Abstract
The homeodomain protein Distal-less-3 (Dlx3) plays a crucial role during embryonic development. This transcription factor is known to be essential for placental formation and to be involved in skin and skeletal organogenesis. In humans, a frameshift mutation in the coding sequence of the DLX3 gene results in an ectodermal dysplasia called Tricho-Dento-Osseous syndrome (TDO). The main features of this autosomal dominant disorder are defects in hair, teeth, and bone. To investigate the functional alterations caused by the mutated DLX3TDO isoform ex vivo, we used tetracycline-inducible osteoblastic and keratinocyte cell lines and calvarial derived osteoblasts in which the expression of DLX3WT and/or DLX3TDO could be regulated and monitored. Immunocytochemical analysis revealed that both DLX3WT and DLX3TDO recombinant proteins are targeted to the nucleus. However, as demonstrated by electrophoresis mobility shift assay, DLX3TDO is not able to bind to the canonical Dlx3 binding site. Furthermore, we demonstrate that the frameshifted C-terminal domain in DLX3TDO is accountable for the loss of DNA binding activity because the C-terminal domain in DLX3WT is not required for DNA binding activity. Although DLX3TDO alone cannot bind to a Dlx3 responsive element, when DLX3WT and DLX3TDO are co-expressed they form a complex that can bind DNA. Concomitant with the inability to bind DNA, DLX3TDO has a defective transcriptional activity. Moreover, the transcriptional activity of DLX3WT is significantly reduced in the presence of the mutated isoform, indicating that DLX3TDO has a dominant negative effect on DLX3WT transcriptional activity.
Distal-less genes code for homeodomain transcription factors known to be widely involved in the patterning of the developing embryo. In mouse and human, there are six Distal-less genes. In the genome, they are organized into three pairs of inverted, convergently transcribed genes, termed Dlx1–2, Dlx3–4, and Dlx5–6 (1, 2). Dlx3 is associated to Dlx4 on chromosome 11 in mouse and on chromosome 17 in human (3).
During early development, Dlx3 is expressed in the placenta where it plays an essential role in the patterning of this tissue. Underscoring the important role of Dlx3, we showed that due to placental failure, Dlx3 null mice are embryonic lethal (4). Interestingly, Dlx3 is the only member of the Distal-less family that has not been detected in the mammalian central nervous system. However, Dlx3 is expressed in the first and second branchial arches and their derivatives including craniofacial bone (5, 6). Later during development, Dlx3 is expressed in structures involving epithelial-mesenchymal interaction such as teeth and hair follicles, and in skin (5, 7). In vitro and in vivo data support a role of Dlx3 in epidermal differentiation. Dlx3 misexpression in the basal layer of the epidermis leads to reduced proliferation, premature differentiation of basal keratinocytes, and reduced stratification of the epidermis (8). More recently, Dlx3 was shown to be directly involved in the regulation of bone differentiation markers (9, 10), and immunohistochemical analyses revealed Dlx3 expression in diverse secretory cells of mineralized tissues, not only in craniofacial bone and tooth (intramembranous ossification) but also in the appendicular skeleton (endochondral ossification) (9, 11).
Apart from Dlx3 expression pattern and ex vivo studies, there is clinical evidence showing that DLX3 plays a crucial role in the patterning of hair, teeth, and bone. Indeed, an ectodermal dysplasia called Tricho-Dento-Osseous (TDO)2 has been linked to a mutation (GGGG deletion) just downstream of the DLX3 homeodomain (12, 13). Ectodermal dyplasia is a general term used for inherited diseases characterized by anomalies in epithelial-mesenchymal-derived organs. TDO is an autosomal dominant disorder characterized by defects in hair (kinky hair), teeth (enamel hypoplasia and taurodontism), and bone (increased thickness and density of craniofacial bone) (14–16). More recently, the appendicular skeleton was also shown to exhibit increased bone density in TDO patients, indicating the importance of functional DLX3 in both intramembranous and endochondral bone formation (17).
Dlx3 is a transcriptional activator with an optimal DNA binding site comprised by a TAATT motif (18). Its transcriptional activity is dependent on two transactivation domains: one in the N-terminal domain and one located just downstream of the homeodomain region (18, 19). A bipartite nuclear localization signal was identified just upstream of the homeodomain (19). The 4G-deletion causing TDO syndrome occurs 3 base pairs downstream of the homeodomain (13), resulting in a C-terminal domain that is frameshifted just after the first codon following the homeodomain and thus exhibits an amino acid sequence that differs from the C-terminal domain in wild type DLX3 (less than 18% homology). The frameshift also results in the truncation of the protein that counts 255 amino acids instead of 287. Because the two established transactivation domains are required for Dlx3 transcriptional activity (18, 19), the absence of the second transactivation domain makes it likely that DLX3TDO is not functional even though it could potentially still bind DNA through the intact homeodomain region. Although Dlx3 knock-out mice are embryonic lethal, heterozygous mice did not present any of the abnormalities characteristic of TDO (4), eliminating haploinsufficiency as a probable cause of the defects. It is likely that the dominant pattern of inheritance in TDO is due to the formation of non-functional complexes involving the mutated transcription factor, which acts through either a dominant-negative or gain of function mechanism.
To investigate the molecular basis of the pathogenicity, we have compared the biochemical properties of DLX3WT and DLX3TDO, and analyzed the effects of the TDO mutation on the function of DLX3. Given that the TDO syndrome is an autosomal dominant disorder, it is essential to analyze the properties of the mutant isoform in the presence of an equivalent amount of its wild type counterpart. Therefore, we used an ex vivo system to co-express DLX3WT and DLX3TDO, and evaluate the effect of the mutant isoform on the function of the wild type factor. In summary, we show that DLX3TDO is targeted to the nucleus, is not able to bind DNA, can interact with DLX3WT and indirectly bind DNA, and is a defective transcription factor exerting a dominant negative effect on DLX3WT transcriptional activity.
MATERIALS AND METHODS
Plasmids—The TDO mutation was generated by site-directed mutagenesis using the QuikChange® Site-directed Mutagenesis Kit (Stratagene) utilizing the following primers: forward, AAACTCTACAAGAACΔAGGTGCCGCTGGAGC and reverse, GCTCCAGCGGCACCTΔGTTCTTGTAGAGTTT (subscript Δ indicates the position of the GGGG deletion). The bidirectional vector pBi-4 was used to simultaneously express the reporter protein EGFP with V5DLX3WT and V5DLX3TDO and also to express V5DLX3WT with FlagDLX3TDO (Fig. 1A), under control of a unique tetracycline responsive element. For non-inducible expression, V5DLX3WT and V5DLX3TDO were subcloned into the pCi-Neo plasmid (Promega). pCMV-GTF3-Δ1–3(V5), a construct expressing a V5-tagged truncated isoform of the general transcription factor 3 (20) was used as a negative control in one of the supershift assays. For the production of bacterial recombinant proteins, V5DLX3WT, V5DLX3TDO, and FlagDLX3TDO were cloned into a pet15b vector in which the expression of the transgene is isopropyl 1-thio-β-d-galactopyranoside-inducible and a His tag is added to the N-terminal part of the protein (pet15b-V5DLX3WT, pet15b-V5DLX3TDO, and pet15b-FlagDLX3TDO).
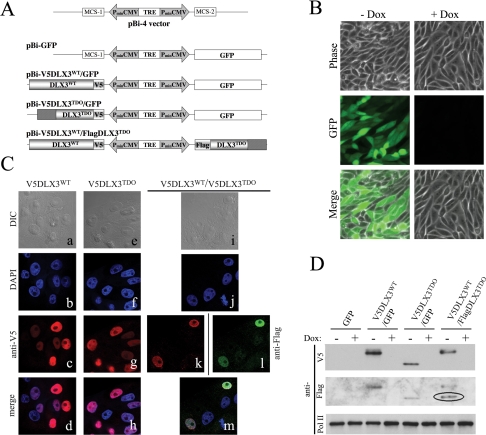
FIGURE 1.
Inducible expression of DLX3WT and/or DLX3TDO in Saos2-Tet-Off cells and nuclear localization. A, constructs developed using the bidirectional plasmid of the tetracycline system (pBi-4) allowing the inducible co-expression of two transgenes under control of a unique tetracycline responsive element. GFP alone (pBi-GFP), V5DLX3WT and GFP (pBi-V5DLX3WT/GFP), V5DLX3TDO and GFP (pBi-V5DLX3TDO/GFP), or V5DLX3WT and FlagDLX3TDO (pBi-V5DLX3WT/FlagDLX3TDO) were subcloned into pBi-4. B, panels showing GFP expression in Saos2-Tet-Off cells transfected with pBi-GFP and grown with or without doxycycline (Dox). The same exposure time was used for -Dox and +Dox. C, immunocytochemical analysis. Saos2-Tet-Off cells were transiently transfected with pBi-V5DLX3WT/GFP (a–d), pBi-V5DLX3TDO/GFP (e–h), or pBi-V5DLX3WT/FlagDLX3TDO (i–m), grown for 48 h on gelatin-coated glass coverslips, and fixed with 4% paraformaldehyde. Immunocytochemical stainings were performed using anti-V5 (c, g, and k) or anti-FLAG (l) as primary antibodies and, respectively, Alexa Fluor 543-conjugated goat anti-mouse IgG and Alexa Fluor 488-conjugated goat anti-rabbit IgG as secondary antibody. 4′,6-Diamidino-2-phenylindole (DAPI) was used to stain nuclei (b, f, and j). Observations and image acquisitions were performed using Zeiss 510 META confocal microscope. Fluorescent images were merged (d, h, and m) and differential interference contrast (DIC) images were acquired to visualize cell shape (a, e, and i). D, Western blot analysis of nuclear extracts. The expression of V5DLX3WT, V5DLX3TDO, and FlagDLX3TDO in Saos2-Tet-Off cells transiently transfected with the pBi constructs (+/-Dox) was analyzed using anti-V5, anti-FLAG, and anti-PolII (loading control). Blotting with anti-FLAG antibody was performed on the same membrane that was blotted with anti-V5 antibody. The band detected with anti-FLAG antibody is circled.
Cell Culture and Transfections—Saos2 human osteosarcoma Tet-Off cells (Clontech) were grown in Dulbecco's modified Eagle's medium (10% fetal bovine serum, 1% penicillin/streptomycin, and 1 μg/ml G418). For transfections, the cells were grown to at least 70% confluence. 2 million cells were used per transfection with pBi-GFP, pBi-V5DLX3WT/GFP, pBi-V5DLX3TDO/GFP, and pBi-V5DLX3WT/FlagDLX3TDO (Amaxa Nucleofactor). The transfected cells were seeded and grown in the absence or presence of 2 μg/ml doxycycline.
PAM212 cells (21) were used to produce a tetracycline-inducible keratinocyte cell line. These cells were stably transfected with a prtTA2-M2/IRES-Neo plamid obtained after subcloning of the rtTA2-M2 cassette (22) into the pCMV-IRES-Neo (Clontech). rtTA-M2 is a mutagenized form of rtTA that shows a lower basal activity and a higher sensitivity to doxycycline (Dox) than the original rtTA (22). The presence of the IRES cassette before the neomycin (Neo) resistance gene allowed coexpression of the rtTA-M2 transactivator and the Neo resistance gene, increasing the chance to select clones that express sufficient amounts of the transactivator in Neo-resistant cells. 20 clones were isolated and functionality of the tet system was screened by transient transfection with pTRE2-luc (expression of luciferase under the control of tetracycline responsive element). Cells were grown in the presence or absence of 2 μg/ml Dox and luciferase activity was estimated. One clone exhibiting a low basal activity of the transactivator in the absence of Dox and a strong induction in the presence of Dox (+Dox/-Dox ratio) was selected for subsequent experiments and named PAM-Tet-On. PAM-Tet-On cells were then transfected with the pBi constructs described earlier using Amaxa Nucleofactor.
MC3T3 cells were maintained in α-minimal essential medium supplemented with 10% fetal bovine serum. They were transfected using FuGENE 6 (Roche).
Production and Purification of Recombinant Proteins in Escherichia coli—Epicurian coli BL21-CodonPlus™-RIL strain of E. coli (Stratagene, La Jolla, CA) was transformed with pet15b-V5DLX3WT, pet15b-V5DLX3TDO, or pet15b-FlagDLX3TDO. Individual colonies were grown at 37 °C until optical density at 600 nm reached 0.5. Cells were then induced with isopropyl 1-thio-β-d-galactopyranoside and grown for 7 h at 30 °C. Cell pellets were resuspended in 1× PBS and sonicated. Cell lysates were clarified by centrifugation at 10,000 × g for 10 min and recombinant proteins were purified using HisTrap™ FF crude kit (GE Healthcare).
Production of Recombinant Proteins Using Reticulocyte Lysate System—In vitro transcribed/translated V5DLX3WT and V5DLX3TDO were produced from pet15b-V5DLX3WT and pet15b-V5DLX3TDO, respectively, using Promega TnT coupled with a rabbit reticulocyte lysate system (Promega).
Immunocytochemistry—Transfected cells were seeded on glass coverslips coated with 0.1% gelatin. 48 h after transfection, cells were washed three times in PBS and fixed with 4% paraformaldehyde in PBS for 15 min at room temperature. Free aldehydes were saturated using 50 mm NH4Cl. A 5-min incubation in 0.2% Triton in PBS was used to permeabilize the cells before blocking unspecific sites using 3% bovine serum albumin in PBS for 1 h. Primary antibodies diluted in blocking solution were applied for 1 h. Primary antibodies used were anti-V5 (Serotec) and anti-FLAG (Cell Signaling). Secondary antibodies diluted in blocking solution were applied for 30 min. Secondary antibodies used were Alexa Fluor® 543-conjugated goat anti-mouse IgG and Alexa Fluor® 488-conjugated goat anti-rabbit IgG (Invitrogen). Nuclei were stained using 4′,6-diamidino-2-phenylindole and coverslips were mounted on glass slides using Mowiol (Calbiochem). Pictures were acquired using a Zeiss 510 META confocal microscope.
Western Blot Analysis—Nuclear extracts isolated using the Nuclear Extract Kit (Active Motif) were run on 4–12% BisTris gels and MOPS SDS running buffer. Proteins were transferred onto polyvinylidene difluoride membranes and blocked in 5% nonfat powdered milk in TBS/Tween at room temperature for 1 h. The blots were probed with primary antibody diluted in 5% nonfat powdered milk in TBS/Tween and then with secondary antibody diluted in TBS/Tween, both at room temperature for 1 h. Primary antibodies used were anti-V5 (1:2000, Serotec), anti-FLAG (1:1000, Sigma), and anti-PolII (1:1000, Santa Cruz). The secondary antibody used was goat anti-mouse horseradish peroxidase (1:3000, Bio-Rad). After application of each antibody, the blots were rinsed three times with TBS/Tween under similar conditions. The blots were developed by ECL (Pierce).
Electrophoresis Mobility Shift Assays—Nuclear extracts were prepared using the Nuclear Extract Kit (Active Motif). Different probes known to bind Dlx3 were used: a probe containing the Dlx3 consensus binding site determined by SELEX (18) (GGGGGATAATTGCTGG), a probe containing the Dlx3 binding site identified in the osteocalcin promoter (9) (GGGCCCCCAATTAGTCCTCCC), and a probe containing the junctional regulatory element present in the promoter of the glycoprotein hormone α subunit expressed in the placenta (23) (GGGGGGTAATTACAGGCCC). These probes were radiolabeled using the High Prime DNA Labeling Kit (Roche) and [γ-32P]dCTP. Nuclear extracts were preincubated in 1× gel shift binding buffer (Promega) for 15 min at 4 °C, with an excess of unlabeled probe for competition assays or with appropriate antibody for supershift assays. As a control for the competition assays, a mutated unlabeled probe was used (GGGGGATGGCCGCTGG). Antibodies used for supershift assays were anti-V5 (Serotec) and anti-FLAG (Sigma). After this preincubation, each sample was supplemented with 5 × 104 dpm of radiolabeled probe and incubated for 30 min at 4 °C. The binding reactions were resolved on 6% DNA retardation gels (Invitrogen). The gels were dried and DNA-protein complexes were visualized by autoradiography. Electrophoretic mobility shift assay (EMSA) using proteins produced with a reticulocyte lysate system and a probe containing a Dlx3 binding site derived from the Runx2 promoter (TTCCATAGACATAATAATGAAGGAAAG) were performed using a procedure that has been described previously (9).
Co-immunoprecipitation—48 h after transfection, GFP was visualized and the cultured cells were rinsed with phosphate-buffered saline, scraped with Nonidet P-40 lysis buffer (50 mm Tris-HCl, pH 7.5, 5 mm EDTA, 150 mm NaCl, 0.5% Nonidet P-40) supplemented with a protease inhibitor mixture (Complete Mini, EDTA-free, Roche) and sonicated. Co-immunoprecipitation (co-IP) was performed using anti-FLAG® M2 Affinity Gel (Sigma). The affinity gel was washed three times in 1× TBS before incubation with cell extract supernatants (Nonidet P-40 lysates) at 4 °C overnight. The affinity gel was then washed three times in 1× TBS before elution of the immunoprecipitated proteins in 2× NuPAGE® LDS Sample Buffer (Invitrogen). Co-immunoprecipitated proteins were analyzed by Western blot using anti-V5 antibody. In some conditions, 10 μg/ml ethidium bromide (Invitrogen) or 10 units of RQ1 DNase (Promega) were added to the cell extracts prior to co-IP. Co-IP with purified bacterial recombinant proteins was performed similarly.
Reporter Assays—To determine the transcriptional activity, a synthetic oligonucleotide containing three tandem copies of the Dlx3 responsive element (DRE; GCGATAATTGCGGCGATAATTGCGGCGATAATTGCG) followed by the hamster sarcoma virus thymidine kinase proximal promoter region was subcloned from the reporter plasmid Dlx3-CAT used in previous studies (18) and inserted into the pGL3-basic vector (pGL3-3XDRE) driving a Firefly luciferase reporter cassette. As a control, an identical construct was prepared in which the central TAATT was changed to TGGCC (pGL3-3XmutDRE). Saos2-Tet-Off cells were transiently co-transfected with one of the pBi constructs, one of the pGL3 constructs, and the pRL-TK vector (Renilla luciferase used for normalization). 24 h after transfection, relative luciferase activity was measured using the Dual Luciferase® Reporter Assay System (Promega). Luciferase assays using a Runx2 promoter construct (-600 LUC) were performed in MC3T3 cells, following procedures as previously described (10).
RESULTS
DLX3WT and DLX3TDO Are Targeted to the Nucleus, but DLX3TDO Is Not Able to Bind to the Dlx3 Consensus Site—The Saos2-Tet-Off cell line (Clontech) was used to compare the biochemical properties of DLX3WT and DLX3TDO ex vivo. As shown in Fig. 1A, expression vectors were developed using the bidirectional plasmid of the tetracycline inducible system (pBi-4), inserting GFP alone as a reporter gene (pBi-GFP), GFP and DLX3WT tagged with a V5 epitope (pBi-V5DLX3WT/GFP), GFP and DLX3TDO tagged with a V5 epitope (pBi-V5DLX3TDO/GFP), or DLX3WT tagged with a V5 epitope and DLX3TDO tagged with a FLAG epitope (pBi-V5DLX3WT/FlagDLX3TDO). Saos2-Tet-Off cells were transiently transfected with the different pBi constructs and grown with or without doxycycline. After 48 h, the transfection efficiency was checked by detection of GFP expression (Fig. 1B). As assayed by fluorescence-activated cell sorter analysis, more than 70% of the cells transfected with pBi-GFP, pBi-V5DLX3WT/GFP, or pBi-V5DLX3TDO/GFP were GFP-positive (data not shown). As expected, the addition of doxycycline to those cells shut down the expression of GFP (Fig. 1B).
Immunocytochemical analysis using anti-V5 and anti-FLAG showed that both DLX3WT and DLX3TDO are located in the nucleus, even when the two isoforms are co-expressed (Fig. 1C). 48 h after transfection, nuclear extracts were harvested and analyzed by Western blot. Using anti-V5 antibody, we detected the expression of V5DLX3WT in cells transfected with pBi-V5DLX3WT/GFP and pBi-V5DLX3WT/FlagDLX3TDO, and V5DLX3TDO in cells transfected with pBi-V5DLX3TDO/GFP (Fig. 1D). Blotting the same membrane with anti-FLAG antibody, we detected the production of FlagDLX3TDO in cells transfected with pBi-V5DLX3WT/FlagDLX3TDO (Fig. 1D). As expected, the addition of doxycycline turned off the expression of all transgenes. Anti-PolII antibody was used to check the equal loading of nuclear extracts.
To determine the ability of DLX3WT and DLX3TDO to bind DNA, EMSA were performed using a consensus probe that was previously shown to bind Dlx3 (18). We first checked the ability of recombinant V5DLX3WT and V5DLX3TDO produced in bacteria to bind the Dlx3 binding site. Different amounts of recombinant protein (between 5 and 40 ng) were used in EMSA. As expected, a protein-DNA complex was formed between V5DLX3WT and its consensus binding site (Fig. 2A, lanes 2–5), whereas no complex was formed with V5DLX3TDO at any concentration (Fig. 2A, lanes 6–9), suggesting that the bacterial recombinant V5DLX3TDO is not able to bind the Dlx3 consensus site in vitro. Another independent assay was performed using recombinant V5DLX3WT and V5DLX3TDO that were in vitro transcribed/translated in a reticulocyte lysate system. The probe used in that assay contains a Dlx3 binding site that was previously identified in the Runx2 promoter (10). The complex formed between V5DLX3WT and the Runx2 promoter probe was competed by an excess of unlabeled probe and supershifted in the presence of anti-V5, which confirms the specificity of the protein-DNA complex formed (Fig. 2B, lanes 3–5). No complex was formed between V5DLX3TDO and the same probe (Fig. 2B, lanes 6–8).
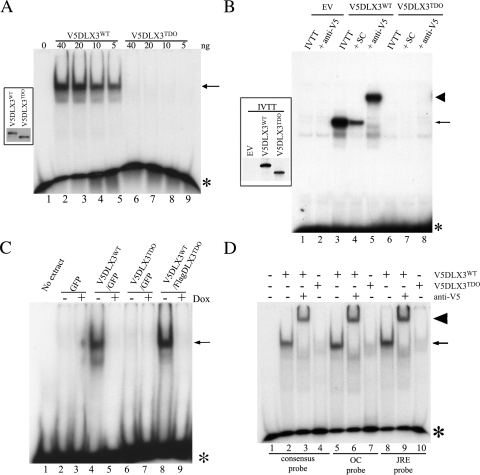
FIGURE 2.
DNA binding capacity of DLX3WT and DLX3TDO. EMSA were performed to compare the capacity of DLX3WT and DLX3TDO to bind DNA. A, recombinant V5DLX3WT and V5DLX3TDO were produced in E. coli, purified, and different amounts of protein were analyzed in EMSA using the Dlx3 consensus probe (GGGGGATAATTGCTGG). The inset on the left-hand side shows a Western blot analysis of both recombinant proteins using anti-V5 antibody. B, V5DLX3WT and V5DLX3TDO were in vitro transcribed/translated (IVTT) from pet15b-V5DLX3WT and pet15b-V5DLX3TDO (in vitro transcribed/translated, reticulocyte lysate system) and analyzed by EMSA using a probe containing a Dlx3 binding site previously identified in the Runx2 promoter. Empty vector (pet15b, EV) was used as a control. Excess unlabeled probe were used for self-competition (SC) and anti-V5 antibody was used for supershift assays. Asterisks, free radiolabeled probe; arrows, protein-DNA complexes; arrowheads, antibody-protein-DNA complexes. C, Saos2-Tet-Off cells transfected with pBi-GFP, pBi-V5DLX3WT/GFP, pBi-V5DLX3TDO/GFP, or pBi-V5DLX3WT/FlagDLX3TDO were grown with or without doxycycline for 48 h. Nuclear extracts were harvested and EMSA was performed using the Dlx3 consensus probe. D, recombinant V5DLX3WT and V5DLX3TDO were analyzed by EMSA using different probes known to bind Dlx3: a probe containing the Dlx3 consensus binding site determined by SELEX (GGGGGATAATTGCTGG), a probe containing the Dlx3 binding site identified in the osteocalcin (OC) promoter (GGGCCCCCAATTAGTCCTCCC), and a probe containing the junctional regulatory element (JRE) present in the promoter of the glycoprotein hormone α subunit expressed in the placenta. Anti-V5 antibody was used for supershift assays.
We then performed EMSA using the nuclear extracts introduced earlier (Fig. 1D). No complex was formed between nuclear extracts from cells transfected with pBi-GFP and the consensus probe (Fig. 2C, lanes 2), whereas a protein-DNA complex was formed with nuclear extracts containing V5DLX3WT (Fig. 2C, lane 4). Nuclear extracts with V5DLX3TDO did not form any complex with the Dlx3 consensus probe (Fig. 2C, lanes 6), confirming that the mutated transcription factor is not able to bind DNA, even in the context of a cell extract. However, a complex was formed when using extracts containing both V5DLX3WT and FlagDLX3TDO (Fig. 2C, lane 8), suggesting that the presence of DLX3TDO does not prevent DLX3WT from binding to the DNA. In all cases, the addition of doxycycline to repress the expression of the transgene resulted in the absence of protein-DNA complex (Fig. 2C, lanes 3, 5, 7, and 9).
We confirmed that DLX3TDO is not able to bind DNA by performing EMSA using different probes known to bind Dlx3 and different sources for the recombinant proteins. Recombinant V5DLX3TDO purified from bacteria did not form any complex with a probe containing either the Dlx3 binding site identified in the osteocalcin promoter or the junctional regulatory element present in the promoter of the glycoprotein hormone α subunit expressed in the placenta (Fig. 2D, lanes 7 and 10). As expected, V5DLX3WT did form a complex with those probes (Fig. 2D, lanes 5 and 8), and the complex was supershifted in the presence of anti-V5 antibody (Fig. 2D, lanes 6 and 9). The same experiment performed using nuclear extracts from Saos2-Tet-Off cells gave the same result (data not shown). Nuclear extracts were also harvested from a keratinocyte cell line in which the expression of V5DLX3WT or V5DLX3TDO was induced. PAM-Tet-On cells were transfected with pBi-V5DLX3WT/GFP or pBi-V5DLX3TDO/GFP and grown with or without doxycycline. Nuclear extracts from these cells were analyzed by EMSA using the Dlx3 consensus probe. V5DLX3TDO was not able to form a complex with the consensus Dlx3 binding site (data not shown).
DLX3WT and DLX3TDO Form a Complex That Is Able to Bind to the DNA—To address the possibility that DLX3TDO could interact with DLX3WT and form a complex, we performed co-immunoprecipitation assays. Extracts from Saos2-Tet-Off cells transfected with pBi-V5DLX3WT/FlagDLX3WT or pBi-V5DLX3WT/FlagDLX3TDO were immunoprecipitated with anti-FLAG-coated beads. Western blot analysis of the immunoprecipitation products using anti-V5 antibody revealed that V5DLX3WT was pulled-down with both FlagDLX3WT and FlagDLX3TDO (Fig. 3A). Extracts from cells transfected with pBi-V5DLX3WT/GFP were used as a control for interaction specificity. When EtBr or DNase were added to the cell extracts before performing the co-IP, V5DLX3WT was still pulled down with both FlagDLX3WT and FlagDLX3TDO, demonstrating that V5DLX3WT is not unspecifically pulled down via interactions with the sheared DNA present in the cell extracts (Fig. 3A). Performing the same experiment with purified recombinant V5DLX3WT and FlagDLX3TDO, we confirmed that the interaction detected in cell extracts is a direct interaction between the two proteins (Fig. 3B). These results confirm that DLX3 is able to interact with itself and with its mutated counterpart. Thus, although DLX3TDO by itself is not able to bind DNA, it can directly interact with DLX3WT and potentially affect its transcriptional activity.
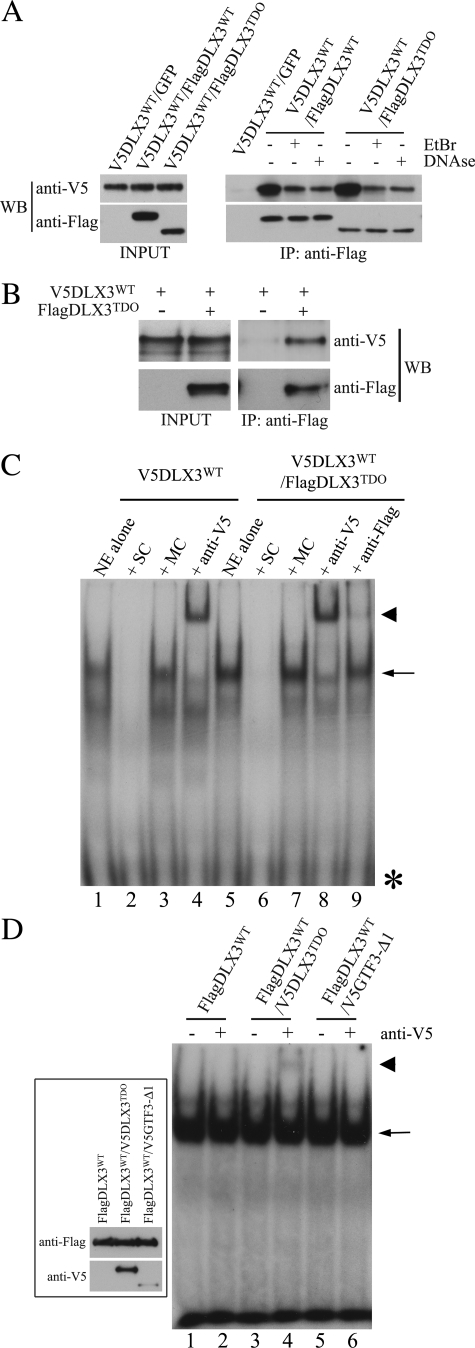
FIGURE 3.
Interaction between DLX3WT and DLX3TDO. A, co-IP using anti-FLAG-agarose beads performed on cell extracts containing V5DLX3WT alone, V5DLX3WT and FlagDLX3WT, or V5DLX3WT and FlagDLX3TDO. Western blot analysis of both inputs and immunoprecipitation products was performed using both anti-V5 and anti-FLAG antibodies. EtBr or DNase were added in some of the samples prior to co-IP. B, co-IP performed on purified bacterial recombinant proteins V5DLX3WT and FlagDLX3TDO, using anti-FLAG-agarose beads. Western blot analysis of inputs and immunoprecipitation products was performed using both anti-V5 and anti-FLAG antibodies. C, EMSA (described in the legend to Fig. 2) competition assays using an excess of non-radioactive consensus probe (self competition, SC) or mutant probe (mutant competition, MC) with nuclear extracts (NE) from Saos2-Tet-Off cells transfected with pBi-V5DLX3WT/GFP and pBi-V5DLX3WT/FlagDLX3TDO. Supershift assays using anti-V5 and anti-FLAG with nuclear extracts from Saos2-Tet-Off cells transfected with pBi-V5DLX3WT/GFP and pBi-V5DLX3WT/FlagDLX3TDO. D, supershift assays using anti-V5 with nuclear extracts from Saos2-Tet-Off cells expressing FlagDLX3WT alone, FlagDLX3WT and V5DLX3TDO, or FlagDLX3WT and V5GTF3–Δ1. The inset on the left-hand side shows a Western blot analysis of recombinant proteins using anti-V5 and anti-FLAG antibodies. Asterisks, free radiolabeled probe; arrows, protein-DNA complexes; arrowheads, antibody-protein-DNA complexes.
We then investigated if a DLX3TDO-DLX3WT complex could bind DNA. We performed supershift assays using anti-V5 and anti-FLAG antibodies on nuclear extracts containing V5DLX3WT alone or both V5DLX3WT and FlagDLX3TDO. Supershift assays using anti-V5 antibody confirmed the presence of DLX3WT in the protein-DNA complexes formed in both conditions (Fig. 3C, lanes 4 and 8). The ability of the anti-FLAG antibody to supershift a protein-DNA complex was first tested using nuclear extracts containing FlagDLX3WT (data not shown). We then used the same antibody to perform supershift assays with nuclear extracts from cells expressing V5DLX3WT and FlagDLX3TDO. Even though most of the protein-DNA complex remained unshifted, we could detect a small proportion of supershifted complex (Fig. 3C, compare lanes 7 and 9). Because DLX3TDO is not able to bind to the probe by itself, these observations suggest the presence of hybrid complexes containing both V5DLX3WT and FlagDLX3TDO. To confirm the partial supershift, we performed the same assay using nuclear extracts containing FLAG-tagged DLX3WT and V5-tagged DLX3TDO, and the supershifts using anti-V5. Nuclear extracts containing FlagDLX3WT alone or FlagDLX3WT with V5GTF3–Δ1, a truncated isoform of the general transcription factor 3 tagged with a V5 epitope (20), were used as negative controls. In all cases, FlagDLX3WT formed a complex with the Dlx3 consensus probe (Fig. 3D, lanes 1, 3, and 5). The addition of anti-V5 antibody resulted in a partial supershift of the protein-DNA complex formed in the presence of FlagDLX3WT and V5DLX3TDO (Fig. 3D, lane 4). No supershift was observed when anti-V5 was added to a mixture containing FlagDLX3WT alone (Fig. 3D, lane 2) or FlagDLX3WT with V5GTF3-Δ1 (Fig. 3D, lane 6). Taken together, these observations indicate that although DLX3TDO is not able to directly bind DNA, it can form a complex with DLX3WT, which is able to bind DNA.
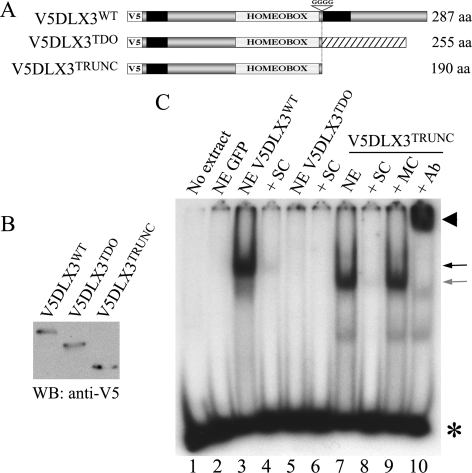
The Presence of the Frameshifted C-terminal Domain in DLX3TDO Is Responsible for Its Inability to Bind DNA—The mutation in the DLX3TDO allele is located just downstream of the homeodomain. As the mutated isoform is not able to bind the Dlx3 consensus sequence, we tested if this was due to the absence of the normal C-terminal domain of DLX3WT. For this purpose, Saos2-Tet-Off cells were transfected with pBi-V5DLX3WT/GFP, pBi-V5DLX3TDO/GFP, or pBi-V5DLX3TRUNC/GFP, a construct containing a truncated form of DLX3 in which a stop codon was inserted immediately downstream of the homeodomain (Fig. 4A). Thus, the truncated isoform lacks the C-terminal sequence that differs between DLX3WT and DLX3TDO. Western blot analysis using anti-V5 antibody was used to confirm the presence of the three different isoforms in the nuclear extracts (Fig. 4B). EMSA was performed to analyze the capability of each isoform to bind the Dlx3 consensus sequence. As shown in Fig. 4C, both V5DLX3WT and V5DLX3TRUNC were able to bind the Dlx3 consensus probe (Fig. 4C, lanes 3 and 7). The complex formed between V5DLX3TRUNC and the probe competed with an excess of unlabeled probe (Fig. 4C, lane 8) but not with an excess of a mutated probe (Fig. 4C, lane 9). The protein-DNA complex was supershifted in the presence of anti-V5 antibody (Fig. 4C, lane 10), demonstrating the presence of V5DLX3TRUNC in the complex. These results determine that the C-terminal domain of DLX3 downstream of the homeodomain is not essential for the binding to DNA. They also strongly support the hypothesis that the frameshifted C-terminal domain in the DLX3TDO isoform is responsible for the inability to bind the DNA.
FIGURE 4.
C-terminal domain and DNA binding capacity. A, V5DLX3TRUNC, a truncated form of V5DLX3, was generated by inserting a stop codon in place of the GGGG that are deleted in the TDO allele. Black boxes indicate the position of transactivation domains. B, Western blot (WB) analysis of nuclear extracts from Saos2-Tet-Off cells transiently transfected with pBi-V5DLX3WT/GFP, pBi-V5DLX3TDO/GFP, or pBi-V5DLX3TRUNC/GFP, using anti-V5 antibody. C, EMSA using the same nuclear extracts and the Dlx3 consensus and mutant probes described in Fig. 3. NE, nuclear extract; SC, self-competitor; MC, mutant competitor; Ab, anti-V5 antibody. Asterisks, free radioactive probe; arrows, protein-DNA complexes; arrowheads, antibody-protein-DNA complexes.
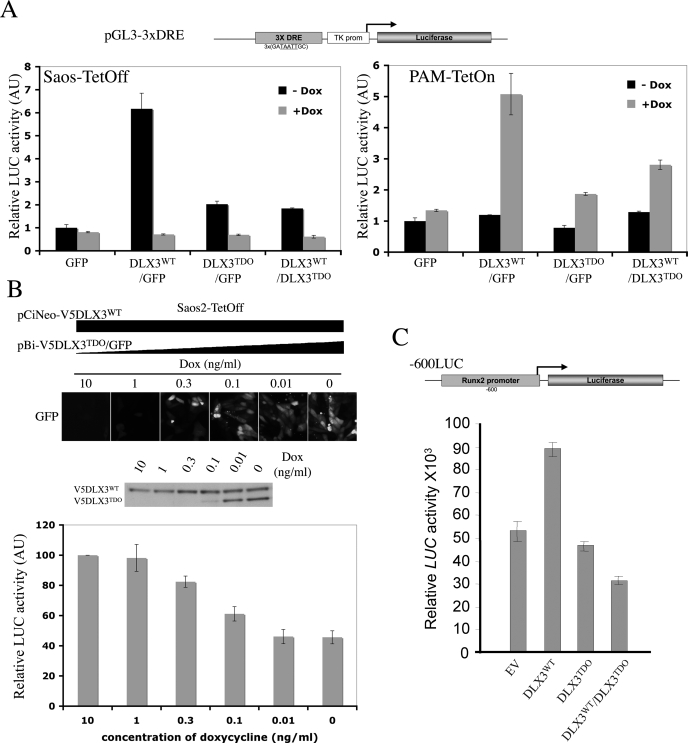
DLX3TDO Has a Defective Transcriptional Activity and Affects the Transcriptional Activity of DLX3WT—Luciferase reporter assays were performed to assess the transcriptional activity of DLX3TDO in vivo and to determine whether DLX3WT transcriptional activity is altered by the presence of DLX3TDO. Saos2-Tet-Off cells were co-transfected with each of the pBi constructs, a reporter construct pGL3-3XDRE (firefly luciferase downstream of three copies of the Dlx3 responsive element, Fig. 5A), and pRL-TK (constitutive expression of Renilla luciferase) for normalization. For each condition, half of the cells were seeded in medium containing 2 μg/ml doxycycline, whereas the other half was seeded in regular medium. 24 h after transfection, cell lysates were harvested and both firefly and Renilla luciferase activities were measured. The ratio between firefly and Renilla luciferase activities was determined and all values were normalized against the measure obtained with pBi-GFP/-Dox (Fig. 5A, left panel). In the presence of DLX3WT, the luciferase activity detected was about 6 times higher than basal activity. The luciferase activity detected in cells expressing DLX3TDO alone was much lower but still 2 times higher than the basal activity. Interestingly, the transcriptional activity detected in the presence of both DLX3WT and DLX3TDO was also significantly reduced compared with that observed with DLX3WT alone. In all cases, the activity was reduced to the basal level when doxycycline was used to repress the expression of the transgenes.
FIGURE 5.
Effect of the TDO mutation on DLX3 transcriptional activity. A, luciferase (LUC) reporter assays performed using each of the pBi constructs, the pGL3-3xDRE reporter construct containing three tandem copies of the Dlx3 responsive element upstream of a firefly luciferase reporter cassette, and pRL-TK (Renilla luciferase). Left panel, Saos2-Tet-Off cells were transfected with those constructs, grown with or without Dox for 24 h, and dual luciferase assays were performed to determine both firefly and Renilla luciferase activities. Relative luciferase activity (firefly/Renilla ratio) was determined in each condition. Right panel, same assay performed on PAM-Tet-On cells. B, luciferase reporter assay performed on Saos2-Tet-Off cells transfected with pCiNeo-DLX3WT (constitutive expression of V5DLX3WT), pBi-V5DLX3TDO/GFP (inducible expression of V5DLX3TDO), pGL3-3xDRE and pRL-TK, and grown with different amounts of Dox (10, 1, 0.3, 0.1, 0.01, or 0 ng/ml) for 24 h. GFP expression was observed under a fluorescent microscope, V5DLX3WT and V5DLX3TDO expression were analyzed by Western blot using anti-V5 antibody, and relative luciferase activity was determined. C, luciferase reporter assays performed on MC3T3 cells using a reporter construct containing the Runx2 promoter upstream of a firefly luciferase reporter cassette (-600LUC). The reporter construct was co-transfected with pCi-Neo (empty vector, EV), pCi-Neo-V5DLX3WT, pCi-Neo-V5DLX3TDO, or with both pCi-Neo-V5DLX3WT and pCi-Neo-V5DLX3TDO. pRL-Null (promoter-less Renilla luciferase vector) was used as a control for normalization. 24 h after transfection, relative luciferase activity was determined.
Luciferase assays were also performed on PAM-Tet-On cells, in the same conditions used for Saos2-Tet-Off cells (Fig. 5A, right panel). Cells transfected with pBi-GFP exhibited a low basal luciferase activity in the absence of doxycycline that was not significantly modified when the expression of GFP was induced by doxycycline. In the absence of doxycycline, the luciferase activity detected in PAM-Tet-On cells transfected with any of the pBi constructs was basal (same as GFP). However, when doxycycline was added to induce the expression of V5DLX3WT, the luciferase activity measured was 5 times higher than the basal level. The induction of V5DLX3TDO expression resulted in a much lower activity that was 2 times higher than basal activity. When the expression of both V5DLX3WT and FlagDLX3TDO was induced, the luciferase activity measured was also significantly lower than that detected in the presence of V5DLX3WT alone and 3 times higher than basal activity. These observations corroborate the data obtained using Saos2-Tet-Off cells.
To confirm the inhibitory effect of the presence of DLX3TDO on DLX3WT transcriptional activity, we performed luciferase reporter assays on cells expressing a constant amount of DLX3WT and increasing amounts of DLX3TDO. Saos2-Tet-Off cells were cotransfected with pCiNeo-V5DLX3WT (constitutive expression under control of the CMV promoter), pBi-V5DLX3TDO/GFP (Tet-inducible expression), pGL3-3xDRE, and pRL-TK. The transfected cells were split in 6 wells containing different concentrations of doxycycline (10, 1, 0.3, 0.1, 0.01, and 0 ng/ml). The luciferase activity measured was maximal in cells grown in the presence of 10 ng/ml doxycycline, which is enough to prevent the expression of V5DLX3TDO, whereas progressive reduction in the amount of doxycycline added resulted in a progressive increase in the expression of V5DLX3TDO and a progressive decrease in luciferase activity (Fig. 5B), demonstrating a dose-dependent inhibitory effect of the presence of DLX3TDO on DLX3WT transcriptional activity. All together, these results suggest that the presence of DLX3TDO negatively affects DLX3WT transcriptional activity in a system where transcription regulation is controlled by three tandem copies of the Dlx3 responsive element.
DLX3TDO Interferes with the Regulation of Runx2 Transcription by DLX3WT—We tested the effect of DLX3TDO on DLX3WT transcriptional activity in the context of the Runx2 promoter that was previously shown to bind and be regulated by Dlx3 (10). MC3T3 cells and a reporter construct containing the 600 bp of the Runx2 promoter upstream of a firefly luciferase cassette (-600LUC) were used for this dual luciferase assay (Fig. 5C). V5DLX3WT and/or V5DLX3TDO were expressed using pCi-Neo-V5DLX3WT and/or pCi-Neo-V5DLX3TDO. The empty vector (EV) was used as a control. The relative luciferase activity measured when V5DLX3WT was expressed was about 80% higher than that detected when cells were transfected with the EV. No significant change in relative luciferase activity was observed in cells overexpressing V5DLX3TDO. Interestingly, when both V5DLX3WT and V5DLX3TDO were co-expressed, the luciferase activity detected was about 30% lower than that measured with the EV. These data support the conclusion that DLX3TDO has a defective transcriptional activity and exerts a dominant negative effect on DLX3WT transcriptional activity.
DISCUSSION
In this study, we characterized the biochemical properties of the mutant DLX3 isoform responsible for TDO syndrome. We show that although DLX3TDO is targeted to the nucleus, it is not able to directly bind DNA. However, DLX3TDO can interact with DLX3WT and form a complex that is capable of binding DNA. Finally, DLX3TDO is a defective transcription factor exerting a dominant-negative effect on DLX3WT transcriptional activity.
The finding that the targeting to the nucleus is not affected by the TDO mutation is consistent with the previous identification of the Dlx3 nuclear localization signal just upstream of the homeodomain (19), a region that is not affected in DLX3TDO. However, the fact that DLX3TDO has no ability to directly bind to canonical homeodomain DNA binding sites was not expected because the frameshift mutation in the DLX3TDO isoform occurs downstream of the homeodomain.
Different frameshift and/or truncation mutations occurring in the C-terminal domain of other homeobox transcription factors have been studied (24–28). In these reports, the mutation exerted consequences on transactivation properties and the DNA binding activity was either unaffected or increased, but never abolished. For DLX3TDO, we show that a frameshift mutation just downstream of the homeodomain prevents DNA binding. Moreover, we demonstrate that the C-terminal domain in DLX3WT is not required for DNA binding, whereas the modified C-terminal domain in DLX3TDO prevents DNA binding. It could be proposed that the frameshifted C-terminal domain of DLX3TDO confers an altered conformation that somewhat interferes with the ability of the third helix of the homeodomain to insert into the major groove of the DNA.
The absence of phenotype in heterozygous Dlx3 knock-out mice (4) suggested that the pathology in TDO was due to a dominant-negative or gain of function effect as opposed to haploinsufficiency, although there was no evidence for either possibility until now. Here we show that the transcriptional activity of DLX3TDO is altered, which is consistent with the fact that it is not able to bind DNA. Interestingly, although the presence of DLX3TDO does not prevent DLX3WT from binding to the DNA, reporter assays demonstrate that DLX3TDO interferes with DLX3WT transcriptional activity. This is clearly evident by the diminished DLX3WT transcriptional activity on bone-related genes, such as Runx2 in the presence of DLX3TDO.
At the time we submitted this manuscript, an ex vivo study of the same mutation was published by Choi et al. (29), where the authors propose that DLX3TDO binds DNA. However, in this study, supershift assays were not performed to confirm the presence of DLX3WT or DLX3TDO in the protein-DNA complexes. Regarding transcriptional activity, Choi et al. (29) show that MC3T3 and C2C12 cells overexpressing DLX3TDO exhibit an increase in osteocalcin promoter activity when compared with cells overexpressing DLX3WT. However, because these reporter assays were performed on stably transfected cells, it is possible that the augmented osteocalcin promoter activity is due to an indirect effect of the long-term expression of DLX3TDO rather than to a direct effect of DLX3TDO on the osteocalcin promoter. Moreover, Choi et al. (29) did not consider the situation where both isoforms are co-expressed. In the conditions of our studies, with short term co-expression of both DLX3WT and DLX3TDO, we demonstrate that DLX3TDO is a defective transcription factor and exerts a dominant-negative effect on DLX3WT transcriptional activity.
There are two classes of dominant-negative mutants based on the capacity to form multimers (30). Considering transcription factors, the first class of dominant-negative mutants comprise dimeric or multimeric proteins whose activity requires oligomerization. The presence of a derivative capable of interacting with the wild type protein but otherwise defective either in its DNA binding activity or transactivation capacity, will form non-functional multimers and affect the activity of the wild type transcription factor. In the second class of dominant negative mutants, the transcription factor is active as a monomer. In that case, the dominant negative effect is due to a competition between the wild type and mutant transcription factors for the binding to the DNA. From the results presented in this work, showing that DLX3WT can form homodimers and can heterodimerize with DLX3TDO, DLX3TDO can be classified into the first class of dominant negative mutants.
Several studies have addressed the functional consequences of frameshift mutations occurring in the C-terminal domain of homeodomain transcription factors forming homodimers (24–28). Mutations in PITX2 that are associated with Axenfeld-Rieger syndrome have been widely characterized. Axenfeld-Rieger syndrome is an autosomal dominant human disorder characterized by ocular anterior chamber anomalies, dental hypoplasia, mild craniofacial dysmorphism, and umbilical stump abnormalities (31). Frameshift and nonsense mutations occurring in the PITX2 C-terminal domain have been detailed (25, 26). The truncated isoform ΔT1261 of the PITX2a gene is able to bind DNA but is unable to interact with its partner Pit-1 thus exhibits a reduced transactivation capacity (25). A second study described a deletion that results in a frameshift starting in codon 122 (D122FS) and in a truncated protein of 153 amino acids (26), and a previously described nonsense mutation that introduces a stop codon in position 133 (W133Stop). Both mutations result in the deletion of a larger part of the C-terminal domain as opposed to the ΔT1261 mutation described above. Both D122FS and W133Stop mutants exhibited an increased DNA binding activity compared with WT PITX2, an increased affinity for WT PITX2 (stronger interaction than PITX2-PITX2), and a higher transactivation capacity. The effect of all these mutations on transcriptional activity are due to post-translational modifications of the PITX2 protein that appear to alter protein conformation (26).
Our results suggest that the DLX3WT-DLX3TDO complex can bind DNA although with a lower affinity than DLX3WT alone. Thus, although DLX3TDO by itself is not able to bind DNA, it can interact with DLX3WT and in this way indirectly bind DNA and influence DLX3WT transcriptional activity. Additional mechanisms that influence homeodomain function may be attributed to modifications in the ability to interact with other homeodomain proteins. Previous studies have shown that Dlx2 and Dlx5 were able to form homodimers and to heterodimerize with Msx1 and Msx2 (32). Furthermore, in the context of osteogenic gene expression, Newberry and collaborators (33) showed that Dlx2 and Msx2 form dimers through the 127–143 residues of Dlx2 to reverse an Msx-dependent repression of the osteocalcin gene.
The present studies show evidence for the molecular consequences of disruption of normal DLX3-mediated transactivation in the presence of the DLX3TDO mutant protein. The challenge is to understand how the effect of DLX3TDO on the activity of DLX3WT accounts for the phenotype of TDO patients. In the recent study published by Choi et al. (29), the authors show that cells overexpressing DLX3TDO exhibit an enhanced increase of alkaline phosphatase activity, mineral deposition, and osteocalcin promoter activity, as compared with the increase observed in cells overexpressing DLX3WT, which is consistent with the increased bone density observed in TDO patients. However, these consequences are not necessarily due to a direct effect of DLX3TDO on the expression of genes that are known to be induced by Dlx3 in bone. An important consideration is understanding that Dlx3 is expressed very early in osteoprogenitor cells and only transiently associated with osteoblast promoters for activation at specific stages of osteoblast differentiation (9, 10). Dlx3 also interacts with other transcriptional regulators, Runx2, Msx2, and Dlx5, and can further modulate expression of target genes (9, 33). Equally plausible is a Dlx3-repressive stage-specific transactivation of target genes or in alternate tissues. Thus, Dlx3 activities are coordinated with other factors and may involve a Dlx3-concerted activation and repression of gene expression to support the temporal expression of target genes during osteoblastogenesis. Osteoblast differentiation is highly regulated by many secreted factors or transcription factor activity in subpopulations of bone cells that either stimulate or restrain maturation to regulate the rate of bone formation and turnover in the adult skeleton. Any one of a large number of regulatory factors could be influenced by non-physiologic expression of DLX3WT or DLX3TDO. The use of mouse models of the TDO syndrome will be an invaluable tool to identify such target genes and clearly elucidate the mechanisms contributing to the hair, teeth, and bone phenotype in TDO syndrome.
Acknowledgments
We thank Dr. S. Yuspa and members of the Laboratory of Cancer Biology and Genetics, NCI; Dr. G. S. Stein and colleagues (UMASS Medical School) for many helpful discussions and suggestions; and Drs. Buonanno and Vullhorst for the GTF3 plasmid.
This work was supported, in whole or in part, by National Institutes of Health Grant R37DE012528 (to J. B. L.) and by the Intramural Research Program of the NIAMS (to M. I. M.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: TDO,Tricho-Dento-Osseous; EMSA,electrophoretic mobility shift assay; EV, empty vector; WT, wild type; GFP, green fluorescent protein; Dox, doxycycline; PBS, phosphate-buffered saline; BisTris, 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol; MOPS, 4-morpholinepropanesulfonic acid; IP, immunoprecipitation; TBS, Tris-buffered saline; CMV, cytomegalovirus; rtTA, reverse tetracycline controlled transactivator.
References
- 1.McGuinness, T., Porteus, M. H., Smiga, S., Bulfone, A., Kingsley, C., Qiu, M., Liu, J. K., Long, J. E., Xu, D., and Rubenstein, J. L. (1996) Genomics 35 473-485 [DOI] [PubMed] [Google Scholar]
- 2.Nakamura, S., Stock, D. W., Wydner, K. L., Bollekens, J. A., Takeshita, K., Nagai, B. M., Chiba, S., Kitamura, T., Freeland, T. M., Zhao, Z., Minowada, J., Lawrence, J. B., Weiss, K. M., and Ruddle, F. H. (1996) Genomics 38 314-324 [DOI] [PubMed] [Google Scholar]
- 3.Sumiyama, K., Irvine, S. Q., Stock, D. W., Weiss, K. M., Kawasaki, K., Shimizu, N., Shashikant, C. S., Miller, W., and Ruddle, F. H. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 780-785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morasso, M. I., Grinberg, A., Robinson, G., Sargent, T. D., and Mahon, K. A. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 162-167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robinson, G. W., and Mahon, K. A. (1994) Mech. Dev. 48 199-215 [DOI] [PubMed] [Google Scholar]
- 6.Depew, M. J., Simpson, C. A., Morasso, M., and Rubenstein, J. L. (2005) J. Anat. 207 501-561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morasso, M. I., Mahon, K. A., and Sargent, T. D. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 3968-3972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morasso, M. I., Markova, N. G., and Sargent, T. D. (1996) J. Cell Biol. 135 1879-1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hassan, M. Q., Javed, A., Morasso, M. I., Karlin, J., Montecino, M., van Wijnen, A. J., Stein, G. S., Stein, J. L., and Lian, J. B. (2004) Mol. Cell. Biol. 24 9248-9261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hassan, M. Q., Tare, R. S., Lee, S. H., Mandeville, M., Morasso, M. I., Javed, A., van Wijnen, A. J., Stein, J. L., Stein, G. S., and Lian, J. B. (2006) J. Biol. Chem. 281 40515-40526 [DOI] [PubMed] [Google Scholar]
- 11.Ghoul-Mazgar, S., Hotton, D., Lezot, F., Blin-Wakkach, C., Asselin, A., Sautier, J. M., and Berdal, A. (2005) Bone 37 799-809 [DOI] [PubMed] [Google Scholar]
- 12.Price, J. A., Wright, J. T., Kula, K., Bowden, D. W., and Hart, T. C. (1998) J. Med. Genet. 35 825-828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Price, J. A., Bowden, D. W., Wright, J. T., Pettenati, M. J., and Hart, T. C. (1998) Hum. Mol. Genet. 7 563-569 [DOI] [PubMed] [Google Scholar]
- 14.Quattromani, F., Shapiro, S. D., Young, R. S., Jorgenson, R. J., Parker, J. W., Blumhardt, R., and Reece, R. R. (1983) Hum. Genet. 64 116-121 [DOI] [PubMed] [Google Scholar]
- 15.Wright, J. T., Roberts, M. W., Wilson, A. R., and Kudhail, R. (1994) Oral Surg. Oral Med. Oral Pathol. 77 487-493 [DOI] [PubMed] [Google Scholar]
- 16.Wright, J. T., Kula, K., Hall, K., Simmons, J. H., and Hart, T. C. (1997) Am. J. Med. Genet. 72 197-204 [DOI] [PubMed] [Google Scholar]
- 17.Haldeman, R. J., Cooper, L. F., Hart, T. C., Phillips, C., Boyd, C., Lester, G. E., and Wright, J. T. (2004) Bone 35 988-997 [DOI] [PubMed] [Google Scholar]
- 18.Feledy, J. A., Morasso, M. I., Jang, S. I., and Sargent, T. D. (1999) Nucleic Acids Res. 27 764-770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bryan, J. T., and Morasso, M. I. (2000) J. Cell Sci. 113 4013-4023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vullhorst, D., and Buonanno, A. (2005) J. Biol. Chem. 280 31722-31731 [DOI] [PubMed] [Google Scholar]
- 21.Yuspa, S. H., Hawley-Nelson, P., Koehler, B., and Stanley, J. R. (1980) Cancer Res. 40 4694-4703 [PubMed] [Google Scholar]
- 22.Urlinger, S., Baron, U., Thellmann, M., Hasan, M. T., Bujard, H., and Hillen, W. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 7963-7968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roberson, M. S., Meermann, S., Morasso, M. I., Mulvaney-Musa, J. M., and Zhang, T. (2001) J. Biol. Chem. 276 10016-10024 [DOI] [PubMed] [Google Scholar]
- 24.Barbacci, E., Chalkiadaki, A., Masdeu, C., Haumaitre, C., Lokmane, L., Loirat, C., Cloarec, S., Talianidis, I., Bellanne-Chantelot, C., and Cereghini, S. (2004) Hum. Mol. Genet. 13 3139-3149 [DOI] [PubMed] [Google Scholar]
- 25.Espinoza, H. M., Ganga, M., Vadlamudi, U., Martin, D. M., Brooks, B. P., Semina, E. V., Murray, J. C., and Amendt, B. A. (2005) Biochemistry 44 3942-3954 [DOI] [PubMed] [Google Scholar]
- 26.Saadi, I., Toro, R., Kuburas, A., Semina, E., Murray, J. C., and Russo, A. F. (2006) Birth Defects Res. 76 175-181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singh, S., Tang, H. K., Lee, J. Y., and Saunders, G. F. (1998) J. Biol. Chem. 273 21531-21541 [DOI] [PubMed] [Google Scholar]
- 28.Yamagata, K., Yang, Q., Yamamoto, K., Iwahashi, H., Miyagawa, J., Okita, K., Yoshiuchi, I., Miyazaki, J., Noguchi, T., Nakajima, H., Namba, M., Hanafusa, T., and Matsuzawa, Y. (1998) Diabetes 47 1231-1235 [DOI] [PubMed] [Google Scholar]
- 29.Choi, S., Song, I., Ryu, O., Choi, S., Hart, P., Wu, W., Shen, R., and Hart, T. (2008) Bone 42 162-171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herskowitz, I. (1987) Nature 329 219-222 [DOI] [PubMed] [Google Scholar]
- 31.Lines, M. A., Kozlowski, K., and Walter, M. A. (2002) Hum. Mol. Genet. 11 1177-1184 [DOI] [PubMed] [Google Scholar]
- 32.Zhang, H., Hu, G., Wang, H., Sciavolino, P., Iler, N., Shen, M. M., and Abate-Shen, C. (1997) Mol. Cell. Biol. 17 2920-2932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Newberry, E. P., Latifi, T., and Towler, D. A. (1998) Biochemistry 37 16360-16368 [DOI] [PubMed] [Google Scholar]