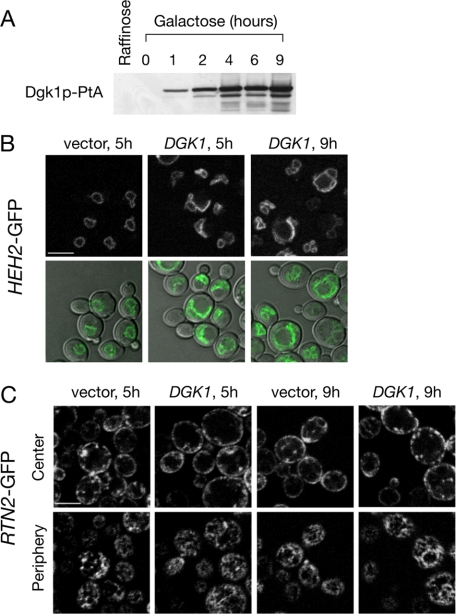
Dgk1p Regulates Nuclear Membrane but Not Cortical ER Membrane Proliferation—We next asked whether the high levels of PA produced by Dgk1p cause membrane proliferation of both ER subcompartments, viz. the cortical ER and the perinuclear ER or nuclear membrane. To address this question, we used a GFP fusion of Rtn2p, a member of the reticulon family of integral membrane proteins that, unlike other ER proteins like Sec63p, localize exclusively to the cortical ER (28, 29). To label the nuclear membrane, we expressed a GFP fusion of Heh2p, an integral inner nuclear membrane protein that does not diffuse to the cortical ER (30). When cells carrying GAL-DGK1 were grown under non-inducing conditions (Fig. 8A), neither the nuclear nor cortical ER was affected.4 Induction of Dgk1p production for 5 h led to irregularly shaped nuclei, often containing one nuclear membrane projection extending into the cytoplasm (Fig. 8B). After 9 h, nuclear expansion was more evident, leading to many cells with two expanded interconnected nuclear compartments. Interestingly, at both time points, no apparent proliferation of the cortical ER membrane was seen. The overall cortical (Fig. 8C, upper panels) and reticular (lower panels) membrane staining of Rtn2p-GFP was not significantly altered in the Dgk1p-overexpressing cells compared with the control cells. Therefore, elevated PA levels, generated by the overexpression of Dgk1p, causes proliferation of the membrane of the nuclear but not cortical ER.
FIGURE 8.
Overexpression of Dgk1p causes proliferation of the nuclear membrane but not cortical ER membrane. A, shown is the time course of the induction of DGK1-PtA. Wild-type cells (RS453) expressing DGK1-PtA under the control of the GAL1/10 promoter in a centromeric plasmid were transferred from raffinose to galactose-containing medium, and protein extracts were prepared at the indicated time points. Protein samples were resolved by SDS-PAGE followed by Western blotting using anti-PtA antibodies. B, cells expressing an HEH2-GFP fusion and DGK1 under the control of the GAL1/10 promoter in a centromeric plasmid were transferred from raffinose to galactose-containing medium for the indicated times. Heh2p-GFP labeling (upper panels) or cell/Heh2p-GFP overlays (lower panels) were visualized at the indicated time points by confocal microscopy. C, cells expressing an RTN2-GFP fusion and DGK1 under the control of the GAL1/10 promoter in a centromeric plasmid were transferred from raffinose to galactose-containing medium for the indicated times. Upper panels, confocal Z-sections corresponding to the cell center; lower panels, confocal sections corresponding to the cell periphery. Scale bars = 5 μm.
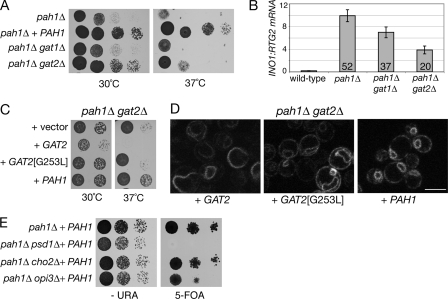
Lack of Biosynthetic PA Enzymes Restores Nuclear Structure in pah1Δ Cells—If the decrease in PA is responsible for restoring a normal nucleus in the pah1Δ dgk1Δ mutant, then other mutations that are known to decrease the rate of the de novo synthesis of PA should have a similar compensatory effect in pah1Δ cells. To test this hypothesis, we deleted the redundant acyltransferase genes GAT1 and GAT2, previously shown to be required for PA production (Ref. 31; see also Fig. 1A), in the pah1Δ mutant. The pah1Δ gat2Δ double mutant grew better than the pah1Δ mutant at both 30 and 37 °C (Fig. 9A) and exhibited a >50% decrease in the transcriptional derepression of the INO1 gene as measured by quantitative reverse transcription-PCR (Fig. 9B). This epistatic effect is due to the lack of Gat2p enzymatic activity, required for PA production, because it can be reproduced using a mutant (G253L) that is predicted to be catalytically dead (Fig. 9C) (32). Notably, the nuclear structure in pah1Δ cells lacking Gat2p activity was significantly improved compared with the pah1Δ cells (Fig. 9D). Therefore, inhibition of upstream steps that reduce PA synthesis is sufficient to decrease the membrane proliferation defects of pah1Δ cells. We next analyzed the effect of inhibiting steps downstream of the PA phosphatase reaction, leading to the production of PE (pah1Δ psd1Δ) or PC (pah1Δ cho2Δ and pah1Δ opi3Δ) in pah1Δ cells. Interestingly, we found that the pah1Δ psd1Δ double mutant is synthetically lethal (Fig. 9E), possibly because of the simultaneous inhibition of both the CDP-DAG (due to psd1Δ) and Kennedy (due to pah1Δ) pathways of phospholipid biosynthesis. Unlike pah1Δ gat2Δ, neither the pah1Δ cho2Δ nor pah1Δ opi3Δ mutant exhibited improved growth (Fig. 9E) or nuclear membrane structure.4 Taken together, these data indicate that high PA levels are a major trigger for the nuclear membrane proliferation in yeast cells.
FIGURE 9.
Genetic interactions of phospholipid biosynthetic mutants with pah1Δ indicate a role for PA in nuclear membrane proliferation. A, deletion of GAT2 partially rescues the growth defects of pah1Δ. Serial dilutions of the indicated strains were spotted on YEPD plates and grown for 2 days at 30 and 37 °C. B, deletion of GAT2 decreases INO1 mRNA levels in the pah1Δ cells. The mRNA levels of INO1 were analyzed in the indicated strains grown in YEPD medium by quantitative real-time PCR. Amplification of each sample was performed in duplicate and normalized to RTG2. Values and errors were calculated from three independent experiments. The values indicated within the bars represent the -fold derepression over the values measured in the wild-type strain. C, the GAT2-dependent rescue depends on its catalytic activity. Serial dilutions of the indicated strains were spotted on synthetic plates and grown for 2 days at 30 and 37 °C. D, lack of active GAT2 partially rescues nuclear membrane proliferation in pah1Δ cells. The pah1Δ gat2Δ mutant expressing SEC63-GFP was transformed with the indicated plasmids, and cells were observed by confocal microscopy. Scale bar = 5 μm. E, deletion of PSD1, CHO2, and OPI3 does not rescue the growth defects of pah1Δ. Serial dilutions of the indicated strains were spotted on -URA (left panel) or 5-fluoroorotic acid (5-FOA; right panel) plates and grown for 2 or 3 days, respectively, at 30 °C.
DISCUSSION
How phospholipids impact on organelle structure remains largely unknown. In this work, we have shown that PA homeostasis is a critical factor controlling nuclear structure in yeast. In two genetic screens looking for novel regulators of PA metabolism, we identified Dgk1p, the first DAG kinase described in yeast. Dgk1p-driven overproduction of PA at the nuclear membrane causes nuclear expansion, without proliferating cortical ER membrane. Thus, the modulation of Dgk1p activity offers a unique model system to study the regulation of nuclear growth.
Dgk1p Is a Novel CTP-dependent DAG Kinase—DAG kinase enzymes have been identified in almost all organisms examined, including bacteria, plants, worms, flies, and mammals. All DAG kinases have a catalytic domain that contains an ATP-binding site with the sequence GXGXXG that is essential for the kinase reaction (22, 23). Dgk1p is an unconventional DAG kinase that uses CTP, instead of ATP, as a phosphate donor and therefore does not exhibit sequence similarity to DAG kinases from other species. Furthermore, Dgk1p contains a short motif recently identified in a family of CTP-dependent phytol and dolichol kinases (33)4. Although the significance of this difference between Dgk1p and the other DAG kinases is not yet clear, it could have important physiological implications. Previous studies have established that elevated levels of cytosolic CTP (due to a defect in CTP feedback inhibition of CTP synthase) lead to increased PA levels, derepression of INO1, and the inositol excretion phenotype (34). Our data raise the possibility that this regulation is mediated by the CTP-driven activation of Dgk1p.
Regulation of PA Homeostasis by Dgk1p—The genetic interactions between DGK1 and PAH1 fit well with the biochemical data on the activity of Dgk1p. Overexpression of a hyperactive Pah1p mutant decreases cellular PA by converting it to DAG, causing the PA-dependent repression of phospholipid biosynthetic genes by Opi1p, and results in inositol auxotrophy (11). This lethality can be rescued by the overexpression of Dgk1p, which catalyzes the reverse reaction and restores PA levels. The fact that the deletion of DGK1 in pah1Δ cells returns PA levels to normal could indicate that, in wild-type cells, Dgk1p produces some of the ER pools of PA that are accessible to Pah1p.
Although the dgk1Δ mutation returns the levels of the other abundant phospholipids, PC and PE, almost to normal in pah1Δ cells, it does not change the defects in phosphatidylinositol and triacylglycerol levels. The fact that dgk1Δ pah1Δ cells have completely normal nuclear structure but still exhibit growth defects at 30 °C and are temperature-sensitive at 37 °C, like pah1Δ cells, suggests that PA and/or PC and PE are responsible for the effects on the nuclear membrane, whereas phosphatidylinositol and triacylglycerol levels contribute more to the toxicity of pah1Δ cells.
Interestingly, not all enzymes involved in the production of PA can rescue pah1Δ when removed. Although deletion of either Gat1p or Gat2p ER acyltransferases has been shown previously to significantly reduce cellular PA levels (31), only gat2Δ can partially rescue the pah1Δ defects. The fact that these two enzymes have distinct fatty acyl substrate specificities (31) raises the intriguing possibility that different PA pools, with respect to acyl chain length and/or saturation, could have different physiological effects. Alternatively, differences in the effects of the gat1Δ and gat2Δ mutations in lipid synthesis downstream of PA (35) could also contribute to their differential rescue of pah1Δ cells.
A Role for PA Metabolism in Nuclear Structure—Overexpression of certain transmembrane enzymes involved in lipid biosynthesis, such as hydroxymethylglutaryl-CoA and cytochrome b5, induces membrane biogenesis in the form of karmellae, stacks of proliferated ER wrapped around the nucleus (36, 37). In both cases, ER membrane proliferation could be induced even when the catalytically inactive proteins were overexpressed (37, 38), indicating that, unlike Dgk1p, membrane proliferation in these cells is not a direct consequence of changes in lipid composition but rather a response to accommodate the excess transmembrane proteins.
Here, we have shown that the DAG kinase activity of Dgk1p is responsible for nuclear membrane proliferation. High PA levels can have two distinct effects on lipid homeostasis and membrane biogenesis that could be responsible for nuclear growth. First, PA acts as a signal that derepresses transcription of key enzymes of the CDP-DAG pathway (Fig. 1A), which provide bulk phospholipids for membrane formation. Indeed, transcriptional derepression of these enzymes is necessary for nuclear membrane proliferation in pah1Δ cells (10). However, constitutive overexpression of the biosynthetic enzymes in cells lacking the repressor OPI1 does not lead to nuclear membrane expansion, suggesting that, apart from the derepression of lipid synthesis in the ER, other events contribute to nuclear membrane expansion (11). The second effect of PA on membranes is due to its conical structure and its bending functions that are implicated in many remodeling events like membrane budding and fission. Changes in the physical properties of the double bilayered nuclear membrane could contribute to its expansion. Interestingly, a role for membrane tubulation in nuclear membrane expansion has been described recently in tissue cultured cells in which expression of membrane-deforming mutants of CTP-phosphocholine cytidylyltransferase-α causes nuclear membrane proliferation (39, 40).
The distinct functions of Dgk1p-driven PA production in the nuclear envelope are also underscored by the fact that overproduction of Dgk1p does not appear to significantly induce cortical ER membrane proliferation, as visualized using a reticulon-GFP fusion. This raises the intriguing question of whether PA and downstream-derived phospholipids are differentially processed in the two subcompartments or whether active Dgk1p concentrates preferentially on the nuclear membrane. Given the significant changes that yeast nuclei undergo during cell division, the temporal and spatial regulation of PA metabolism by Dgk1p and Pah1p could play crucial roles in this process. Future studies will further address the role of Dgk1p in nuclear structure and phospholipid biosynthesis.
Supplementary Material
Acknowledgments
We thank Dr. Aaron Neiman for plasmids and Drs. N. Ktistakis, F. Reggiori, and H. Santos-Rosa for comments on the manuscript.
Author's Choice—Final version full access.
This work was supported, in whole or in part, by National Institutes of Health Grant GM-28140 from the United States Public Health Service (to G. M. C.). This work was also supported by a Wellcome Trust Career Development Fellowship in Basic Biomedical Science (to S. S). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
This article was selected as a Paper of the Week.
Footnotes
The abbreviations used are: ER, endoplasmic reticulum; PA, phosphatidate; DAG, diacylglycerol; GFP, green fluorescent protein; PE, phosphatidylethanolamine; PC, phosphatidylcholine; PtA, protein A; RFP, red fluorescent protein.
S. Siniossoglou, unpublished data.
References
- 1.van Meer, G., Voelker, D. R., and Feigenson, G. W. (2008) Nat. Rev. Mol. Cell Biol. 9 112-124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fagone, P., Sriburi, R., Ward-Chapman, C., Frank, M., Wang, J., Gunter, C., Brewer, J. W., and Jackowski, S. (2007) J. Biol. Chem. 282 7591-7605 [DOI] [PubMed] [Google Scholar]
- 3.Vance, J. E., Pan, D., Vance, D. E., and Campenot, R. B. (1991) J. Cell Biol. 115 1061-1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hetzer, M. W., Walther, T. C., and Mattaj, I. W. (2005) Annu. Rev. Cell Dev. Biol. 21 347-380 [DOI] [PubMed] [Google Scholar]
- 5.Byers, B., and Goetsch, L. (1975) J. Bacteriol. 124 511-523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burke, B., and Ellenberg, J. (2002) Nat. Rev. Mol. Cell Biol. 3 487-497 [DOI] [PubMed] [Google Scholar]
- 7.Leitch, A. R. (2000) Microbiol. Mol. Biol. Rev. 64 138-152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mounkes, L., Kozlov, S., Burke, B., and Stewart, C. L. (2003) Curr. Opin. Genet. Dev. 13 223-230 [DOI] [PubMed] [Google Scholar]
- 9.Han, G.-S., Wu, W. I., and Carman, G. M. (2006) J. Biol. Chem. 281 9210-9218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Santos-Rosa, H., Leung, J., Grimsey, N., Peak-Chew, S., and Siniossoglou, S. (2005) EMBO J. 24 1931-1941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O'Hara, L., Han, G.-S., Peak-Chew, S., Grimsey, N., Carman, G. M., and Siniossoglou, S. (2006) J. Biol. Chem. 281 34537-34548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carman, G. M., and Henry, S. A. (2007) J. Biol. Chem. 282 37293-37297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loewen, C. J., Gaspar, M. L., Jesch, S. A., Delon, C., Ktistakis, N. T., Henry, S. A., and Levine, T. P. (2004) Science 304 1644-1647 [DOI] [PubMed] [Google Scholar]
- 14.Han, G.-S., Siniossoglou, S., and Carman, G. M. (2007) J. Biol. Chem. 282 37026-37035 [DOI] [PubMed] [Google Scholar]
- 15.Nasmyth, K., and Reed, S. I. (1980) Proc. Natl. Acad. Sci. U. S. A. 77 2119-2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siniossoglou, S., Peak-Chew, S. Y., and Pelham, H. R. (2000) EMBO J. 19 4885-4894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Müller, M. O., Meylan-Bettex, M., Eckstein, F., Martinoia, E., Siegenthaler, P. A., and Bovet, L. (2000) J. Biol. Chem. 275 19475-19481 [DOI] [PubMed] [Google Scholar]
- 18.Carman, G. M., and Fischl, A. S. (1992) Methods Enzymol. 209 305-312 [DOI] [PubMed] [Google Scholar]
- 19.Kosodo, Y., Imai, K., Hirata, A., Noda, Y., Takatsuki, A., Adachi, H., and Yoda, K. (2001) Yeast 18 1003-1014 [DOI] [PubMed] [Google Scholar]
- 20.Belendiuk, G., Mangnall, D., Tung, B., Westley, J., and Getz, G. S. (1978) J. Biol. Chem. 253 4555-4565 [PubMed] [Google Scholar]
- 21.Kelley, M. J., and Carman, G. M. (1987) J. Biol. Chem. 262 14563-14570 [PubMed] [Google Scholar]
- 22.Luo, B., Regier, D. S., Prescott, S. M., and Topham, M. K. (2004) Cell. Signal. 16 983-989 [DOI] [PubMed] [Google Scholar]
- 23.Sakane, F., Imai, S., Kai, M., Yasuda, S., and Kanoh, H. (2007) Biochim. Biophys. Acta 1771 793-806 [DOI] [PubMed] [Google Scholar]
- 24.Szkopinska, A., Nowak, L., Swiezewska, E., and Palamarczyk, G. (1988) Arch. Biochem. Biophys. 266 124-131 [DOI] [PubMed] [Google Scholar]
- 25.Han, G.-S., O'Hara, L., Siniossoglou, S., and Carman, G. M. (2008) J. Biol. Chem. 283 20443-20453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakanishi, H., de los Santos, P., and Neiman, A. M. (2004) Mol. Biol. Cell 15 1802-1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zeniou-Meyer, M., Zabari, N., Ashery, U., Chasserot-Golaz, S., Haeberlé, A. M., Demais, V., Bailly, Y., Gottfried, I., Nakanishi, H., Neiman, A. M., Du, G., Frohman, M. A., Bader, M. F., and Vitale, N. (2007) J. Biol. Chem. 282 21746-21757 [DOI] [PubMed] [Google Scholar]
- 28.Voeltz, G. K., Prinz, W. A., Shibata, Y., Rist, J. M., and Rapoport, T. A. (2006) Cell 124 573-586 [DOI] [PubMed] [Google Scholar]
- 29.De Craene, J. O., Coleman, J., Estrada de Martin, P., Pypaert, M., Anderson, S., Yates, J. R., III, Ferro-Novick, S., and Novick, P. (2006) Mol. Biol. Cell 17 3009-3020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.King, M. C., Lusk, C. P., and Blobel, G. (2006) Nature 442 1003-1007 [DOI] [PubMed] [Google Scholar]
- 31.Zheng, Z., and Zou, J. (2001) J. Biol. Chem. 276 41710-41716 [DOI] [PubMed] [Google Scholar]
- 32.Lewin, T. M., Wang, P., and Coleman, R. A. (1999) Biochemistry 38 5764-5771 [DOI] [PubMed] [Google Scholar]
- 33.Shridas, P., and Waechter, C. J. (2006) J. Biol. Chem. 281 31696-31704 [DOI] [PubMed] [Google Scholar]
- 34.Ostrander, D. B., O'Brien, D. J., Gorman, J. A., and Carman, G. M. (1998) J. Biol. Chem. 273 18992-19001 [DOI] [PubMed] [Google Scholar]
- 35.Zaremberg, V., and McMaster, C. R. (2002) J. Biol. Chem. 277 39035-39044 [DOI] [PubMed] [Google Scholar]
- 36.Wright, R., Basson, M., D'Ari, L., and Rine, J. (1988) J. Cell Biol. 107 101-114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vergères, G., Yen, T. S., Aggeler, J., Lausier, J., and Waskell, L. (1993) J. Cell Sci. 106 249-259 [DOI] [PubMed] [Google Scholar]
- 38.Parrish, M. L., Sengstag, C., Rine, J. D., and Wright, R. L. (1995) Mol. Biol. Cell 6 1535-1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lagace, T. A., and Ridgway, N. D. (2005) Mol. Biol. Cell 16 1120-1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gehrig, K., Cornell, R. B., and Ridgway, N. D. (2008) Mol. Biol. Cell 19 237-247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nikawa, J., and Yamashita, S. (1984) Eur. J. Biochem. 143 251-256 [DOI] [PubMed] [Google Scholar]
- 42.Kyte, J., and Doolittle, R. F. (1982) J. Mol. Biol. 157 105-132 [DOI] [PubMed] [Google Scholar]
- 43.Wimmer, C., Doye, V., Grandi, P., Nehrbass, U., and Hurt, E. C. (1992) EMBO J. 11 5051-5061 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.