Abstract
Protein sumoylation is a regulated process that is important for the health of human and yeast cells. In budding yeast, a subset of sumoylated proteins is targeted for ubiquitination by a conserved heterodimeric ubiquitin (Ub) ligase, Slx5–Slx8, which is needed to suppress the accumulation of high molecular weight small ubiquitin-like modifier (SUMO) conjugates. Structure-function analysis indicates that the Slx5–Slx8 complex contains multiple SUMO-binding domains that are collectively required for in vivo function. To determine the specificity of Slx5–Slx8, we assayed its Ub ligase activity using sumoylated Siz2 as an in vitro substrate. In contrast to unsumoylated or multisumoylated Siz2, substrates containing poly-SUMO conjugates were efficiently ubiquitinated by Slx5–Slx8. Although Siz2 itself was ubiquitinated, the bulk of the Ub was conjugated to SUMO residues. Slx5–Slx8 primarily mono-ubiquitinated the N-terminal SUMO moiety of the chain. These data indicate that the Slx5–Slx8 Ub ligase is stimulated by poly-SUMO conjugates and that it can ubiquitinate a poly-SUMO chain.
Sumoylation regulates a diverse set of cellular processes and is essential for viability in budding yeast (1). Sumoylation resembles ubiquitination in that the C terminus of SUMO2 is conjugated to lysine residues of target proteins. In yeast, this occurs through the sequential activity of an activating (E1) enzyme (Aos1/Uba2), a conjugating (E2) enzyme (Ubc9), and one of several SUMO ligases (E3) (e.g. Mms21 and Siz2) (2–4). Although a large class of Ub E3 ligases are RING-domain proteins (5), SUMO E3 ligases often contain a variant domain known as SP-RING (3, 6). Sumoylation can be reversed by the Ulp1 and Ulp2 isopeptidases, which catalyze the cleavage of SUMO polypeptides from target proteins (7, 8).
In yeast, recombinational DNA repair depends on sumoylation. It has been known for some time that DNA damage tolerance is compromised in yeast strains with defects in Ubc9, Mms21, Ulp1, or Ulp2 (7, 9–11). More recently, cells lacking the Srs2 anti-recombinase have been shown to require the Ulp1 isopeptidase for viability (12), and cells defective in UBC9 and MMS21 accumulate Rad51-dependent cruciform structures during DNA replication (13). In the best characterized cases, SUMO has been shown to link Srs2 to proliferating cell nuclear antigen (14, 15) and to be conjugated to Rad52 (16, 17).
Sumoylation is distinct from ubiquitination in that target proteins are typically modified by single SUMO moieties that result in mono- or multisumoylated products as opposed to targets bearing poly-SUMO chains (1). Nonetheless, poly-SUMO conjugates are commonly observed in in vitro sumoylation reactions (3, 18, 19), and they accumulate in yeast cells deficient in the Ulp2 SUMO protease or Ub-mediated proteolysis (20, 21). The polymerization of SUMO in yeast (called Smt3) occurs preferentially through its three N-terminal lysine residues (lysines 11, 15, and 19) that are found in a domain of 20 amino acids (aa) unique to SUMO (18–20). The function of such poly-sumoylation is unknown, although in the case of ulp2Δ cells, it appears to be toxic (20).
SLX5 and SLX8 are required for the viability of yeast cells lacking the Sgs1 DNA helicase (22). These genes encode a heterodimeric Ub ligase that links sumoylation to recombinational DNA repair (21, 23–25). On their own, slx5Δ and slx8Δ null mutants display similar phenotypes, including slow growth, sensitivity to hydroxyurea (HU), and increased rates of gross chromosomal rearrangements and mitotic recombination (22, 26, 27). Like mutants deficient in sumoylation, slx5Δ and slx8Δ mutants display a clonal lethality that is dependent on the 2μ circle (9, 28, 29), and this can be suppressed by eliminating genes in the RAD51-independent recombination pathway (26). The SLX5 and SLX8 genes were also isolated in a screen for suppressors of a mot1–301 allele, along with mutations in most of the enzymes involved in the sumoylation pathway (30). Importantly, there is an accumulation of sumoylated proteins in slx5Δ and slx8Δ cells that correspond to high molecular weight SUMO conjugates (21, 24, 30).
The Slx5–Slx8 complex interacts directly with SUMO via SUMO interacting motifs in each subunit (21, 24, 28, 31). The complex appears to be functionally conserved in Schizosaccharomyces pombe, where the related RING-finger proteins Rfp1 and Rfp2 also interact with SUMO and Slx8 to suppress the build-up of sumoylated proteins (21, 24, 32–34). The idea that the Ub ligase activity of the complex is directed toward sumoylated targets is supported by the finding that substrates containing a SUMO sequence are preferentially ubiquitinated (24, 32, 33).
In this study we investigated the specificity of the Slx5–Slx8 Ub ligase toward sumoylated substrates. We found that its activity on free SUMO or a multisumoylated test substrate was low, whereas ubiquitination of a poly-sumoylated substrate was high. Characterization of this reaction revealed that Slx5–Slx8 primarily mono-ubiquitinated the N-terminal end of the SUMO chain. Although there is no known physiological role for poly-sumoylation, we found increased levels of these conjugates in cells lacking the Sgs1 DNA helicase. Thus, it appears that one essential function of the Slx5–Slx8 Ub ligase is to ubiquitinate poly-sumoylated proteins that arise in sgs1Δ cells.
EXPERIMENTAL PROCEDURES
Yeast Strains and Plasmids—Standard methods and media were used for the propagation, transformation, and culturing of Saccharomyces cerevisiae (35). Strain JD194 (MATα ura3Δ5 his3–11,15 leu2–3,112 pre1–1 pre2–2) was kindly provided by Dr. Kiran Madura. Additional genotypes and plasmid construction details are available on request.
Expression and Purification of Recombinant Proteins—GST- and His6-tagged proteins were produced in Escherichia coli BL21-RIL cells (Stratagene) using the T7 expression system of Studier (36). His6-tagged proteins were expressed and purified as described previously (37). GST-tagged proteins were expressed similarly but purified following lysis in Buffer A (25 mm Tris-HCl (pH 7.5), 10% glycerol, 1 mm EDTA, 0.01% Nonidet P-40, 1 mm dithiothreitol, and 0.1 mm phenylmethylsulfonyl fluoride) containing 200 mm NaCl and protease inhibitors. The extract was applied to a 1-ml GST-TRAP column (GE Healthcare), washed with 20 ml of Buffer A containing 200 mm NaCl, and eluted with the same buffer containing 10 mm glutathione. Peak protein fractions were dialyzed in Buffer A containing 100 mm NaCl. Additional versions of GST–Smt3 and GST-Ub fusion proteins were constructed with protease 3C and cAMPdPK consensus sites downstream of the GST domain. Following treatment with cAMPdPK and γ-[32P]ATP, proteins were re-bound to glutathione beads and washed free of ATP prior to eluting the radiolabeled protein via protease 3C cleavage. Expression and purification of Ulp1UD (38) has been described (28).
SUMO Binding Assay—Physical interactions between His6/FLAG-tagged Smt3 (HF-Smt3) (2) and GST-Slx5 or GST-Slx8 were detected following incubation on ice for 1 h in a final volume of 0.1 ml using Buffer A with 50 mm NaCl as the incubation buffer. This reaction was then diluted with 0.3 ml of incubation buffer and mixed with 20 μl of glutathione beads (40 μl of 50% slurry) for 1 h at 4 °C. The beads were recovered by low speed spin and washed three times with incubation buffer. Bound proteins were eluted with 25 μl of SDS sample buffer and detected by immunoblotting as described before (37).
In Vitro Sumoylation Assay—The standard sumoylation reaction was performed in the presence of 20 mm HEPES (pH 7.5), 5 mm MgCl2, 2 mm ATP, 5 μm ZnSO4, and 0.1 mm dithiothreitol. Unless otherwise stated, reactions were incubated at 30 °C for 60 min and contained 2 nm Aos1/Uba2, 30 nm Ubc9, 100 nm Siz2-V5, and 2 μm HF-Smt3-G98 in a total volume of 20 μl. Immunoblotting was carried out essentially as described (37) except that proteins were transferred to polyvinylidene difluoride or a 0.22-μm nitrocellulose membrane. Where indicated, mature HF-Smt3-G98 was substituted with HF-Smt3-G97 or HF-Smt3-G98A mutants, whereas latter experiments employed Smt3 and variants that were simply His6-tagged.
In Vitro Ubiquitination Assay—Ubiquitination was performed using the same buffer conditions as the sumoylation assay. Reactions were incubated at 30 °C for 15 min and contained 2 nm Uba1, 30 nm Ubc5, 10 nm unlabeled ubiquitin, and 350,000 cpm 32P-Ub in a total volume of 30 μl. The products of this reaction were resolved by SDS-PAGE (typically 15% acrylamide). The gel was then fixed and visualized on a PhosphorImager (Amersham Biosciences).
RESULTS
Multiple SUMO Binding Domains Are Required for Slx5–Slx8 Function—To locate potential SUMO-binding domains within the two subunits of the Ub ligase, we fused portions of the N and C termini of Slx5 or Slx8 to the C terminus of GST. Purified recombinant proteins were then assayed for their ability to bind SUMO following incubation with glutathione beads. In contrast to GST alone, GST-Slx5 bound strongly to SUMO (Fig. 1A). Two small N-terminal truncations (ΔN100 and ΔN200) showed diminishing affinity to SUMO, whereas deletion of 300 aa or more eliminated this activity. Analysis of C-terminal deletions revealed a strong SUMO-binding domain within the first 100 aa of Slx5, although additional mapping identified a second larger domain between aa 101 and 300 (Fig. 1B). Thus, the first 300 aa of Slx5 contains at least two SUMO-binding domains. When the N-terminal deletions were examined for their ability to complement sgs1Δ synthetic lethality, a correlation was found between loss of SUMO binding and loss of complementation (Fig. 1A). And consistent with its role in promoting Ub ligase activity (23), the RING domain of Slx5 was required to complement sgs1Δ synthetic lethality. However, deletion of a single N-terminal SUMO-binding domain was tolerated in sgs1Δ cells. A similar analysis revealed a SUMO binding activity in Slx8 that was localized to the C-terminal 111 aa (Fig. 1C). As with Slx5, removal of the one SUMO-binding domain in Slx8 was tolerated in sgs1Δ cells. Consistent with the mapping of multiple SUMO interacting motifs within these subunits (21, 24), we conclude that there are at least three SUMO-binding domains within the Slx5–Slx8 Ub ligase.
FIGURE 1.
Identification of SUMO-binding domains in Slx5 and Slx8. A, right panel, the SLX5 ORF or fragments corresponding to the indicated N- and C-terminal truncations were cloned downstream of the SLX5 promoter in vector pRS415 (LEU2/CEN/ARS). These constructs were then transformed into strain JMY1464 [(sgs1Δ slx5Δ plus pJM500 (SGS1/URA3/ADE3)]. Transformants were resuspended at a concentration of A600 = 3.0, and following 10-fold serial dilution, ∼5 μl was pinned onto solid synthetic complete media (SD) lacking leucine (-LEU) or containing 5-fluoro-orotic acid to select against pJM500. Plates were photographed following 3 days of growth at 30 °C. Left panel, the indicated proteins were fused to the C terminus of GST and purified from bacteria. GST fusions or GST alone (1 μg) were then incubated on ice with 1 μg of HF-Smt3, and bound proteins were detected by glutathione bead pulldown assay and immunoblotting with anti-FLAG antibody. B, fine structure mapping of the N-terminal SUMO-binding domains of Slx5 was carried out as in A. Shading is used in the schematic to indicate the minimal SUMO-binding domains. C, the SLX8 open reading frame or fragments corresponding to the indicated truncations were assayed as in A except that strain VCY1524 [(sgs1Δ slx8Δ plus pJM500 (SGS1/URA3/ADE3)] was used as host.
The Severity of slx5 and slx8 Phenotypes Correlates with in Vivo Sumoylation Levels—To correlate the sumoylation defects of slx5 and slx8 mutants with their other phenotypes, we characterized a collection of point and truncation alleles with respect to their effect on in vitro Ub ligase activity, accumulation of hyper-sumoylated proteins, HU sensitivity, and sgs1Δ synthetic lethality. Among point mutation alleles that map to the RING domains of Slx5 and Slx8, four were shown to eliminate Ub ligase activity in vitro (slx5-6, slx5-8, slx8-1, and slx8-3) and those displayed lethality in the sgs1Δ background (Fig. 2A) (23, 37)). Those retaining Ub ligase activity (slx5-5, slx5-7, and slx8-2) displayed intermediate complementation in the sgs1Δ background (Fig. 2A) (37). To assay hyper-sumoylation, a denatured extract from each strain was immunoblotted with α-Smt3 antibody. All mutations that were synthetically lethal with sgs1Δ showed extreme hyper-sumoylation, whereas the intermediate alleles displayed partially elevated sumoylation levels (Fig. 2B). In these and subsequent experiments the 5% stacking gel was retained on the blot, because it has been shown that highly poly-sumoylated proteins are retained in the stacking gel (20). Interestingly, removing one SUMO-binding domain (slx5-ΔN100, -ΔN200, and slx8-ΔN200) resulted in intermediate levels of hyper-sumoylation, whereas deleting an additional domain (slx5-ΔN300 and slx5-ΔN400) or a RING domain (slx5-ΔC126 and slx8-ΔC74) produced a null phenotype. Consistently, strains with high sumoylation levels grew poorly on HU (Fig. 2C) and required SGS1 to survive (Fig. 2B). Strains with slightly elevated sumoylation levels grew like wild type on HU and survived loss of SGS1. We conclude that the in vivo sumoylation levels and HU sensitivities of slx5 and slx8 alleles correlate well with their sgs1Δ synthetic-lethal phenotypes.
FIGURE 2.
Synthetic-lethal phenotype of SLX5 and SLX8 mutations correlates with the accumulation of hyper-sumoylated proteins. A, alleles of SLX5 containing the indicated RING-finger mutations were tested for complementation of sgs1Δ slx5Δ synthetic lethality as in Fig. 1. B, the indicated SLX5 or SLX8 alleles were integrated into strain JMY1699 (slx5Δ) or SIY778 (slx8Δ), and N-ethylmaleimide extracts were analyzed for Smt3-protein conjugates by 8% SDS-PAGE and immunoblotting using antibodies against Smt3 (upper panel) or Rfa1 (lower panel). A bracket marks the stacking gel. Also shown is the alleles' ability to complement the relevant slxΔ sgs1Δ synthetic lethality. To determine sgs1Δ viability, strains JMY1464 or VCY1525 [(sgs1Δ slx8Δ plus pJM500 (SGS1/URA3/ADE3)] were transformed with LEU2/CEN/ARS plasmids carrying the indicated SLX5 or SLX8 alleles and evaluated for growth on plates containing 5-fluoro-orotic acid. Symbols: +, good growth; +/–, slow growth; –, no growth. C, strains JMY1699 or SIY778 were transformed with LEU2/CEN/ARS plasmids carrying the indicated alleles, were assayed for growth as in Fig. 1A but in the presence or absence of 0.1 m hydroxyurea. The plates were photographed after 3 days of growth at 30 °C.
Sumoylated Siz2 Is Ubiquitinated by Slx5–Slx8—To examine the specificity of the Slx5–Slx8 ubiquitin ligase toward sumoylated proteins, we took advantage of the fact that the SUMO E3 ligase Siz2, which is known to be sumoylated in vivo (39), exhibits auto-sumoylation in vitro. Sumoylated Siz2 was then used as a substrate for in vitro ubiquitination. To validate this system, sumoylation reactions were carried out with epitope-tagged Siz2 (Siz2-V5) and a variety of mutant SUMO substrates. These products were analyzed on duplicate immunoblots that were probed with α-V5 or α-Smt3 (Figs. 3, A and B). Using mature HF-Smt3-G98 as substrate (2), we observed two major species of Siz2 products (Fig. 3A, lane 1). One product migrated as a doublet at 125–150 kDa and was judged to be conjugated to one or more SUMO groups. The upper band is most likely multisumoylated, because it co-migrated with the single product obtained with Smt3-aR (Fig. 3A, lane 4). All nine lysine residues have been mutated to arginine in Smt3-aR, so it is unable to form SUMO chains (20). The second product was a highly poly-sumoylated form of Siz2, which displayed a characteristic mobility just entering the stacking gel (Fig. 3, A and B, lane 1). As expected, Smt3-G97 (lacking the essential C-terminal glycine) gave no sumoylated product, because it cannot form covalent attachments to target proteins (Fig. 3, A and B, lane 2), and Smt3-G98A produced both multi- and poly-sumoylated Siz2 in addition to short multimers of Smt3. Finally, synthesis of these products required functional Smt3, E1, E2 and Siz2 itself (Fig. 3, A and B, lanes 5–8). We conclude that Siz2 has multiple sites for Smt3 attachment, and that it can catalyze the formation of poly-SUMO chains.
FIGURE 3.
Poly-sumoylated Siz2 is preferentially ubiquitinated by Slx5–Slx8. A, standard sumoylation assays were performed in the presence of the indicated Smt3 variants or in the absence of the indicated component. Siz2-V5 products were analyzed by SDS-PAGE and immunoblotting with antibodies against the V5 epitope. B, duplicate sumoylation reactions were analyzed by SDS-PAGE and immunoblotting with antibodies against Smt3. Oligomeric chains of Smt3 are indicated by S2-S5. Note that all Smt3 proteins were N-terminally tagged with His6-FLAG, except for Smt3-aR, which was tagged with His6 only. C, sumoylation reactions were first performed in the presence of wt HF-Smt3 (lanes 1 and 5–14), the indicated HF-Smt3 mutants (lanes 2 and 3), or no Smt3 (lane 4). The reaction products were then subjected to a standard ubiquitination assay in the presence of 32P-Ub and either 10 nm Slx5–Slx8 (lanes 1–6), or 0, 10 pm, 30 pm, 100 pm, 300 pm, 1 nm, 3 nm, or 10 nm Slx5–Slx8 (lanes 7–14). The Ub E1 or E2 was omitted where indicated (lanes 5 and 6).
To detect Ub E3 ligase activity, Siz2 sumoylation products were incubated with Uba1, Ubc5, ATP, and the Slx5–Slx8 dimer as described (23), together with radiolabeled ubiquitin (32P-Ub) as substrate. Following a 15-min incubation, the products were analyzed by SDS-PAGE and autoradiography. As shown in Fig. 3C, samples containing sumoylated Siz2 produced a labeled band near the well of the gel (lanes 1–4). In addition to this band was a background smear of auto-ubiquitinated products such as poly-Ub chains (Ubn) in the resolving gel. The major signal that was trapped in the stacking gel was dependent on Ub E1, Ub E2, and Slx5–Slx8 (lanes 5–7). Importantly, the level of signal was responsive to a titration of Slx5–Slx8 as expected for an Ub E3 ligase activity (Fig. 3C, lanes 7–14). Moreover, products were obtained using Slx5–Slx8 at concentrations as low as 30 pm, which indicates a significant affinity relative to the levels (∼3 nm) needed for robust auto-ubiquitination (23).
Poly-SUMO Chains Are a Preferred Substrate of Slx5–Slx8—Because the ubiquitinated products shown in Fig. 3C could only just enter the stacking gel, it was not possible to determine the structure of these molecules. We therefore considered the following possibilities: (a) the signal in the well could be due to the attachment of Ub chains onto mono- or multisumoylated Siz2 or (b) the signal could be due to one or more Ub residues ligated to poly-sumoylated Siz2 (Siz2-(Smt3)n). These possibilities were distinguished by repeating the assay using radiolabeled Ub mutants in which some or all of its conjugable lysines had been mutated to arginine. As shown in Fig. 4A (lanes 1–4), the ubiquitination signal was unchanged regardless of whether the 32P-Ub was wt, contained Lys-48 as its only lysine (K48O), contained Lys-63 only (K63O), or contained no lysines (aR). The fact that the product obtained with 32P-Ub-aR, which is unable to form chains, is indistinguishable from wt 32P-Ub suggests that the substrate is not mono- or multisumoylated Siz2, which, if mono-ubiquitinated, should migrate into the resolving gel (Fig. 3, A and B). Ubiquitination was also strictly dependent on the synthesis of poly-SUMO chains, because neither wild-type nor mutant 32P-Ubs can be ligated onto Siz2 following sumoylation with Smt3-aR (Fig. 4A, lanes 5–9). We conclude that poly-sumoylated Siz2 is a preferred substrate of Slx5–Slx8.
FIGURE 4.
Poly-SUMO chains are targeted for ubiquitination. A, Siz2 sumoylation reactions were first performed in the presence of wt HF-Smt3 (+), no Smt3 (–), or H6-Smt3-aR (aR). The products were subsequently ubiquitinated in the presence of 32P-Ub and 10 nm Slx5–Slx8 prior to analysis by SDS-PAGE and autoradiography. Ub was either wt or contained the indicated mutations. B, standard sumoylation reactions were performed in the presence of 4 μm HF-Smt3, and the products were subjected to ubiquitination with wt 32P-Ub as above. The products of this reaction were then subjected to a 1-h incubation with recombinant Ulp1UD at the following concentrations: 0, 10 fm, 100 fm, 1 pm, 10 pm, 100 pm, and 1 nm (lanes 1–7, respectively). As a marker for HF-Smt3-Ub*, free HF-Smt3 was subjected to a standard ubiquitination reaction in the presence 100 nm Slx5–Slx8 (lane 8). Bands were identified based on the relative molecular weights of HF-Smt3 (20 kDa) and Ub (7 kDa). The bands migrating at ∼107 and 73 kDa represent mono-ubiquitinated Slx5 and Slx8, as indicated. The molecular weight markers are identical to those in Fig. 3C.
The ability of Siz2-(Smt3)n to serve as a preferred ubiquitination substrate leaves open the question of whether Ub is ligated directly to the target protein (Siz2) or to the SUMO chain itself. To answer this question, ubiquitination reactions were first performed on Siz2-(Smt3)n in the presence of wt 32P-Ub. Subsequently, the products were treated with the Ulp1UD SUMO protease, which cleaves Smt3 from modified proteins and proteolyzes Smt3 chains into monomers. As shown in Fig. 4B, titration of Ulp1UD resulted in the disappearance of the Siz2-(Smt3)n-Ub* signal from the stacking gel, and the appearance of a major radiolabeled product at 27 kDa (Fig. 4B, lanes 5–7). The identification of this band as Smt3-Ub* is based on its predicted molecular weight (HF-Smt3 + 32P-Ub = 20 + 7 kDa) as well as the following control. We attempted to ubiquitinate monomeric SUMO by incubating Uba1, Ubc5, ATP, and 32P-Ub together with free HF-Smt3 and Slx5–Slx8. Monomeric HF-Smt3 was a poor substrate in this reaction; however, some ubiquitinated forms of HF-Smt3 were observed, including diminishing amounts of di- and tri-Ub conjugates (supplemental Fig. S1). The primary mono-ubiquitinated product, Smt3-Ub*, co-migrated with the Ulp1UD-proteolytic product at 27 kDa (Fig. 4B, lane 8), which migrates as a doublet. The source of this doublet is unknown; however, we suspect that proteolysis may be imprecise or that the two bands represent isomers in which Ub is conjugated to different lysine residues of Smt3.
The relatively small amounts of di-ubiquitinated Smt3 (Fig. 4B, lanes 5–7) indicate that this E3 ligase primarily mono-ubiquitinates its substrates. This is consistent with two test proteins used previously (23) and the prominent Slx5–Slx8 auto-ubiquitination products visible in Fig. 4. The Ulp1UD titration resulted in a transient product at 47 kDa (Fig. 4B, lane 4) consistent with the expected (Smt3)2-Ub* intermediate. Larger intermediate products, such as (Smt3)3-Ub* were probably not observed due to rapid proteolysis of the SUMO chain, however we cannot rule out the possibility that Smt3-Ub is a preferred substrate of Ulp1UD. In addition, we did not detect a 54-kDa band (representing (Smt3-Ub*)2) that would be expected from a SUMO chain saturated with mono-Ub residues. Along with the release of the major 27-kDa band was a band at 112 kDa (Fig. 4B, lanes 5–7). This band is consistent with mono-ubiquitinated Siz2-V5 (i.e. 105 plus 7 kDa), which indicates that Slx5–Slx8 also displays activity against the target protein. Taken together, we conclude that Slx5–Slx8 is activated by poly-SUMO chains and that it mono-ubiquitinates both Siz2 and SUMO.
Slx5–Slx8 Ubiquitination Requires the N-terminal Lysines of Smt3—The above experiment raised the question of where on the SUMO chain the Ub was conjugated. We addressed this question by first determining which of the nine lysines of Smt3 were required for efficient ubiquitination by Slx5–Slx8. Johnson and colleagues have previously shown that Smt3 preferentially polymerizes through its three N-terminal lysine residues (lysines 11, 15, and 19), although a SUMO E3 ligase is capable of promoting chain formation through alternative lysine residues when the primary sites have been mutated to arginine (20). We therefore prepared sumoylated Siz2 using an array of Smt3 mutants in which individual lysines, or groups of lysines, had been mutated to arginine. A portion of each reaction (one-third) was analyzed by immunoblotting with either anti-V5 (Fig. 5A) or anti-Smt3 (Fig. 5B). The accumulation of high molecular weight signal in these blots indicates that poly-sumoylation of Siz2 occurs using any of the mutant Smt3 proteins, except for Smt3-aR (Fig. 5, A and B, lanes 9). The remaining portion of the sumoylated reaction product was ubiquitinated by Slx5–Slx8 using 32P-Ub (Fig. 5C). Surprisingly, most of these poly-sumoylated proteins were efficiently ubiquitinated, including Siz2-(Smt3-CT5KR)n, which lacks the five C-terminal lysine residues of Smt3. Smt3s lacking one or two of the N-terminal lysine residues were also ubiquitinated following polymerization (Fig. 5C, lanes 4–7). However, chains of Smt3 lacking all three N-terminal lysines (Smt3–3KR) were altered in their ubiquitination (Fig. 5C, lane 8). Although immunoblotting showed that bulk Siz2-(Smt3–3KR)n product appeared to just enter the stacking gel like chains prepared with wt Smt3 (Fig. 5, A and B, compare lanes 1 and 8), there was less ubiquitinated product at the well, more signal spread throughout the stacking gel, and more intense background signal in the 150- to 250-kDa range of the resolving gel. This increased signal in the resolving gel was also observed with Smt3-aR and, as shown below, appears to be due to ongoing sumoylation of auto-ubiquitinated Slx5 and Slx8 subunits. The enhanced sumoylation of reaction components has previously been noted in cases where poly-sumoylation is inhibited using mutant SUMOs (20).
FIGURE 5.
The N-terminal lysines of Smt3 are important for ubiquitination by Slx5–Slx8. A, standard sumoylation reactions were performed except that the duration of the incubation was 45 min and included His6-Smt3 (WT), the indicated His6-Smt3 mutant proteins, or no Smt3 (lane 10). In lane 2, ATP was excluded from the sumoylation reaction. One-third of each reaction was analyzed by SDS-PAGE and immunoblotting with anti-V5. Unmodified Siz2 is indicated with an arrow. B, one-third of each of the above reactions was immunoblotted with antibodies against Smt3. C, one-third of each of the above reactions was subjected to ubiquitination in the presence of 30 nm Slx5–Slx8 and standard levels of Uba1 and Ubc5.
To confirm the above result, Siz2-(Smt3–3KR)n was ubiquitinated with a titration of Slx5–Slx8 (Fig. 6A). Compared with wt Siz2-(Smt3)n (Fig. 3C), Siz2-(Smt3–3KR)n failed to generate a discrete band near the well, even at the highest level of Ub ligase, and it produced background signal in the resolving gel that was similar to that obtained with Siz2-Smt3-aR (Fig. 6B). Thus, the same lysine residues that are primarily used for SUMO polymerization (lysines 11, 15, and 19) are important for ubiquitination by Slx5–Slx8. A variety of these labeled ubiquitinated reaction products were then subjected to proteolysis by Ulp1UD (Fig. 6C). Treatment of Siz2-Smt3–3KR and Siz2-Smt3-CT5KR ubiquitination products with Ulp1UD reduced the material present in the stack and the higher molecular weight region of the resolving gel. It also generated bands corresponding to Smt3-Ub* (23 kDa; Fig. 6C, lanes 5 and 7), although these were less intense than that obtained with wt Smt3. In all cases, including Smt3-aR, Ulp1UD treatment also released Siz2-Ub* (112 kDa). However, the level of Siz2-Ub* that was released was no greater than that obtained in the absence of sumoylation (Fig. 6C, lanes 10 and 11). Also, the enhanced sumoylation of Slx5–Slx8 subunits with Smt3 mutants that fail to polymerize well (Smt3–3KR and Smt3-aR), did not stimulate their auto-ubiquitination (Fig. 6C, lanes 5 and 7). Thus, Slx5–Slx8-dependent ubiquitination preferentially targets poly-SUMO chains and is sensitive to the loss of the three N-terminal lysines of SUMO. Because of their role in SUMO conjugation, these residues are expected to be most readily available on the terminal SUMO moiety of the poly-SUMO chain.
FIGURE 6.
Characterization of alternative poly-SUMO chains ubiquitinated by Slx5–Slx8. A, consecutive sumoylation-ubiquitination reactions were performed exactly as in Fig. 3C except that the following Smt3 derivatives were used: wt HF-Smt3 (lane 1), HF-Smt3-G97 (lane 2), HF-Smt3-G98A (lane 3), no Smt3 (lane 4), or His6-Smt3–3KR (lanes 6–15). B, consecutive sumoylation-ubiquitination reactions were performed exactly as above except that His6-Smt3-aR was used as substrate in lanes 6–15. C, consecutive sumoylation-ubiquitination reactions were untreated or cleaved with 10 nm Ulp1UD as in Fig. 4B, except that the initial sumoylation reactions were performed in the presence of the following Smt3 proteins: His6-Smt3 (WT), His6-Smt3–3KR (3KR), His6-Smt3-CT5KR (CT5KR), His6-Smt3-aR (aR), or no Smt3 (None).
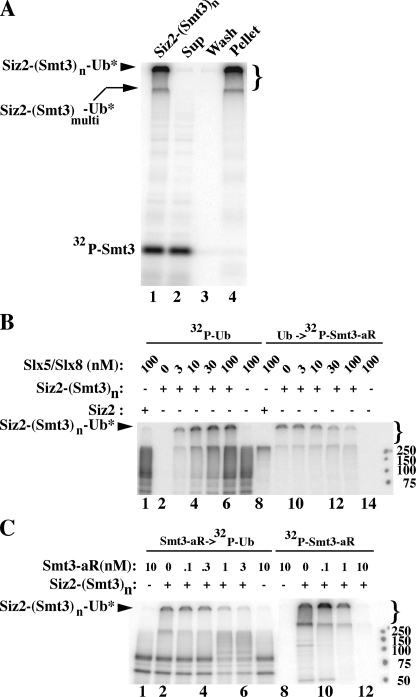
The Terminal SUMO Moiety Is Targeted for Ubiquitination by Slx5–Slx8—If lysine residues 11, 15, or 19 of the terminal SUMO moiety are targeted for ubiquitination by Slx5–Slx8, then ubiquitination should interfere with further SUMO chain elongation. To test this idea, the Siz2-(Smt3)n chains were purified away from any free Smt3 used in its preparation. We reasoned that the very large molecular weight of the Siz2-(Smt3)n product may allow it to be purified by ultracentrifugation. To validate this method, we prepared poly-sumoylated Siz2 using 32P-Smt3 and subjected the products to centrifugation at 113,000 × g. The pellet was then washed, repelleted, and resuspended before loading equivalent samples from each step on an SDS gel for analysis by autoradiography. As shown in Fig. 7A, the Siz2-(Smt3)n product was successfully purified away from the 32P-Smt3. We then prepared unlabeled poly-sumoylated Siz2 using cold Smt3 and subjected it to the same purification scheme. The product was then ubiquitinated with increasing concentrations of Slx5–Slx8 using either 32P-Ub (as a control) or cold Ub. The products of the cold ubiquitination were then subjected to a new round of sumoylation using 32P-Smt3-aR. Consistent with previous results, purified Siz2-(Smt3)n was efficiently labeled with 32P-Ub as the concentration of Slx5–Slx8 was increased in the control reactions (Fig. 7B, lanes 2–6 and supplemental Fig. S2A) while unmodified Siz2 was weakly labeled (Fig. 7B, lane 1). In the absence of ubiquitination, Siz2-(Smt3)n was capable of being labeled with 32P-Smt3-aR (Fig. 7B, lane 9). However, increasing ubiquitination of the purified Siz2-(Smt3)n led to the inhibition of further SUMO chain elongation using 32P-Smt3-aR (Fig. 7B, lanes 10–13). We conclude that ubiquitination of poly-SUMO chains inhibits further growth of the SUMO chain.
FIGURE 7.
Ubiquitination of poly-SUMO chains blocks further sumoylation. A, radiolabeled Siz2-(Smt3*)n was formed in a 45-min sumoylation reaction containing 1 μm His6-Smt3 and 32P-Smt3. The products were centrifuged at 113,000 × g, the pellet was washed with reaction buffer lacking ATP, pelleted again, and resuspended in 20 mm HEPES (pH 7.5), 300 mm NaCl, 0.02% Nonidet P-40, 10% glycerol, 1 mm EDTA, 1 mm dithiothreitol, and 0.1 mm phenylmethylsulfonyl fluoride. The indicated fractions from this regimen were analyzed by SDS-PAGE and autoradiography. Each lane represents one-tenth of the original reaction. B, unlabeled Siz2-(Smt3)n was prepared by centrifugation as above from a scaled-up reaction. Portions of the purified material were then subjected to 30-min ubiquitination reactions containing 10 nm Uba1, 100 nm Ubc5, 50 nm unlabeled Ub, and 350,000 cpm 32P-Ub in the presence of the indicated concentrations of Slx5–Slx8 to monitor ubiquitination (lanes 1–8). Alternatively, portions of Siz2-(Smt3)n were first subjected to a 30-min ubiquitination reaction containing 10 nm Uba1, 100 nm Ubc5, and 2 μm Ub in the presence of the indicated concentrations of Slx5–Slx8. These products were then subjected to a 30-min sumoylation reaction containing 2 nm Aos1/Uba2, 30 nm Ubc9, 1 μm Smt3-aR, and 350,000 cpm 32P-Smt3-aR (lanes 9–14). Following this incubation, the reaction products were analyzed by SDS-PAGE and autoradiography. In lanes 1 and 8 unmodified Siz2 was used in place of Siz2-(Smt3)n. C, purified Siz2-(Smt3)n or mock (–) was sumoylated for 30 min with the indicated concentration of His6-Smt3-aR in the presence of 10 nm Aos1/Uba2 and 100 nm Ubc9. The products were then ubiquitinated for 20 min with 250,000 cpm 32P-Ub and 5 nm Uba1, 50 nm Ubc4, 50 nm Slx5–Slx8, plus 20 nm unlabeled Ub (lanes 1–7). As control, the sumoylation step was monitored by incubating Siz2-(Smt3)n or mock (–) under the same conditions but with 250,000 cpm 32P-Smt3-aR and the indicated concentrations of unlabeled His6-Smt3-aR (lanes 8–12).
The above experiment suffers from the possibility that ubiquitination inhibits Siz2 activity. More importantly, the experiment does not rule out the possibility that Ub is also added to internal sites within the SUMO chain. To test this, we performed the reverse experiment: purified Siz2-(Smt3)n was first incubated under sumoylation conditions with increasing concentrations of unlabeled Smt3-aR, and the products were then ubiquitinated with 32P-Ub. As shown in Fig. 7C (lanes 1–7), increasing concentrations of Smt3-aR specifically inhibited ubiquitination of Siz2(Smt3)n: background signal resulting from auto-ubiquitination of Slx5–Slx8 and other factors was unaffected by Smt3-aR (Fig. 7C and supplemental Fig. S2B). In control reactions Siz2-(Smt3)n efficiently incorporated 32P-Smt3-aR (lanes 8–12). Thus, ubiquitination of poly-SUMO chains was dependent on a SUMO chain whose terminal SUMO moiety could be modified.
Highly Sumoylated Proteins Accumulate in Specific Strain Backgrounds—The requirement for the Slx5–Slx8 Ub ligase in sgs1Δ strains may be related to the requirement for Ulp1 in srs2Δ strains (12). For example, both factors may act to suppress the accumulation of sumoylated proteins. To test whether these DNA repair mutants displayed altered patterns of sumoylated proteins, total denatured protein was isolated from several strains (sgs1Δ, srs2Δ, and slx5Δ) and analyzed by immunoblot. As shown in Fig. 8A, sgs1Δ and srs2Δ cells contained an excess of hyper-sumoylated proteins that was similar to that found in slx5Δ strains. In contrast, the post-replication repair mutant rad6Δ did not accumulate these proteins (data not shown). The abundance of hyper-sumoylated proteins in these two recombinational repair mutants suggests that Slx5–Slx8 or Ulp1, respectively, are needed to suppress the accumulation of toxic levels of these modified proteins.
FIGURE 8.
Poly-sumoylated proteins accumulate in the absence of SGS1, SRS2, or proteasome function. A, total yeast extracts were prepared by the trichloroacetic acid method and resolved by 10% SDS-PAGE prior to immunoblotting and staining with antibodies against Smt3. The strains are JMY2472 (WT), NJY2402 (slx5Δ), NJY506 (slx5Δ), JMY1460 (sgs1Δ) JMY2472 (WT), JMY286 (srs2Δ), and NJY379 (srs2Δ). B, shown is the blot in A stained with Ponceau Red to reveal protein loading. C, extracts from the following strains were prepared and immunoblotted with antibodies against Smt3 as above: W303–1 a (WT), SIY778 (slx8Δ), and JMY1699 (slx5Δ) grown at 30 °C. Strain JDY194 (pre1-1 pre2-2) was grown at 25 °C (lanes 4 and 7), at 30 °C for 2 or 4 h (lanes 5 and 6), or at 37 °C for 2 or 4 h (lanes 8 and 9).
To test the idea that Ub-mediated proteolysis controls the abundance of sumoylated proteins, total denatured protein was isolated from a temperature-sensitive 20 S proteasome mutant strain (pre1-1 pre2-2) and immunoblotted with antibody against Smt3 (Fig. 8C). At the permissive temperature, the pre1 pre2 mutant had wt levels of sumoylated proteins (Fig. 8C, lanes 4 and 7), but after increasing time at the semi-permissive temperature (30 °C) or the non-permissive temperature (37 °C), there was an increased abundance of sumoylated proteins in the 100- to 250-kDa range similar to that seen in the slx5Δ and slx8Δ strains (Fig. 8C, lanes 6, 8, and 9). As observed using other conditions to limit Ub-mediated proteolysis (21), evidence of long poly-SUMO chains was visible in the stacking gel following 4 h at 37 °C. Thus, the hyper-sumoylation observed in slx5Δ and slx8Δ cells may result from the failure of poly-sumoylated proteins to be degraded by the proteasome.
DISCUSSION
The main finding of this study is that the Slx5–Slx8 Ub ligase is activated by poly-SUMO conjugates. Activation by this substrate is consistent with the preferential interaction between Slx5–Slx8 and poly-SUMO conjugates (21), and with the presence of multiple SUMO-binding domains in Slx5–Slx8. We suggest that these multiple SUMO-binding domains are needed to interact with polymerized SUMO moieties. Indeed, the SUMO-binding domain located in the N terminus of Slx5 and the one in Slx8 have been shown to contain well conserved amino acid sequence motifs that match the SUMO interacting motif consensus (21, 24). We have previously reported that Slx5 and Slx8 can promote SUMO chain formation in the presence of Aos1/Uba2 and Ubc9, but their RING domains were not required for this activity (28). The identification of multiple SUMO-binding domains may explain this in vitro result. For example, the Slx5–Slx8 heterodimer, or Slx5 alone, may bind multiple SUMO moieties in a way that would increase their local concentration and their conjugation efficiency by Ubc9. Further, the multiple SUMO-binding domains of Slx5 may confine adjacent SUMO polypeptides to the same orientation found in a SUMO chain. Consistent with this idea, Slx5 is significantly more active in promoting high molecular weight SUMO chains than is Slx8 (28).
Siz2 was chosen as a test substrate because it is known to be sumoylated in vivo (39) and because auto-sumoylated Siz2 was expected to carry an assortment of SUMO modifications. Based on the fact that most in vivo sumoylation is monomeric, we expected that a single SUMO polypeptide would be sufficient to bind the ligase to the target so that it could be ubiquitinated directly. Instead, we found that the Ub ligase not only prefers the poly-sumoylated form of Siz2, but that it modifies its N-terminal SUMO moiety. This conclusion is supported by the lack of ubiquitination of Siz2 that has been modified by Smt3-aR, which is a good model of a mono-sumoylated substrate. An alternative explanation for this result is that Smt3-aR is not recognized efficiently by Slx5–Slx8 or that Siz2 is a poor receptor for Ub. However, Smt3-aR can partially complement the smt3Δ null mutation in vivo (20), and Siz2 must contain at least one site for ubiquitination, because it was ubiquitinated independent of its sumoylation status. Although in vivo evidence for mixed SUMO-Ub chains has not yet been obtained, it may not be unexpected given that the spectrum of Ub modifications is extremely diverse, including Ub chains with mixed linkages (40).
Several results are consistent with the idea that the Slx5–Slx8 Ub ligase specifically targets three N-terminal lysine residues of SUMO. First, the inhibition of ubiquitination by capping SUMO chains with Smt3-aR demonstrates that there is little or no internal ubiquitination of the SUMO chain. For ubiquitination to occur at an internal SUMO moiety, a poly-SUMO chain with Lys-15 linkages, for example, would have to be ubiquitinated as close as four residues away from the inter-SUMO linkage (at Lys-11 or Lys-19), which is likely to be sterically excluded. Second, if a SUMO chain were saturated with Ub at these sites, then we should have observed a unique intermediate following Ulp1 cleavage, (Smt3-Ub)2, which we have never seen. Lastly, SUMO chain elongation is inhibited by ubiquitination, suggesting that Ub is competing for these N-terminal residues.
The recent discovery of similar SUMO-targeted Ub ligases in fission yeast suggests that mechanisms to eliminate sumoylated proteins are highly conserved and functionally important (21, 32, 33). In the case of Slx5–Slx8, it remains to be determined whether its targets are limited to poly-SUMO conjugates. Moreover, it will be interesting to determine whether these conjugates are biologically active or just toxic artifacts. In this regard we find it interesting that yeast Smt3 can tolerate the mutation of many of its lysine residues to arginine, individually and collectively, but mutating all lysines (Smt3-aR) confers a sickness to otherwise wt cells (20). Although it is reasonable to suspect that Smt3-aR is structurally defective due to its many mutations, it remains possible that some of its defect is due to an inability to form poly-SUMO chains. Thus, it will be interesting to determine whether the accumulation of hyper-sumoylated proteins that we observed in the absence of SGS1 or SRS2 represents a functional response to their DNA repair defects. And regardless of whether these hyper-sumoylated proteins are functionally important or simply toxic intermediates, a major unanswered question is why Slx5–Slx8 is specifically needed in the absence of SGS1. Unlike Slx5–Slx8, which represents a module in the DNA integrity network (41), loss of other SUMO regulators such as Ulp2, Siz1, or Siz2 does not result in lethality in the absence of SGS1.3 A potential explanation for this specificity is that Slx5–Slx8 ubiquitinates poly-SUMO chains that have a particular length or type of linkage. Alternatively, Slx5–Slx8 may show specificity due to its localization to DNA replication foci (26).
Presented in Fig. 9 is a working model to accommodate the above findings, including the accumulation of poly-sumoylated proteins and their destruction by Slx5–Slx8. In this model replicative DNA damage, which is exacerbated in the absence of the Sgs1 DNA helicase, leads to the “activation” of one or more recombinational repair proteins by sumoylation. Although poly-sumoylation is not a major feature of SUMO function (20) it may be a transient step following recombinational repair, perhaps to eliminate or further modify the repair protein. In support of this possibility we have found that smt3-aR cells are sensitive to DNA damage by methyl methane sulfonate.4 In the absence of the Slx5–Slx8 Ub ligase, the poly-sumoylated protein remains active in stimulating recombination. In wt cells, we propose that the Slx5–Slx8 Ub ligase promotes Ubc4/5-dependent mono-ubiquitination of lysines 11, 15, or 19 on the terminal SUMO residue. Future studies will be needed to address the question of whether extension of the Ub chain is necessary for proteasomal targeting and whether the redundant Uls1 E3 ligase plays a role in that reaction (21). However, until a biologically relevant substrate for the Slx5–Slx8 Ub ligase is identified, we cannot rule out the possibility that the ligase ubiquitinates its sumoylated substrate directly.
FIGURE 9.
Model for the generation of poly-sumoylated proteins and their Ub-dependent proteasomal degradation. In response to replicative DNA damage, a DNA repair protein is activated by mono-sumoylation (S). This damage is expected to be exacerbated in the absence of the Sgs1 DNA helicase. Poly-SUMO chain formation may alter the activity of the repair protein or act as a first step in its destruction. The Slx5–Slx8 Ub ligase, together with Ubc4 or Ubc5, modifies the SUMO chain at its growing end to limit its length or to direct it to the proteasome following Ub chain elongation via an unknown mechanism.
Supplementary Material
Acknowledgments
We thank Mark Hochstrasser, Erica Johnson, Kiran Madura, and Drew Vershon for providing strains, plasmids, antibodies, and other reagents. We also thank Kiran Madura for critical reading of the manuscript, and Jackie Fung and Suzanne Shanower for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant GM071268. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
Footnotes
The abbreviations used are: SUMO, small ubiquitin-like modifier; HU, hydroxyurea; SIM, SUMO interacting motif; GST, glutathione S-transferase; wt, wild type; Ulp1UD, Ulp1 Ulp domain; SD, synthetic complete medium; E1, ubiquitin-activating enzyme; E2, ubiquitin carrier protein; E3, ubiquitin-protein isopeptide ligase; aa, amino acid(s); aR, protein in which all lysine residues are changed to arginine; Ub,ubiquitin.
J. R. Mullen and S. J. Brill, unpublished results.
J. R. Mullen and S. J. Brill, unpublished result.
References
- 1.Johnson, E. S. (2004) Annu. Rev. Biochem. 73 355–382 [DOI] [PubMed] [Google Scholar]
- 2.Johnson, E. S., Schwienhorst, I., Dohmen, R. J., and Blobel, G. (1997) EMBO J. 16 5509–5519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson, E. S., and Gupta, A. A. (2001) Cell 106 735–744 [DOI] [PubMed] [Google Scholar]
- 4.Ardley, H. C., and Robinson, P. A. (2005) Essays Biochem. 41 15–30 [DOI] [PubMed] [Google Scholar]
- 5.Lorick, K. L., Jensen, J. P., Fang, S., Ong, A. M., Hatakeyama, S., and Weissman, A. M. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 11364–11369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hochstrasser, M. (2001) Cell 107 5–8 [DOI] [PubMed] [Google Scholar]
- 7.Li, S. J., and Hochstrasser, M. (2000) Mol. Cell Biol. 20 2367–2377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li, S. J., and Hochstrasser, M. (1999) Nature 398 246–251 [DOI] [PubMed] [Google Scholar]
- 9.Zhao, X., and Blobel, G. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 4777–4782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maeda, D., Seki, M., Onoda, F., Branzei, D., Kawabe, Y., and Enomoto, T. (2004) DNA Repair (Amst.) 3 335–341 [DOI] [PubMed] [Google Scholar]
- 11.Hoege, C., Pfander, B., Moldovan, G. L., Pyrowolakis, G., and Jentsch, S. (2002) Nature 419 135–141 [DOI] [PubMed] [Google Scholar]
- 12.Soustelle, C., Vernis, L., Freon, K., Reynaud-Angelin, A., Chanet, R., Fabre, F., and Heude, M. (2004) Mol. Cell Biol. 24 5130–5143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Branzei, D., Sollier, J., Liberi, G., Zhao, X., Maeda, D., Seki, M., Enomoto, T., Ohta, K., and Foiani, M. (2006) Cell 127 509–522 [DOI] [PubMed] [Google Scholar]
- 14.Papouli, E., Chen, S., Davies, A. A., Huttner, D., Krejci, L., Sung, P., and Ulrich, H. D. (2005) Mol. Cell 19 123–133 [DOI] [PubMed] [Google Scholar]
- 15.Pfander, B., Moldovan, G. L., Sacher, M., Hoege, C., and Jentsch, S. (2005) Nature 436 428–433 [DOI] [PubMed] [Google Scholar]
- 16.Torres-Rosell, J., Sunjevaric, I., De Piccoli, G., Sacher, M., Eckert-Boulet, N., Reid, R., Jentsch, S., Rothstein, R., Aragon, L., and Lisby, M. (2007) Nat. Cell Biol. 9 923–931 [DOI] [PubMed] [Google Scholar]
- 17.Sacher, M., Pfander, B., Hoege, C., and Jentsch, S. (2006) Nat. Cell Biol. 8 1284–1290 [DOI] [PubMed] [Google Scholar]
- 18.Bencsath, K. P., Podgorski, M. S., Pagala, V. R., Slaughter, C. A., and Schulman, B. A. (2002) J. Biol. Chem. 277 47938–47945 [DOI] [PubMed] [Google Scholar]
- 19.Tatham, M. H., Jaffray, E., Vaughan, O. A., Desterro, J. M., Botting, C. H., Naismith, J. H., and Hay, R. T. (2001) J. Biol. Chem. 276 35368–35374 [DOI] [PubMed] [Google Scholar]
- 20.Bylebyl, G. R., Belichenko, I., and Johnson, E. S. (2003) J. Biol. Chem. 278 44113–44120 [DOI] [PubMed] [Google Scholar]
- 21.Uzunova, K., Gottsche, K., Miteva, M., Weisshaar, S. R., Glanemann, C., Schnellhardt, M., Niessen, M., Scheel, H., Hofmann, K., Johnson, E. S., Praefcke, G. J., and Dohmen, R. J. (2007) J. Biol. Chem. 282 34167–34175 [DOI] [PubMed] [Google Scholar]
- 22.Mullen, J. R., Kaliraman, V., Ibrahim, S. S., and Brill, S. J. (2001) Genetics 157 103–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ii, T., Fung, J., Mullen, J. R., and Brill, S. J. (2007) Cell Cycle 6 2800–2809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xie, Y., Kerscher, O., Kroetz, M. B., McConchie, H. F., Sung, P., and Hochstrasser, M. (2007) J. Biol. Chem. 282 34176–34184 [DOI] [PubMed] [Google Scholar]
- 25.Mullen, J. R., Kaliraman, V., and Brill, S. J. (2000) Genetics 154 1101–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burgess, R. C., Rahman, S., Lisby, M., Rothstein, R., and Zhao, X. (2007) Mol. Cell Biol. 27 6153–6162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang, C., Roberts, T. M., Yang, J., Desai, R., and Brown, G. W. (2006) DNA Repair (Amst.) 5 336–346 [DOI] [PubMed] [Google Scholar]
- 28.Ii, T., Mullen, J. R., Slagle, C. E., and Brill, S. J. (2007) DNA Repair (Amst.) 6 1679–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen, X. L., Reindle, A., and Johnson, E. S. (2005) Mol. Cell Biol. 25 4311–4320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang, Z., Jones, G. M., and Prelich, G. (2006) Genetics 172 1499–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uetz, P., Giot, L., Cagney, G., Mansfield, T. A., Judson, R. S., Knight, J. R., Lockshon, D., Narayan, V., Srinivasan, M., Pochart, P., Qureshi-Emili, A., Li, Y., Godwin, B., Conover, D., Kalbfleisch, T., Vijayadamodar, G., Yang, M., Johnston, M., Fields, S., and Rothberg, J. M. (2000) Nature 403 623–627 [DOI] [PubMed] [Google Scholar]
- 32.Sun, H., Leverson, J. D., and Hunter, T. (2007) EMBO J. 18 4102–4112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prudden, J., Pebernard, S., Raffa, G., Slavin, D. A., Perry, J. J., Tainer, J. A., McGowan, C. H., and Boddy, M. N. (2007) EMBO J. 18 4089–4101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kosoy, A., Calonge, T. M., Outwin, E. A., and O'Connell, M. J. (2007) J. Biol. Chem. 282 20388–20394 [DOI] [PubMed] [Google Scholar]
- 35.Rose, M. D., Winston, F., and Hieter, P. (1990) Methods in Yeast Genetics, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- 36.Studier, F. W., Rosenberg, A. H., Dunn, J. J., and Dubendorff, J. W. (1990) Methods Enzymol. 185 60–89 [DOI] [PubMed] [Google Scholar]
- 37.Yang, L., Mullen, J. R., and Brill, S. J. (2006) Nucleic Acids Res. 34 5541–5551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li, S. J., and Hochstrasser, M. (2003) J. Cell Biol. 160 1069–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hannich, J. T., Lewis, A., Kroetz, M. B., Li, S. J., Heide, H., Emili, A., and Hochstrasser, M. (2005) J. Biol. Chem. 280 4102–4110 [DOI] [PubMed] [Google Scholar]
- 40.Kirkpatrick, D. S., Hathaway, N. A., Hanna, J., Elsasser, S., Rush, J., Finley, D., King, R. W., and Gygi, S. P. (2006) Nat. Cell Biol. 8 700–710 [DOI] [PubMed] [Google Scholar]
- 41.Pan, X., Ye, P., Yuan, D. S., Wang, X., Bader, J. S., and Boeke, J. D. (2006) Cell 124 1069–1081 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.