Abstract
The nonsterile environment of the oral cavity facilitates substantial proteolytic processing, not only of resident salivary proteins but also of dietary proteins. To gain insight into whole saliva enzymatic processes, the in vivo generated peptides in this oral fluid were subjected to nano-flow liquid chromatography electrospray ionization tandem mass spectrometry. The 182 peptides identified were predominantly derived from acidic and basic proline-rich proteins, statherin, and histatins. The proteolytic cleavages in the basic proline-rich proteins occurred preferentially after a Gln residue with predominant specificity for the tripeptide Xaa-Pro-Gln, where Xaa in the P3 position was mostly represented by Lys. Using the synthetic substrates Lys-Pro-Gln-pNA and Gly-Gly-Gln-pNA, the overall Km values were determined to be 97 ± 7.7 and 611 ± 28 μm, respectively, confirming glutamine endoprotease activity in whole saliva and the influence of the amino acids in positions P2 and P3 on protease recognition. The pH optimum of Lys-Pro-Gln-pNA hydrolysis was 7.0, and the activity was most effectively inhibited by antipain and 4-(2-aminoethyl) benzenesulfonyl fluoride, was metal ion-dependent, and not inhibited by cysteine protease inhibitors. A systematic evaluation of enzyme activities in various exocrine and nonexocrine contributors to whole saliva revealed that the glutamine endoprotease is derived from dental plaque and likely microbial in origin. The P1 site being occupied by a Gln residue is a nonarchetype with respect to known proteases and indicates the presence of novel glutamine-specific endoprotease(s) in oral fluid.
Proteolytic digestion of proteins is a process that is common to various human body fluids. Such proteolytic activities are in particular associated with fluids that are part of or are released into the gastrointestinal canal. The functional importance of these processes is to convert the digestible macromolecules into forms that can subsequently be absorbed in distal portions of the digestive tract. Fluids such as gastric and pancreatic juice secrete an impressive battery of enzymes that includes amylase, pepsin, trypsin, and chymotrypsin that are specifically geared toward this function of extensive degradation of starch and proteins. It is of interest that proteolytic processing actually starts in the oral cavity, which is the “port of entry” of the gastrointestinal tract. This initial phase of proteolysis in the gastrointestinal system has long been ignored because there are no specific organs designed for secreting an arsenal of proteases such as, for example, in the pancreas. Nevertheless, multiple studies have established that oral fluid displays abundant proteolytic activity that may represent a hitherto unappreciated physical component of digestive activity. Although the significance of oral fluid proteolysis on the initiation of food digestion has not been fully addressed, its proteolytic effect on salivary proteins is being increasingly recognized (1–6). Alterations imposed by proteolytic enzymes on the structure and function of resident salivary proteins could have both primary and secondary functional effects. The primary effects would be related to functions in the oral cavity, and secondary effects to functions further downstream in the alimentary canal.
The predominant contributors to oral fluid, also called whole saliva (WS),2 are derived from the parotid and the submandibular/sublingual glands. Through traditional biochemical analyses, the structural characteristics of the proteins secreted by these glands have been established, generating the fundamental basis of the major salivary secretome (7). The most abundant salivary secretory proteins in these secretions combined are mucous glycoproteins 1 and 2 encoded by MUC5B and MUC7, respectively (8, 9), amylase encoded by AMY1 (10), immunoglobulins, in particular sIgA, acidic proline-rich proteins (PRPs) encoded by PRH1 and PRH2 (11), basic PRPs encoded by PRB1 to PRB4 (11), and PBII (SMR3B) (12), agglutinin encoded by DMBT1 (13), cystatins encoded by CST1 to CST5 (14–16) histatins encoded by HIS1 and HIS2 (17), and statherin encoded by STATH (18, 19). Each of these proteins furthermore appears in families comprising numerous polymorphic isoforms displaying a high sequence homology (7, 20). The properties of these proteins have been well established and comprise lubrication, acid neutralization, and antimicrobial activities, functions that are pertinent to but not limited to the oral cavity.
When the sterile salivary glandular secretions are released into the oral environment, mixing occurs with the nonexocrine constituents of WS. These constituents comprise a variety of host and bacterial cells and their products as well as a serum-like gingival crevicular transudate. The nonexocrine components must contribute substantially to the enzymatic activity of whole saliva given the much lower proteolytic activities of pure salivary glandular secretions (4, 6, 21, 22). Saliva is continuously being secreted (∼500–1500 ml/day), thus providing a steady supply of newly synthesized salivary proteins to the oral cavity. Because of the enzymatic environment encountered upon secretion, some proteins may only be present transiently and for a very short time in their native form in WS. Analysis of the peptidome and degradomics approaches will provide insight into the enzymes involved in such proteolytic processes. In the present study we characterized the origin of naturally occurring peptides in WS and defined a novel class of saliva-associated glutamine-specific endoproteases.
EXPERIMENTAL PROCEDURES
Collection of Oral Samples—Informed consent was obtained from all subjects according to protocols approved by the Institutional Review Board at Boston University. Seven donors ranging in age from 24 to 40 each provided ∼1 ml of unstimulated WS, which had naturally accumulated in the oral cavity between swallowings. For in vivo WS peptidome analysis, protease activity was abolished immediately upon expectoration by collecting WS in tubes placed on ice containing 1 mm PMSF and 2.5 mm EDTA (final concentrations). WS was further fractionated as described below. For studies on WS enzyme activities, masticatory-stimulated WS was collected in the absence of these inhibitors. To remove cellular debris and other particulate matter from the collected WS samples, the samples were centrifuged immediately after collection for 5 min at 14,000 × g at 4 °C in an Eppendorf centrifuge. Submandibular/sublingual secretion (SMSL) was obtained with a custom-fitted device positioned over the orifice of Wharton's duct, and gustatory-stimulated parotid secretion (PS) was collected using a Lashly cup placed under a negative pressure over the Stensen's duct. For the collection of minor gland secretion, the lower lip mucosal surfaces were exposed and dried. Secretion droplets that formed over time over the minor gland ducts were collected with a microtiter pipette and diluted in saliva ion buffer containing 50 mm KCl, 1 mm K2HPO4, 1 mm CaCl2, and 0.1 mm MgCl2 (pH 6.5). Buccal epithelial cells (BEC) were collected by gently scraping the oral buccal epithelial surfaces with a plastic spatula. Supragingival plaque samples were collected from interproximal dental spaces with an explorer 24 h after refraining from oral hygiene. BEC and dental plaque were suspended in saliva ion buffer, vortexed, and centrifuged for 5 min at 14,000 × g to obtain the supernatants. The protein concentrations in all samples were determined by measurement of the absorbance at 215 nm as described (23).
Fractionation of WS—An aliquot of 0.5 ml of WS supernatant from each of the seven donors was pooled into a 10-ml tube containing a filter with a molecular mass cut-off of 5 kDa (Millipore, Billerica, MA). After centrifugation for 30 min at 1,000 × g in a refrigerated Sorvall table top centrifuge (Thermo Fisher Scientific, Waltham, MA), the filtrate containing the proteins/peptides with molecular masses <5 kDa was collected and lyophilized (sample A). The retentate containing the residual peptides and proteins/protein complexes with molecular masses >5 kDa were mixed with acetonitrile in a 1:2 (v/v) ratio as described (24). Upon the addition of acetonitrile, a precipitate of the larger proteins formed, and the supernatant containing the small molecular mass constituents was separated from the pellet by centrifugation at 14,000 × g and lyophilized. Both the filtrate and the supernatant of the retentate were dissolved in 20 μl of loading buffer containing 95% milliQ water, 5% acetonitrile, and 0.25% formic acid.
Liquid Chromatography Electrospray Ionization MS/MS—Mass spectrometric analysis of the peptide samples was carried out using a nano-scale reversed phase HPLC capillary column (75 μm × 10 cm) that was created in-house by packing 5-μm C18 spherical silica beads (Micron Bioresource, Inc., Auburn, CA) into a fused silica capillary with a flame-drawn tip. Samples dissolved in loading buffer were each applied in triplicate to the column (3 μl/injection). A gradient was formed using increasing concentrations of solvent B (97.5% acetonitrile, 0.1% formic acid) over a 55-min time period at a flow rate of ∼200 nl/min. The gradient was provided by a surveyor MS Pump Plus (Thermo-Finnigan, San Jose, CA). Mass spectrometric analysis of the eluted peptides was carried out on a LTQ linear ion trap (Thermo-Finnigan), which was operated in the positive-ion mode. Data-dependent acquisition methods were initiated by a survey MS scan in the range of m/z 390–1500, followed by MS/MS analysis of selected peptide ions.
MS Data Analysis—The obtained MS/MS spectra were searched against human Uniprot protein databases (Swiss Prot and TrEMBL, Swiss Institute of Bioinformatics, Geneva, Switzerland) and against a salivary protein data base containing the 40 most abundant salivary proteins. The selection of proteins for this data base was based on reported measures of abundance of these proteins. Hypothetical proteins (mostly immunoglobulins) and other repeated entries for immunoglobulins were excluded. Searches were performed using SEQUEST software (Bioworks Browser 3.3.1, Thermo-Finnigan, San Jose, CA) with the following SEQUEST parameters: 1) no specific protease enzyme, 2) DeltaCN ≤ 0.1, 3) Peptide probability ≤ 0.5, and 4) XCorr score ≥ 2.2 and 3.5 for Z = 2 and 3, respectively. When data were queried against the salivary protein data base, the following peptide modifications were applied: N-terminal pyroglutamate (pyrrolidone carboxylic acid) formation (–17 Da), serine/threonine phosphorylation (+80 Da), and serine/threonine dehydration (–18 Da). Peptides passing the filter criteria from three consecutive analyses of the two peptide samples were combined. The additional inclusion criteria applied were: at least two different peptides identified per protein and at least two identifications of the same peptide in a total of six MS analyses.
Gln Cleavage Calculations—The number of unique cleavages after a glutamine residue in the PRB sequences was calculated by manually querying each identified peptide against the intact PRP sequences (PRP-2, PRB1, PRB2, PRB3, PRB4S, PRB4L, and IB-8a) using the search option in Microsoft Word. These searches also yielded the amino acids in positions P2 and P3 for each unique peptide. Multiple peptides resulting from identical cleavage events, either at the N or C terminus, were counted once. The percentage of cleavages after a Gln residue was calculated as (total number of unique cleavages after a Gln residue/total number of unique cleavages)*100%.
Hydrolysis of Synthetic Substrates—Lys-Pro-Gln-para-nitroanilide (KPQ-pNA) and Gly-Gly-Gln-para-nitroanilide (GGQ-pNA) were commercially obtained (Quality Controlled Biochemicals, Hopkinton, MA). The peptides were dissolved in Me2SO (ThermoFisher Scientific, Waltham, MA) to an approximate concentration of 20 mm. Accurate concentrations were determined from absorbance measurements (see below) of completely hydrolyzed substrate in WS supernatant. To determine the kinetic parameters of KPQ-pNA and GGQ-pNA hydrolysis in WS supernatant, various concentrations of the substrates were added to 200 μl of pooled WS supernatant. Hydrolysis was followed spectrophotometrically at 405 nm at 37 °C in a Genios microtiter plate reader (Tecan, Mannedorf, Switzerland). Measurements were conducted every 3 min during the initial rate period, which was established to be 0–30 min. The initial velocity (Vi) rates during this period were determined for each KPQ-pNA and GGQ-pNA concentration using Deltasoft Software (Hillsborough, NJ), and a specific molar extinction coefficient (ε) of para-nitroanilide of 10.0 × 10–3 m–1 cm–1 at a path length of 0.55 cm. The data were plotted in a linear Lineweaver-Burk plot to obtain the overall kinetic parameters Vmax and Km, and the theoretical enzyme saturation curves were calculated from the linear equation. To establish the optimum pH of KPQ-pNA cleavage, the pH of 0.5 ml of WS supernatant aliquots was adjusted with either 5 m HCl or 1 m NaOH to pH 4, 5, 6, 7, 8, 9, and 10 prior to the addition of 85 μm KPQ-pNA. Experiments were also carried out in the presence of a series of enzymatic inhibitors (Sigma; see Table 3 for details). For those experiments, WS supernatant (pH 7.0) was incubated for 15 min at 37 °C with the inhibitor prior to the addition of KPQ-pNA.
TABLE 3.
Evaluation of inhibitors of KPQ-pNA hydrolysis in WS supernatant
| Inhibitor | Target enzymes | Concentration used | Inhibitiona |
|---|---|---|---|
| % | |||
| Serine protease inhibitors | |||
| AEBSF | Serine proteases | 1 mm | 81.5 ± 16.9 |
| Aprotinin | Serine proteases | 7.5 μm | No inhibition |
| Benzamidine HCl | Serine proteases | 1.5 mm | No inhibition |
| TLCK | Serine proteases | 25 μm | No inhibition |
| Trypsin inhibitor (TI) | Trypsin, chymotrypsin, plasmin | 2.5 μm | No inhibition |
| Pancreatic TI | Trypsin, chymotrypsin | 1.0 μm | No inhibition |
| Chymostatin | Chymotrypsin | 250 μm | No inhibition |
| 6-Aminohexanoic acid | Chymotrypsin, lysine carboxypeptidase | 1 mm | No inhibition |
| Leupeptin | Serine and thiol proteases | 0.5 mm | No inhibition |
| Antipain | Serine/cysteine proteases | 0.8 mm | 87.8 ± 13.0 |
| Cysteine protease inhibitors | |||
| E-64 | Cysteine proteases | 300 μm | No inhibition |
| N-Ethylmaleimide | Cysteine proteases | 2 mm | No inhibition |
| 2-PDS | Cysteine proteases | 15 μm | No inhibition |
| Metallo protease inhibitors | |||
| EDTA | Metallo proteases | 1 mm | 65.3 ± 14.1 |
| Phosphoramidon | Metallo proteases | 180 μm | 30.1 ± 3.8 |
| Bestatin | Metallo aminopeptidases | 0.5 mm | No inhibition |
| Aspartyl protease inhibitors | |||
| Pepstatin | Aspartyl peptidases | 7 μm | No inhibition |
The percentage of inhibition was determined from the ratio of the initial velocities (Vi) of KPQ-pNA hydrolysis in the presence and absence of inhibitor. Inhibitors that caused less than 10% inhibition are listed as causing no inhibition.
RP-HPLC of Saliva Samples—Aliquots of 100 μl of pooled PS, unstimulated WS supernatant and stimulated WS supernatant were each mixed with 900 μl of buffer A (0.1% trifluoroacetic acid). HPLC analysis was conducted on a HPLC model 715 (Gilson, Middleton, WI) using a C-18 column (TSK-GEL 5 μm, ODS-120T, 4.6 × 250 mm; TOSOHaas, Montgomeryville, PA). The proteins were eluted using a linear gradient from 0 to 55% buffer B containing 80% acetonitrile and 0.1% trifluoroacetic acid over a 74-min time interval at a flow rate of 1.0 ml/min. The eluate was monitored at 219 nm and analyzed using Unipoint software, version 3.30 (Gilson).
Gel Electrophoresis—The cationic proteins in the saliva samples were analyzed by 15% cationic PAGE as previously described (25). Anionic proteins were separated on a 7.5% Ornstein Davis gel (26, 27). The cationic gels were stained with 0.1% (w/v) Coomassie Brilliant Blue R-250 in 10% (v/v) acetic acid and 40% (v/v) methanol, whereas the Ornstein-Davis gels were stained with 0.5% Amido Black in 7% acetic acid. The gels were destained in the same solutions not containing the respective dyes until the band intensity was optimal relative to the background staining.
RESULTS
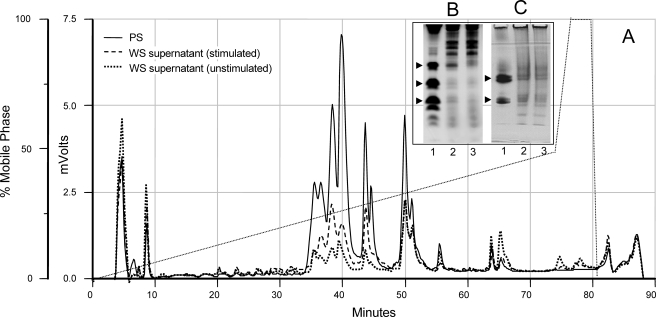
Extent of Proteolysis in the Oral Cavity—From a biological point of view, an important question is to what extent salivary secretory proteins are actually degraded in WS, or what are the peptide levels relative to the intact protein. To this end, studies were undertaken to compare the levels of histatins, basic PRPs and acidic PRPs concentrations in glandular secretions with those in WS. These proteins have been demonstrated to be among the most sensitive to WS-associated proteolysis in prior studies (21, 22, 28, 29). Fig. 1A shows the C18 RP-HPLC chromatographic comparison of proteins present in equal volumes of PS, stimulated WS supernatant, and unstimulated WS supernatant. Intact acidic PRPs, histatins, and statherin elute between 35 and 55 min using the gradient applied (30). The absorbance in this region is substantially lower in WS than in PS (see Fig. 4) or in SMSL secretion (30). Masticatory-stimulated WS was more similar to PS than unstimulated WS, consistent with the fact that masticatory stimulation increases PS output and reduces the residence time of proteins in the proteolytic environment of the oral cavity. For a more accurate assessment of histatin and acidic PRP levels, the same type of saliva samples were applied to cationic PAGE (Fig. 1B) and anionic PAGE (Fig. 1C), respectively. Our previous studies have shown that the electrophoretic mobility of these proteins is not affected by WS constituents (2, 6). From peak area calculations and densitometric analysis of these and other PAGE experiments, it was estimated that histatin levels are on average 5–10-fold lower in WS compared with PS, and acidic PRP levels are 2–3-fold lower.3 This signifies that these proteins are mostly present in a fragmented state in WS.
FIGURE 1.
RP-HPLC chromatogram and PAGE analyses of PS and WS samples. Gustatory-stimulated PS, unstimulated WS, and masticatory-stimulated WS were collected from four subjects. Within each group, equal volumes (100 μl) were pooled and subjected to C18 RP-HPLC (A). The samples from one representative subject were analyzed by cationic PAGE (B), or anionic PAGE (C). Lane 1, PS; lane 2, unstimulated WS supernatant; lane 3: stimulated WS supernatant. Arrows in B and C refer to histatin and acidic PRP proteins, respectively.
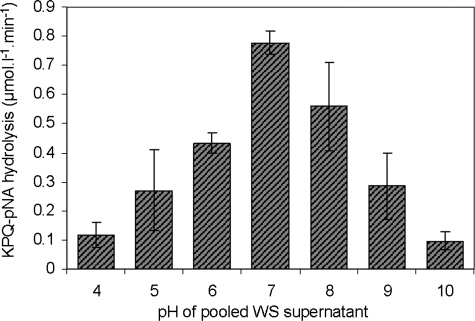
FIGURE 4.
Hydrolysis of KPQ-pNA by whole saliva proteases at various pH. Pooled WS supernatant was adjusted to the indicated pH values with HCl or NaOH, and the initial rates of hydrolysis of KPQ-pNA (85 μm) were determined from the increase in absorbance at 405 nm at 37 °C. The data represent the average of three independent experiments, normalized for activity at pH 7 (unadjusted WS supernatant).
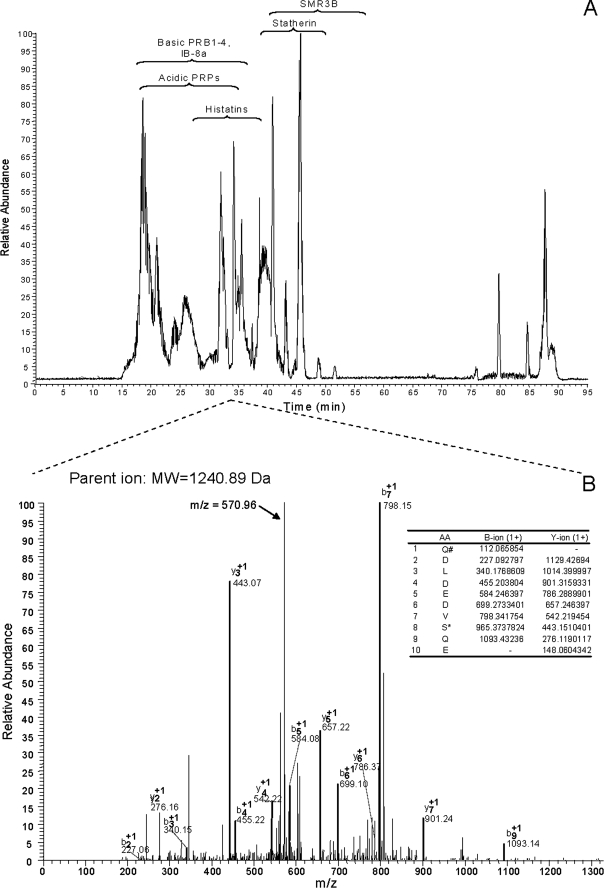
The WS Peptidome—Upon WS sample processing, two peptide-containing fractions were obtained that were each analyzed in triplicate by liquid chromatography electrospray ionization MS/MS. The base-peak chromatograms contained multiple peptide peaks, as exemplified in Fig. 2A. The raw data were initially queried against the Human Unitprot data base to identify all possible human-derived peptides in WS. Utilizing filter criteria as described under “Experimental Procedures,” the main peptides found were derived from acidic PRPs, the basic PRPs, statherin, and histatin 1. To enhance peptide identifications, subsequent searches were carried out with a smaller data base comprising the 40 most abundant salivary proteins and selecting various peptide modifications (see “Experimental Procedures”). The query yielded a 3-fold increase in the number of identified peptides compared with searches in the UniProt data base. In total 182 different peptides were identified in the two peptide samples analyzed (Table 1). Of these, 28 peptides were derived from acidic proline-rich proteins (listed collectively in SwissProt as PRPC), 45 peptides were from basic proline-rich protein PRB1 (listed in Swissprot as PRP1), 10 peptides were from PRB2, 10 peptides were from PRB3, 18 peptides were from PRB4 (the S and L isoforms), 33 peptides were from SMR3B, 4 were peptides from IB-8a, 8 were peptides from histatins, and 26 were peptides from statherin. The percentage of sequence coverage was highest for SMR3B (100%), showing that the peptides found of this protein spanned the entire SMR3B sequence. The second highest sequence coverage was observed for IB-8a (93%), followed by PRB1 (89%), PRPC (89%), PRB2 (88%), statherin (77%), PRB3 (60%), histatins (50%), PRB4S (45%), and PRB4L (32%). For the glycosylated PRPs (PRB3 and PRB4), lower numbers of peptides were recovered, along with a lower percentage of sequence coverage.
FIGURE 2.
Base peak chromatogram of peptides in present in WS supernatant (A) and MS/MS analysis of one of the peptide ions in present in this sample (B). Indicated in the MS/MS spectrum are the observed b- and y-ions and the theoretical b- and y-ions (inset table). The spectrum contained a major unassigned peak with an m/z value of 570.96, which represents the dephosphorylated, doubly charged parent ion (molecular weight = 1240.89). The peptide identified corresponded to the phosphorylated N-terminal 10 residues of PRP-2, QDLDEDVSQE, containing a carboxylated N-terminal glutamine residue and a phosphorylated serine residue.
TABLE 1.
Essential peptides of the whole saliva peptidomea
a Whole saliva supernatant from seven subjects was fractionated into two peptide preparations, which were each analyzed three times by liquid chromatography electrospray ionization MS/MS. Underlined sequences (XPQ) and underlined and bolded sequences (KPQ) represent apparent glutamine endoprotease recognition sequences.
b Peptide could be derived from basic PRP1, PRB2, or IB-8a gene products.
c Peptide could be derived from basic PRP1 or PBR2 gene products.
d Peptide could be derived from basic PBR2 or IB-8a gene products.
e Peptide could be derived from basic PRB4S or PRB4L gene products.
Because of the high sequence homology between basic PRP1, PRB2, and IB-8a, some peptides could have derived from either one of these three proteins (indicated with b, c, and d superscripts in Table 1). Similarly, most of the peptides listed under PRPC, could have been derived from any of the acidic proline-rich protein family members PRP-1, PRP-2, PIF-s, Pa, and Db, which show an exceptionally high sequence homology (31–34). The inability of proper assignment of peptides to a particular parent protein points to a more fundamental problem in mass spectrometry when highly homologous protein isoforms are concerned. Irrespective of the parent protein, the peptides constituting the WS peptidome are to be considered functional elements for a variety of biological processes (6).
The identification of most fragments was straightforward, because many did not contain post-translational modifications. Fig. 2B shows an example of a more complicated identification, representing the MS/MS spectrum of a parent peptide with a molecular mass of 1240.89 Da. Based on the b- and y-ion pattern, this peptide was identified as Q# DLDEDVS*QE, where Q denotes a pyroglutamate residue, and S* indicates a phosphorylated serine. The calculated average mass of this peptide is 1177.14 – 17.0 + 80.0 = 1240.14, closely matching the deconvoluted mass of the identified peptide. Noticeable in the MS/MS spectrum of this fragment is a major, unassigned peak with an m/z value of 570.96. This fragment corresponds to the dephosphorylated, doubly charged parent ion having a calculated m/z value of (1240.89 – 98.0 + 2.0)/2 = 572.44. Such dephosphorylated parent ions in the MS/MS spectrum are typical and characteristic for phosphorylated peptides and help in their identification (35, 36). Interestingly, phosphorylated N-terminal peptides from histatin 1 and statherin were not identified in the expectorated WS sample, possibly because of their relatively low abundance and high affinity for enamel tooth surfaces (37–39).
Identification of Lys-Pro-Gln as a Prominent Cleavage Site—Upon closer inspection of the amino acid sequences of the acidic and basic PRPs peptides, a consistent proteolytic cleavage site after a glutamine residue was observed. Although acidic and basic PRPs are rich in Gln residues, the number of unique cleavages after Gln residues was much higher than could be expected based on the number of Gln residues in the PRP protein sequences (Table 2). For example, in the acidic PRPs (PRP-2) peptides, 65% of the cleavages occurred after a Gln residue, whereas only 23% of this protein is comprised of Gln residues. Even more strikingly, on average 59% of the unique cleavages in basic PRPs (PRB1, 2, 3, 4S, 4L, and IB-8a) were observed after a Gln residue, whereas the Gln content in these proteins is only ∼16%. When the cleavage specificities were further analyzed, it was noted that among the 92 unique Gln cleavages, 78 occurred after the tri-amino sequence XPQ, where X was either a lysine, proline, arginine, glutamine, or asparagine residue. In 37 of the 78 XPQ cleavages, X equaled Lys. Although the KPQ tri-amino acid sequence is a common motif in the proline-rich protein sequences, it appears that this peptide represents a preferred cleavage site for salivary proteases.
TABLE 2.
Glutamine counts and cleavage events
| Protein | Total amino acids | Total Gln | Total Gln | Total cleavagesa | Cleavages after Gln | Cleavages after Gln |
|---|---|---|---|---|---|---|
| % | % | |||||
| PRP-2 | 150 | 35 | 23 | 31 | 20 | 65 |
| PRB1 | 376 | 62 | 17 | 59 | 33 | 56 |
| PRB2 | 382 | 60 | 16 | 18 | 9 | 50 |
| PRB3 | 293 | 42 | 14 | 16 | 8 | 50 |
| PRB4S | 232 | 34 | 15 | 20 | 11 | 55 |
| PRB4L | 276 | 43 | 16 | 8 | 6 | 75 |
| IB-8a | 126 | 20 | 16 | 7 | 5 | 71 |
The total number of unique cleavage events and the total number of unique cleavages after a Gln residue were determined from the peptides listed in Table 1 and the primary amino acid sequences of the respective proteins.
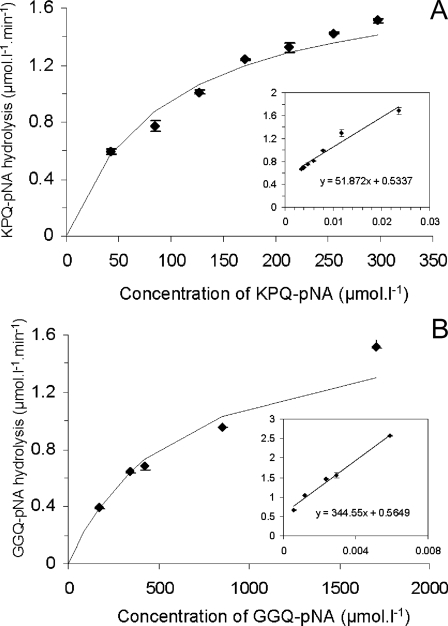
Hydrolysis of KPQ-pNA and GGQ-pNA—To further probe into the unique apparent glutamine-specific endoprotease activity in WS supernatant, two synthetic para-nitroanalide substrates, KPQ-pNA and GGQ-pNA, were obtained. KPQ-pNA was selected based on the above considerations, and GGQ-pNA was chosen to study the importance of the amino acids in positions P2 and P3. GGQ is only present in acidic PRPs and apparently not a favorite cleavage site (Table 1). The KPQ-pNA substrate was efficiently hydrolyzed in WS supernatant with Km values of 97 ± 7.7 μm and a Vmax of 1.8 ± 0.03 μmol liter–1 min–1 (Fig. 3A). GGQ-pNA hydrolysis displayed similar Vmax values (1.8 ± 0.04 μmol liter–1 min–1), but the Km was significantly higher (611 ± 28 μm) (Fig. 3B). Control experiments performed in WS supernatant placed on ice and incubations for 24 h in the absence of saliva did not result in substrate hydrolysis (data not shown). Further studies employing the KPQ-pNA substrate showed that the enzyme(s) display optimal activity at pH 7 (Fig. 4). To determine the class of enzyme(s) involved, the enzymatic activity toward KPQ-pNA was evaluated in the presence of a variety of serine, cysteine, metallo, and aspartyl protease inhibitors (Table 3). The most effective component was antipain (87.8% inhibition), which inhibits papain, trypsin, and plasmin to a lesser extent. The second most effective agents were the serine protease inhibitors 4-(2-aminoethyl) benzenesulfonyl fluoride (81.5%) and PMSF (data not shown). The lack of inhibition by (2S,3S)-3-(N-{(S)-1-[N-(4-guanidinobutyl)carbamoyl]3-methylbutyl}carbamoyl)oxirane-2-carboxylic acid, N-ethylmaleimide, and 2-pyridyl disulfide excludes the participation of proteases belonging to the class of cysteine proteases. The partial inhibition by phosphoramidon may suggest the involvement of metallo proteases. The inhibitory effect of EDTA (65.3%) is pointing toward the importance of metal ions for activity. Indeed, metallo as well as some serine proteases have been reported to be metal ion-dependent (40, 41). The hydrolysis of KPQ-pNA could be completely abolished by the combination of PMSF and EDTA (data not shown).
FIGURE 3.
Overall kinetic parameters of whole saliva enzymatic activity toward glutamine-containing substrates. A, enzyme saturation curve and Lineweaver-Burk plot (inset) obtained using KPQ-pNA as the substrate. B, enzyme saturation curve and Lineweaver-Burk plot (inset) obtained with GGQ-pNA. Substrate hydrolysis (Vi) was determined by measuring the increase in absorbance at 405 nm during the initial phase of the reaction (0–30 min). Represented are the means and standard deviation of two experiments each performed in duplicate.
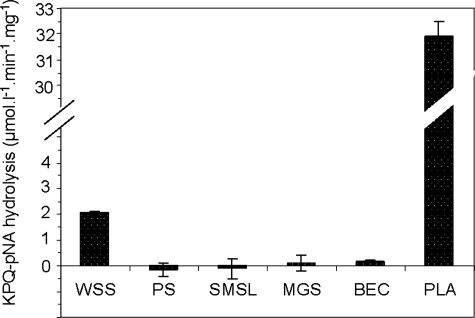
Determination of the Source of Glutamine Endoprotease Activity—An effort was made to determine the origin of the newly discovered enzymatic activity by measuring KPQ-pNA hydrolysis in various exocrine and nonexocrine contributors to WS. Glandular secretions derived from the major glands (SMSL and PS) as well as from minor lip mucosal glands were evaluated. Potential nonexocrine sources examined were BEC and supra gingival dental plaque. The results presented in Fig. 5 showed that PS, SMSL, minor gland secretion, and BEC were devoid of glutamine endoprotease activity. On the other hand, the KPQ-pNA hydrolysis rate in dental plaque supernatant far exceeded that observed in WS supernatant, clearly illustrating that the glutamine endoprotease activity in WS supernatant is derived from the oral microbial biofilm.
FIGURE 5.
Determination of the source of glutamine endoprotease activity in WS supernatant. Hydrolysis of KPQ-pNA (85 μm) was examined in whole saliva supernatant (WSS), in the major glandular secretions PS and SMSL, in minor gland secretion (MGS), in Buccalepithelial cell supernatant (BEC) and in dental plaque supernatant (PLA) collected from one subject. Hydrolysis rates were normalized for protein concentration differences in the respective oral specimens. Shown are the averages of experiments performed in triplicate.
DISCUSSION
Protein Digestion in the Oral Cavity—The contribution of salivary proteins to the protein fraction entering the gastrointestinal canal is substantial. The protein concentration in WS is on average 2–4 mg/ml. With a total output of saliva of ∼1 liter/day, 2–4 g of salivary proteins are released into the gastrointestinal system on a daily basis. With an average dietary protein intake of 0.8 g/kg/day, salivary proteins constitute ∼4–8% of all proteins to be digested. The peptidome data presented provide compelling evidence that their “digestion” is initiated in the oral cavity. The importance of glutamine endoprotease activity in whole saliva resides not only in its role in the degradation of salivary proteins but possibly also of glutamine-containing dietary proteins.
The Major Constituents of the Whole Saliva Peptidome—PRPs, statherin, and histatins appear in this study to be the essential constituents of the whole saliva peptidome. It might be considered surprising that no peptides were identified from highly abundant salivary proteins such as amylase and mucous glycoproteins. We cannot exclude that the peptidome was not analyzed to completion for methodological reasons. It should be noted, however, that only the small molecular mass peptides were detected in the current liquid chromatography electrospray ionization MS/MS approach, taking into account an m/z range of 390–1500 Da and the fact that most peptides passing the Xcorr filter criteria are doubly or triply charged. Furthermore, the in vivo residence time of intact glandular salivary proteins in the proteolytic environment of the oral cavity is quite short. With an average resting (unstimulated) salivary flow rate of 12–18 ml/h and an oral fluid volume of ∼1 ml (42), it can be calculated that oral fluid is renewed every 3–5 min. The peptides generated in this short time span must be derived from proteins that are highly susceptible to proteolysis.
Functional Significance of Oral Fluid Proteolysis—The sequence overlap in PRPs and their proteolytic susceptibilities could have significant functional implications. The release of an identical peptide from different parent proteins by proteolytic enzymes will lead to a rapid increase in the molar concentrations of that peptide. For example, peptide Q.GPPPQGGRPQ.G is present in all acidic PRP isoforms. Other identified peptides such as Q.GPPPQGGNQPQGPPPPPGKPQ.G are contained in multiple copies within one protein (PRB1). The latter peptide is also present in other PRB isoforms (PRB1, PRB2, and IB-8a). Moreover, a peptide with sequence P.PPPGKPQ.G (listed under PRPC) is present in multiple copies in acidic as well as in most basic PRPs. It can be postulated that peptides that are originally contained in multiple copies within a larger salivary protein and/or are present in multiple proteins exhibit important biological functions and that the functional role of WS proteolysis is to release these peptides. Moreover, the generation of a high number of peptides all with a C-terminal glutamine residue must be of biological significance.
Proteolytic Enzymes in Whole Saliva—It is of interest that there are a number of proteases associated with human salivary glands that are responsible for post-translational processing of some salivary proteins (43, 44). One of the protein families partially fragmented in the gland are the basic PRPs. The primary gene products PRB1, PRB2, and PRB4 show extensive proteolytic processing, but the resulting degradation fragments are characterized by cleavage after RXXR↓ (45) and not after glutamine residues. The absence of glutamine endoprotease activity associated with the biosynthesis of basic PRP peptides in the gland corroborates with our observations that such Gln cleavages only occur in the whole saliva environment and not in the primary salivary secretions. The overriding KPQ↓ activity noticed in dental plaque clearly points toward the oral microbial biofilm as the source of glutamine endoproteases. The oral microbiome consists to date of 619 different species, and at least some of these are well known to produce proteolytic enzymes (46–48). Although many different amino peptidase substrates have been explored in the search for important WS proteases (3), the glutamine endoprotease was not identified, possibly because of its unusual enzymatic cleavage specificity. Our study was based foremost on the MS characterization of the major naturally occurring salivary protein substrates and, combined with synthetic enzymatic substrates, has enabled the identification of this novel enzyme.
Biological Significance of Salivary Glutamine-specific Endoprotease Activity—The results revealed a remarkable specificity in the cleavage pattern of acidic as well as of basic PRPs. Preferential cleavage of KPQ↓ was observed as compared with GGQ↓, emphasizing the importance of the N-terminal flanking residues for substrate binding. Importantly, the cleavage specificity with the P1 site occupied by a Gln residue is a nonarchetype and does not belong to any of the known protease class(es). One group of proteins that are cleaved at Gln or polyQ sites are the inhibitory serpins, which are naturally occurring serine protease inhibitors. Serpins have been described in many eukaryotic systems and are also part of the salivary proteome, but serpins with a reactive P1 Gln residue have been identified exclusively in a few plant species such as barley, rye, and wheat grains (49, 50). It has been suggested that the abundant serpins are likely to participate in the protection of the grain storage proteins against as yet unidentified digestive proteases from insects, pests, or microbial pathogens. An important biological implication of the unique cleavage sites in acidic and basic PRPs identified in this study, where P1 is Gln, P2 is Pro, and P3 is X, and the dominant cleavage site is after Gln, is that such peptides may act as potent serpin-like proteins, in the oral cavity or elsewhere in the gastrointestinal tract, by “trapping” both microbial pathogen-derived or host-derived serine proteases. The role of such saliva-derived peptides in modulating enzymatic activity in the gastrointestinal system would represent an entirely new function of the basic proline-rich proteins, the function of which remains largely elusive to date.
This work was supported, in whole or in part, by National Institutes of Health Grants DE05672, DE07652, DE18132, and DE16699. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: WS, whole saliva; BEC, buccal epithelial cell supernatant; MS, mass spectrometry; MS/MS, tandem MS; PMSF, phenylmethanesulfonyl fluoride; pNA, para-nitroanilide; PRP, proline-rich protein; PS, parotid secretion; RP, reversed phase; HPLC, high performance liquid chromatography; SMSL, submandibular/sublingual secretion.
M. Compese, X. Sun, J. A. Bosch, F. G. Oppenheim, and E. J. Helmerhorst, observations.
References
- 1.Chauncey, H. H. (1961) J. Am. Dent. Assoc. 63 360–368 [DOI] [PubMed] [Google Scholar]
- 2.Helmerhorst, E. J., Alagl, A. S., Siqueira, W. L., and Oppenheim, F. G. (2006) Arch. Oral Biol. 51 1061–1070 [DOI] [PubMed] [Google Scholar]
- 3.Makinen, K. K. (1966) Acta Odontol. Scand. 24 579–604 [DOI] [PubMed] [Google Scholar]
- 4.Payne, J. B., Iacono, V. J., Crawford, I. T., Lepre, B. M., Bernzweig, E., and Grossbard, B. L. (1991) Oral Microbiol. Immunol. 6 169–176 [DOI] [PubMed] [Google Scholar]
- 5.Helmerhorst, E. J. (2007) Ann. N. Y. Acad. Sci. 1098 454–460 [DOI] [PubMed] [Google Scholar]
- 6.Helmerhorst, E. J., and Oppenheim, F. G. (2007) J. Dent. Res. 86 680–693 [DOI] [PubMed] [Google Scholar]
- 7.Oppenheim, F. G., Salih, E., Siqueira, W. L., Zhang, W., and Helmerhorst, E. J. (2007) Ann. N. Y. Acad. Sci. 1098 22–50 [DOI] [PubMed] [Google Scholar]
- 8.Desseyn, J. L., Aubert, J. P., Van Seuningen, I., Porchet, N., and Laine, A. (1997) J. Biol. Chem. 272 16873–16883 [DOI] [PubMed] [Google Scholar]
- 9.Bobek, L. A., Tsai, H., Biesbrock, A. R., and Levine, M. J. (1993) J. Biol. Chem. 268 20563–20569 [PubMed] [Google Scholar]
- 10.Nishide, T., Nakamura, Y., Emi, M., Yamamoto, T., Ogawa, M., Mori, T., and Matsubara, K. (1986) Gene (Amst.) 41 299–304 [DOI] [PubMed] [Google Scholar]
- 11.Maeda, N., Kim, H. S., Azen, E. A., and Smithies, O. (1985) J. Biol. Chem. 260 11123–11130 [PubMed] [Google Scholar]
- 12.Isemura, S. (2000) J. Biochem. (Tokyo) 127 393–398 [DOI] [PubMed] [Google Scholar]
- 13.Prakobphol, A., Xu, F., Hoang, V. M., Larsson, T., Bergstrom, J., Johansson, I., Frangsmyr, L., Holmskov, U., Leffler, H., Nilsson, C., Boren, T., Wright, J. R., Stromberg, N., and Fisher, S. J. (2000) J. Biol. Chem. 275 39860–39866 [DOI] [PubMed] [Google Scholar]
- 14.Bobek, L. A., Aguirre, A., and Levine, M. J. (1991) Biochem. J. 278 627–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saitoh, E., Isemura, S., Sanada, K., and Ohnishi, K. (1991) Biomed. Biochim. Acta 50 599–605 [PubMed] [Google Scholar]
- 16.Saitoh, E., Isemura, S., Sanada, K., and Ohnishi, K. (1992) Agents Actions Suppl. 38 340–348 [DOI] [PubMed] [Google Scholar]
- 17.Sabatini, L. M., and Azen, E. A. (1989) Biochem. Biophys. Res. Commun. 160 495–502 [DOI] [PubMed] [Google Scholar]
- 18.Sabatini, L. M., Carlock, L. R., Johnson, G. W., and Azen, E. A. (1987) Am. J. Hum. Genet. 41 1048–1060 [PMC free article] [PubMed] [Google Scholar]
- 19.Sabatini, L. M., Ota, T., and Azen, E. A. (1993) Mol. Biol. Evol. 10 497–511 [DOI] [PubMed] [Google Scholar]
- 20.Azen, A. E., Amberger, E., Fisher, S., Prakobphol, A., and Niece, R. L. (1996) Am. J. Hum. Genet. 58 143–153 [PMC free article] [PubMed] [Google Scholar]
- 21.Baum, B. J., Bird, J. L., Millar, D. B., and Longton, R. W. (1976) Arch. Biochem. Biophys. 177 427–436 [DOI] [PubMed] [Google Scholar]
- 22.Minaguchi, K., Madapallimattam, G., and Bennick, A. (1988) Biochem. J. 250 171–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arneberg, P. (1971) Scand. J. Dent. Res. 79 60–64 [DOI] [PubMed] [Google Scholar]
- 24.Merrell, K., Southwick, K., Graves, S. W., Esplin, M. S., Lewis, N. E., and Thulin, C. D. (2004) J. Biomol. Tech. 15 238–248 [PMC free article] [PubMed] [Google Scholar]
- 25.Baum, B. J., Bird, J. L., Millar, D. B., and Longton, R. W. (1977) Comp. Biochem. Physiol. A 56 115–120 [DOI] [PubMed] [Google Scholar]
- 26.Ornstein, L. (1964) Ann. N. Y. Acad. Sci. 121 321–349 [DOI] [PubMed] [Google Scholar]
- 27.Davis, B. J. (1964) Ann. N. Y. Acad. Sci. 121 404–427 [DOI] [PubMed] [Google Scholar]
- 28.Inzitari, R., Cabras, T., Rossetti, D. V., Fanali, C., Vitali, A., Pellegrini, M., Paludetti, G., Manni, A., Giardina, B., Messana, I., and Castagnola, M. (2006) Proteomics 6 6370–6379 [DOI] [PubMed] [Google Scholar]
- 29.Castagnola, M., Inzitari, R., Rossetti, D. V., Olmi, C., Cabras, T., Piras, V., Nicolussi, P., Sanna, M. T., Pellegrini, M., Giardina, B., and Messana, I. (2004) J. Biol. Chem. 279 41436–41443 [DOI] [PubMed] [Google Scholar]
- 30.Flora, B., Gusman, H., Helmerhorst, E. J., Troxler, R. F., and Oppenheim, F. G. (2001) Protein Expr. Purif. 23 198–206 [DOI] [PubMed] [Google Scholar]
- 31.Oppenheim, F. G., Hay, D. I., and Franzblau, C. (1971) Biochemistry 10 4233–4238 [DOI] [PubMed] [Google Scholar]
- 32.Wong, R. S., and Bennick, A. (1980) J. Biol. Chem. 255 5943–5948 [PubMed] [Google Scholar]
- 33.Wong, R. S., Hofmann, T., and Bennick, A. (1979) J. Biol. Chem. 254 4800–4808 [PubMed] [Google Scholar]
- 34.Hay, D. I., Bennick, A., Schlesinger, D. H., Minaguchi, K., Madapallimattam, G., and Schluckebier, S. K. (1988) Biochem. J. 255 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salih, E. (2005) Mass Spectrom. Rev. 24 828–846 [DOI] [PubMed] [Google Scholar]
- 36.Villen, J., Beausoleil, S. A., Gerber, S. A., and Gygi, S. P. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 1488–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Al-Hashimi, I., and Levine, M. J. (1989) Arch. Oral Biol. 34 289–295 [DOI] [PubMed] [Google Scholar]
- 38.Li, J., Helmerhorst, E. J., Corley, R. B., Luus, L. E., Troxler, R. F., and Oppenheim, F. G. (2003) Oral Microbiol. Immunol. 18 183–191 [DOI] [PubMed] [Google Scholar]
- 39.Li, J., Helmerhorst, E. J., Yao, Y., Nunn, M. E., Troxler, R. F., and Oppenheim, F. G. (2004) Arch. Oral Biol. 49 379–385 [DOI] [PubMed] [Google Scholar]
- 40.Basak, A., Toure, B. B., Lazure, C., Mbikay, M., Chretien, M., and Seidah, N. G. (1999) Biochem. J. 343 29–37 [PMC free article] [PubMed] [Google Scholar]
- 41.Ishihara, M., Kawanishi, A., Watanabe, H., Tomochika, K., Miyoshi, S., and Shinoda, S. (2002) Microbiol. Immunol. 46 298–303 [DOI] [PubMed] [Google Scholar]
- 42.Dawes, C. (2004) Caries Res. 38 236–240 [DOI] [PubMed] [Google Scholar]
- 43.Chan, M., and Bennick, A. (2001) Eur. J. Biochem. 268 3423–3431 [DOI] [PubMed] [Google Scholar]
- 44.Messana, I., Cabras, T., Pisano, E., Sanna, M. T., Olianas, A., Manconi, B., Pellegrini, M., Paludetti, G., Scarano, E., Fiorita, A., Agostino, S., Contucci, A. M., Calo, L., Picciotti, P. M., Manni, A., Bennick, A., Vitali, A., Fanali, C., Inzitari, R., and Castagnola, M. (2008) Mol. Cell. Proteomics 7 911–926 [DOI] [PubMed] [Google Scholar]
- 45.Stubbs, M., Chan, J., Kwan, A., So, J., Barchynsky, U., Rassouli-Rahsti, M., Robinson, R., and Bennick, A. (1998) Arch. Oral Biol. 43 753–770 [DOI] [PubMed] [Google Scholar]
- 46.Potempa, J., Banbula, A., and Travis, J. (2000) Periodontol. 2000 24 153–192 [DOI] [PubMed] [Google Scholar]
- 47.Slots, J. (1981) J. Clin. Microbiol. 14 288–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wikstrom, M. B. (1983) Appl. Environ. Microbiol. 45 393–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hejgaard, J. (2005) Biol. Chem. 386 1319–1323 [DOI] [PubMed] [Google Scholar]
- 50.Vora, H., McIntire, J., Kumar, P., Deshpande, M., and Khosla, C. (2007) Biotechnol. Bioengineer. 98 177–185 [DOI] [PubMed] [Google Scholar]