Abstract
Protein Z (PZ) is a multidomain vitamin K-dependent plasma protein that functions as a cofactor to promote the inactivation of factor Xa (fXa) by PZ-dependent protease inhibitor (ZPI) by three orders of magnitude. To understand the mechanism by which PZ improves the reactivity of fXa with ZPI, we expressed wild-type PZ, PZ lacking the γ-carboxyglutamic acid domain (GD-PZ), and a chimeric PZ mutant in which both Gla and EGF-like domains of the molecule were substituted with identical domains of fXa. The ZPI binding and the cofactor function of the PZ derivatives were characterized in both binding and kinetic assays. The binding assay indicated that all PZ derivatives interact with ZPI with a similar dissociation constant (KD) of ∼7 nm. However, the apparent KD for the chimeric PZ-mediated ZPI inhibition of fXa was elevated 6-fold on PC/PS vesicles and its capacity to function as a cofactor to accelerate the ZPI inhibition of fXa was also decreased 6-fold. The cofactor activity of GD-PZ was dramatically impaired; however, the deletion mutant exhibited a normal cofactor function in solution. A chimeric activated protein C mutant containing the Gla domain of fXa was susceptible to inhibition by ZPI in the presence of PZ. These results suggest that: (i) the ZPI interactive site of PZ is located within the C-terminal domain of the cofactor and (ii) a specific interaction between the Gla domains of PZ and fXa contributes ∼6-fold to the acceleration of the ZPI inhibition of fXa on phospholipid membranes.
Protein Z (PZ)2 is a vitamin K-dependent coagulation glyco-protein, which has a genetic organization that is identical to that of factor Xa (fXa) and other similar vitamin K-dependent coagulation factors (1, 2). Thus, PZ has an N-terminal γ-carboxyglutamic acid (Gla) domain that is followed by two epidermal growth factor (EGF)-like domains (light chain homologue) and a C-terminal pseudo catalytic domain (heavy chain homologue) (1, 2). Unlike fXa and other vitamin K-dependent coagulation proteases, two of the catalytic triad residues of the C-terminal domain have not been conserved in the homologous regions of PZ (1, 2). Thus, PZ has no catalytic function, but instead it binds to PZ-dependent protease inhibitor (ZPI) with a high affinity, thereby promoting the ZPI inhibition of fXa in the presence of PC/PS vesicles and Ca2+ by approximately three orders of magnitude (3–5). ZPI is a 72-kDa serpin with a plasma concentration of 2.6–2.9 μg/ml that appears to interact with the active site pocket of fXa by a covalent mechanism similar to that observed with antithrombin and most other inhibitory serpins (4–6). However, unlike antithrombin, which can inhibit all coagulation proteases, ZPI is a specific inhibitor of fXa and factor XIa (4, 7, 8), though recent results have indicated that it may also inhibit factor IXa (9). ZPI may play a critical role in the regulation of the coagulation cascade because its deficiency is associated with venous thromboembolic diseases (10, 11). ZPI by itself is a poor inhibitor of fXa, unless it forms a complex with PZ on membrane phospholipids in the presence of Ca2+ (4). However, the inactivation of factor XIa by ZPI is independent of a cofactor (7, 8).
The mechanism by which PZ functions as a cofactor to accelerate the reactivity of fXa with ZPI on negatively charged membrane surfaces is not known nor is the PZ domain(s) that contains the ZPI interactive site(s) of the cofactor. To investigate the role of the light chain and heavy chain homologues of PZ in interaction with ZPI and to provide further insight into the mechanism of the cofactor function of PZ, we developed an expression/purification system for PZ and expressed a recombinant wild type (rPZ), a deletion mutant lacking its Gla domain (GD-PZ) and a chimeric form of PZ in which the light chain homologue of the molecule has been substituted with the same regions of factor X (exons encoding Gla, hydrophobic stack, EGF1 and EGF2 domains). The cofactor function of the PZ derivatives were characterized in both binding and inhibition assays using recombinant ZPI. The results indicate that the chimeric PZ interacts with ZPI with a normal affinity in the direct binding assay in the absence of PC/PS vesicles; it however, exhibits ∼6-fold weaker affinity for the inhibitor in the fXa inhibition assay on the negatively charged phospholipid vesicles in the presence of Ca2+. The cofactor activity of the chimeric PZ in the ZPI inhibition of fXa was also decreased 6-fold under the same conditions. The cofactor activity of GD-PZ was dramatically impaired on PC/PS vesicles, though the mutant exhibited a normal cofactor activity in solution. Interestingly, an activated protein C chimera containing the Gla domain of fXa reacted with ZPI in the presence, but not in the absence of PZ on PC/PS vesicles in the presence of Ca2+. We conclude that the ZPI interactive site of PZ is located within the C-terminal heavy chain homologue of the cofactor. Furthermore, PZ interacts with the Gla domain of fXa to facilitate the formation of a ternary complex, and the association of the serpin with the protease on the negatively charged membrane surface.
EXPERIMENTAL PROCEDURES
Construction, Mutagenesis, and Expression of Recombinant Proteins—A PZ cDNA containing three extraneous point mutations (obtained from Open Biosystems, Huntsville, AL) was subcloned into the HindIII and XbaI restriction enzyme sites of the pRc/RSV mammalian expression vector (Invitrogen, Carlsbad, CA) and converted to the wild-type form by repairing the mutations by PCR mutagenesis methods. To facilitate the purification of PZ, the sequence of a 12-residue epitope for a Ca2+-dependent monoclonal antibody, HPC4, was also ligated to the 3′-end of the cDNA immediately before the native stop codon of the cofactor. The accuracy of the PZ cDNA in the expression vector was confirmed by sequencing the entire cDNA. The PZ chimera in which the exons encoding the light chain homologue were replaced with the same exons encoding the light chain of fXa (Gla, hydrophobic stack, and both EGF-like domains) (PZ-fX/LC) was prepared by the PCR mutagenesis approach using the same HPC4-tagged PZ cDNA sequence. The expression vectors, which contain a neomycin gene for the G418 resistant selection, were transferred to human embryonic kidney (HEK-293) cells, several G418-resistant clones were selected and examined for PZ expression by an ELISA using the HPC4 monoclonal antibody and a polyclonal anti-PZ antibody obtained from Hematologic Technologies Inc. (Essex Junction, VT). A clone positive for the expression of each PZ derivative was identified, expanded, and 20 liters of cell culture supernatant were collected, concentrated, and purified by immunoaffinity chromatography using the HPC4 antibody linked to Affigel 10 (Bio-Rad) as described (12). The HPC4 eluates were further chromatographed on a Mono Q column using a gradient of 0.1–1 m NaCl as described (13). The recombinant proteins from single preparations were frozen at –80 °C in small aliquots until use. Because of a very poor yield in the mammalian expression system, the Gla-domainless PZ (GD-PZ lacking residues 1–47) was expressed in the InsectSelect system (Invitrogen) using the pIZ/V5-His expression vector transfected to sf21 insect cells according to the manufacturer's instruction. The construction strategy was such that the sequence of His tag in the vector remained downstream of the stop codon for GD-PZ; thus it was not incorporated into the sequence of the mutant cofactor as we described previously for the expression of other proteins in this system (14). The expression, purification, and characterization of recombinant ZPI in HEK-293 cells has been described previously (15). The expression and purification of fXa lacking the EGF1 domain (fXa-desEGF1) or both Gla and EGF1 domains (E2-fXa) has been described (16). The activated protein C mutants in which either the Gla domain or both the Gla and the first EGF-like domains of the protease were replaced with the corresponding regions of fXa (APC-fX/Gla and APC-fX/Gla/EGF1) were expressed in the same expression system as described (17).
Human plasma protein Z (PZ) and fXa were purchased from Hematologic Technologies Inc. (Essex Junction, VT). Phospholipid vesicles containing 80% phosphatidylcholine and 20% phosphatidylserine (PC/PS) were prepared as described (18). The chromogenic substrates, S2765 and Spectrozyme PCa (SpPCa) were purchased from American Diagnostica (Green-wich, CT).
Inhibition Assays—The time course and concentration dependence of fXa inhibition by ZPI was studied both in the absence and presence of PZ derivatives on PC/PS vesicles under pseudo-first-order conditions as described (15). In the absence of PZ, fXa derivatives (1 nm) were incubated with ZPI (100–250 nm) on PC/PS vesicles (20 μm) in 0.1 m NaCl, 0.02 m Tris-HCl, pH 7.5, 0.1% polyethylene glycol 8000 (PEG 8000), 0.1 mg/ml bovine serum albumin (BSA), and 5 mm Ca2+ (TBS/Ca2+) for 30–120 min in 50-μl volumes in 96-well polystyrene plates at room temperature as described (15). In the presence of PZ, the reaction conditions were the same except that fXa was incubated with ZPI (5–40 nm) in complex with a saturating concentration of PZ derivatives (100 nm) for 15 s to 10 min on PC/PS vesicles (20 μm) in the same TBS buffer system. The inactivation reactions were stopped by the addition of 50 μl of S2765 (0.2 mm final) in TBS containing 50 mm EDTA, and the remaining enzyme activity was measured with a Vmax Kinetics Microplate Reader (Molecular Devices, Menlo Park, CA) at 405 nm. The cofactor concentration dependence of the ZPI inhibition of fXa indicated that PZ concentrations are saturating under these conditions. The observed pseudo-first-order (kobs) rate constants were calculated from a first-order rate equation and the second-order rate constants (k2) were calculated from the slope of linear plots of kobs values versus PZ-ZPI complex concentrations as described (15). The apparent dissociation constants of PZ derivatives for interaction with ZPI were estimated from the hyperbolic dependence of kobs values on PZ concentrations in the presence of a fixed concentration of ZPI (5 nm) on PC/PS in TBS/Ca2+.
Interaction with ZPI—The affinity of PZ derivatives for interaction with ZPI was evaluated by an ELISA-based binding assay using 96-well flat microtiter plates coated with a polyclonal antibody (2 μg/ml) to ZPI in TBS containing 1 mm CaCl2 overnight at 4 °C. After washing plates three times with the same TBS buffer containing 0.05% Tween 20, they were incubated with 1% BSA in TBS/Ca2+ for 2 h at room temperature. After the plates were washed three times, they were incubated with ZPI (2 μg/ml) for 1 h followed by washing and incubation with PZ derivatives (0–200 nm in TBS/Ca2+ containing 0.1% BSA) for another 1 h. After the plates were washed again, they were incubated with a goat anti-PZ polyclonal antibody (1 μg/ml) for 1 h. Then, the plates were washed and incubated with rabbit anti-goat IgG (KPL, MD, 1:1000) for 1 h. After washing, the plates were incubated with 2,2′-azino-di(3-ethylbenzthiazo-line-6-sulfonate) (ABTS; KPL, Gaithersburg, MD). Colorimetric analysis was performed by measuring absorbance values at 405 nm as described above. All treatments were performed in duplicate and repeated at least twice. In a variation of this assay, the microtiter plate was directly coated with ZPI (2 μg/ml), and the binding of PZ derivatives to the serpin was evaluated as described above.
Competitive Binding—The competitive effect of the GD-PZ mutant on fXa inhibition by ZPI in the presence of wild-type PZ was studied using the same discontinuous assay method described above. In this case, the inhibition of fXa (0.5 nm) by ZPI (5 nm) in the presence of pPZ (10 nm) was monitored as a function of increasing concentrations of GD-PZ (0–1000 nm) in TBS/Ca2+ on PC/PS vesicles (20 μm). Following 2–5 min incubation at room temperature, 50 μl of S2765 was added to each well, and the remaining activity of fXa was measured as described above. Under these experimental conditions, ∼80% of the enzyme activity in the absence of the competitor was inhibited. The apparent dissociation constant was determined from the non-linear regression analysis of the saturable GD-PZ concentration dependence for recovery of the enzyme activity monitored by the hydrolysis of the chromogenic substrate at 405 nm as described above.
RESULTS AND DISCUSSION
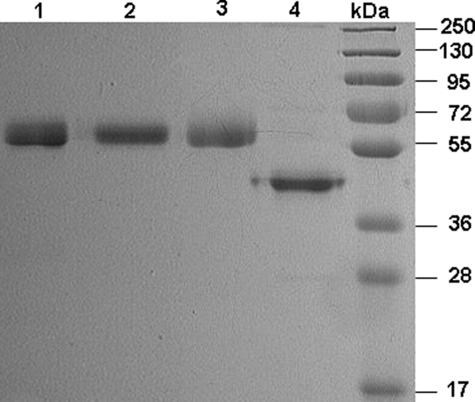
Expression and Purification of Recombinant Proteins—Recombinant PZ derivatives were expressed in HEK-293 (rPZ and PZ-fX/LC chimera) or insect cells (GD-PZ) in fusion with the 12-residue epitope for the Ca2+-dependent monoclonal HPC4 antibody to facilitate their purification by immunoaffinity chromatography as described under “Experimental Procedures.” Thus, following passage of the supernatants through the immobilized HPC4 antibody, the substitution of Ca2+ with EDTA in the wash buffer was sufficient to elute the recombinant proteins from the antibody column as described (15). The HPC4 eluates for rPZ and PZ-fX/LC chimera were further purified by an anion exchange chromatography employing a Mono Q column as described (13). SDS-PAGE analysis under non-reducing conditions suggested that the expressed proteins have been purified to homogeneity and both rPZ and PZ-fX/LC chimera migrate as single bands with expected molecular masses of ∼62 kDa (Fig. 1, lanes 2 and 3), the same value observed for the plasma-derived PZ (lane 1). The GD-PZ mutant expressed in the insect cells migrated with an apparent molecular mass of ∼45 kDa under the same conditions (Fig. 1, lane 4).
FIGURE 1.
SDS-PAGE analysis of PZ derivatives under non-reducing conditions. Lane 1, plasma-derived PZ (pPZ); lane 2, recombinant PZ (rPZ); lane 3, PZ-fX/LC; lane 4, GD-PZ; lane 6, molecular mass standards in kDa.
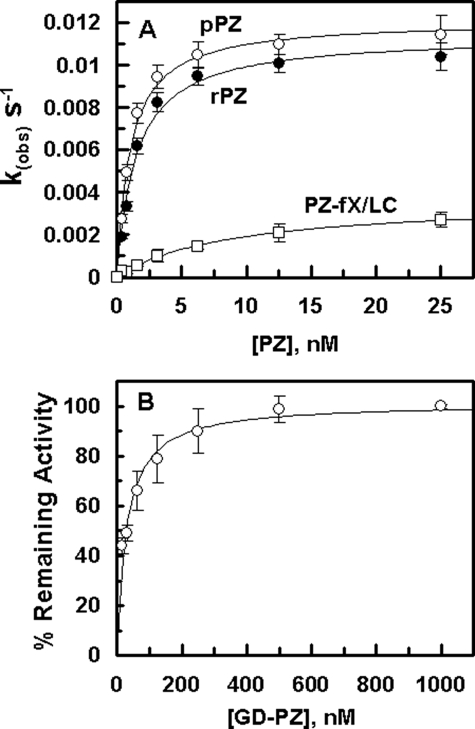
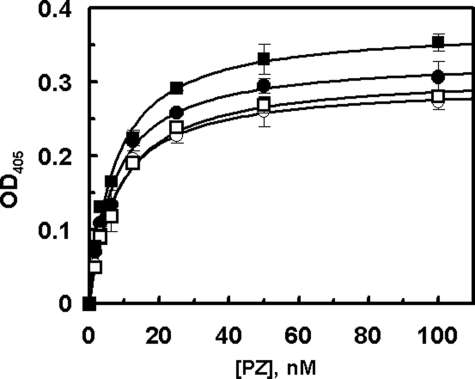
Interaction with ZPI—The PZ concentration dependence of the ZPI inhibition of fXa on PC/PS vesicles suggested that both pPZ and rPZ exhibit similar cofactor activity, thus promoting the ZPI inhibition of fXa with Kd(app) values of 1.1 nm and 1.4 nm, respectively, on PC/PS vesicles in the presence of calcium (Fig. 2A). Relative to rPZ, the corresponding value for the PZ-fX/LC chimera (8.7 nm) was elevated ∼6-fold (Fig. 2A). In contrast to the chimera, the cofactor activity of GD-PZ was dramatically impaired (see below) and thus the data with this mutant could not be included in this figure. Nevertheless, the GD-PZ mutant functioned as an effective competitor of fXa inhibition by the PZ-ZPI complex on PC/PS vesicles with a Kd(app) value of 29 ± 5 nm (Fig. 2B), suggesting that the ZPI interactive site is located within GD-PZ. The interaction of PZ derivatives with ZPI was also evaluated by an ELISA binding assay in the absence of PC/PS vesicles, but the presence of Ca2+. Interestingly, all PZ derivatives (pPZ, rPZ, PZ-fX/LC, and GD-PZ) interacted with ZPI with similar dissociation constants of 7–8 nm in this assay (Fig. 3). The results of the binding assays were essentially identical whether or not the interactions were analyzed with ZPI being directly bound to the ELISA plate or the serpin was first captured on the plate by a polyclonal antibody to ZPI. These results suggest that all recombinant PZ derivatives have been folded properly and that the ZPI interactive site of PZ is located within the C-terminal heavy chain homologue of the cofactor. We also conducted the binding assays in the presence of 1 mm EDTA. In this case, however, no interaction for ZPI was detected with any of the PZ derivatives. Whether the lack of binding was due to a Ca2+ requirement for the PZ-ZPI interaction was not investigated.
FIGURE 2.
Dependence of kobs values on the concentration of PZ derivatives in ZPI inhibition of fXa and the competitive effect of GD-PZ on fXa inhibition by the PZ-ZPI complex. A, the kobs values for the ZPI (5 nm) inhibition of fXa (0.5 nm) in the presence of increasing concentrations of pPZ (○), rPZ (•), and PZ-fX/LC chimera (□) were determined on PC/PS vesicles (20 μm) in TBS/Ca2+ at room temperature by an amidolytic activity assay described under “Experimental Procedures.” The non-linear regression analysis of data yielded Kd(app) values of 1.1 ± 0.1 nm for pPZ, 1.5 ± 0.2 nm for rPZ, and 8.7 ± 0.9 nm for PZ-fX/LC. B, the competitive effect of GD-PZ on inhibition of fXa (0.5 nm) was monitored in the presence of ZPI (5 nm) and pPZ (10 nm) on PC/PS vesicles (20 μm) in TBS/Ca2+ at room temperature by an amidolytic activity assay. The non-linear regression analysis of data yielded Kd(app) values of 29 ± 5 nm for the GD-PZ inhibition of the PZ cofactor activity. Data are derived from averages of 2–3 independent measurements from single batches of protein preparations.
FIGURE 3.
The binding of PZ derivatives to ZPI. An ELISA-based binding assay using a polyclonal anti-ZPI (2 μg/ml) as the capture antibody was used to evaluate the affinity of PZ derivatives for interaction with the serpin as described under “Experimental Procedures.” The symbols are: pPZ (○), rPZ (•), PZ-fX/LC chimera (□), and GD-PZ (▪). The non-linear regression analysis of data according to a hyperbolic equation yielded KD values of 7.5 ± 0.5 nm for pPZ, 7.1 ± 0.1 nm for rPZ, 8.4 ± 0.1 nm for PZ-fX/LC, and 7.1 ± 0.2 nm for GD-PZ. Data are derived from averages of at least 2–3 independent measurements.
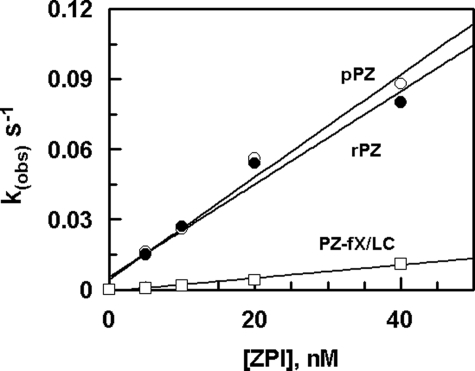
Cofactor Effects of PZ Derivatives in the ZPI Inhibition of fXa—The second-order association rate constants for the PZ-mediated ZPI inhibition of fXa were determined from the slope of the linear plots of kobs values in the presence of saturating concentrations of PZ derivatives (Fig. 4), which are presented in Table 1. The second-order association rate constant for the ZPI inhibition of fXa ((2.1 ± 0.2) × 106) in the presence of the plasma-derived PZ was essentially similar to the same value observed in the presence of rPZ ((1.8 ± 0.2) × 106), suggesting that both PZ and rPZ promote the ZPI inhibition of fXa to a similar extent of ∼2000-fold. Relative to rPZ, the cofactor activity of the PZ-fX/LC chimera was decreased 6-fold, which correlates well with the 6-fold elevated Kd(app) observed for the mutant in the inhibition assay presented in Fig. 2. The basis for the lower cofactor activity of the PZ chimera appears to be due to the loss of an interaction between the Gla domains of fXa and PZ on PC/PS vesicles, because an fXa chimera containing the Gla domain of activated protein C (APC) also reacted with ZPI with ∼3-fold lower k2 value (Table 1). Further support for the existence of a specific interaction between the Gla domains of PZ and fXa was provided by the observation that the APC chimeras containing either the Gla or both the Gla and EGF1 domains of fXa reacted with ZPI in the presence of PZ on PC/PS vesicles (Table 1). In the absence of PZ no reactivity for the APC chimeras was detected under the same experimental conditions. Wild-type APC did not react with ZPI in either the absence or presence of PZ on PC/PS vesicles. These results suggest that the C-terminal heavy chain homologue of PZ contains the interactive-site for ZPI with the N-terminal Gla domain of the cofactor also interacting with fXa, thereby contributing to optimal ZPI inhibition of fXa on the negatively charged phospholipid membrane. In agreement with this hypothesis, the PZ-mediated ZPI inhibition of the fXa mutant lacking the first EGF1 domain exhibited an ∼4-fold decreased k2 value, as this result has also been reported previously (15). This impairment in the interaction is not likely due to the loss of a specific interaction of the EGF1 domain of fXa with PZ because both wild-type fXa and an fXa mutant lacking both the Gla and EGF1 domains (E2-fXa) exhibited similar reactivity with ZPI in complex with either PZ or GD-PZ in the absence of PC/PS vesicles in TBS/Ca2+ (data not presented). Furthermore, similar k2 values were obtained for the reaction of fXa with both wild-type PZ (5.0 ± 0.5 × 103 m–1 s–1) and GD-PZ (6.2 ± 0.1 × 103 m–1 s–1) in solution, suggesting an ∼6-fold cofactor effect for GD-PZ in the ZPI inhibition reactions. These results indicate that the assembly of both PZ and fXa on PC/PS vesicles is essential for the effective ZPI inhibition of fXa. However, analysis of results with Gla domain chimeric proteases suggest that most of the cofactor effect of the PZ Gla domain can be recapitulated by Gla domains of other vitamin K-dependent coagulation proteins. A specific interaction between the Gla domains of PZ and fXa makes a modest 5–6-fold contribution to the acceleration of the reaction.
FIGURE 4.
Concentration dependence of the ZPI inhibition of fXa in the presence of PZ derivatives on PC/PS vesicles. The kobs values for the ZPI inhibition of fXa (0.5 nm) in the presence of PZ derivatives: pPZ (○), rPZ (•), and PZ-fX/LC chimera (□) (100 nm each) were determined on PC/PS vesicles (20 μm) in TBS/Ca2+ at room temperature by an amidolytic activity assay described under “Experimental Procedures.” The k2 values were calculated from the slopes of the straight lines and presented in Table 1. Data are derived from averages of at least 2–3 independent measurements.
TABLE 1.
Second-order rate (k2) constants for the ZPI inhibition of fXa derivatives in the absence and presence of PZ derivatives on PC/PS vesicles
The second-order rate constants (in m–1s–1) in the absence of PZ were determined from the remaining activities of fXa derivatives (1 nm each) after incubation with ZPI (100–250 nm) for 30–120 min in TBS/Ca2+ at room temperature by an amidolytic activity assay described under “Experimental Procedures.” The values in the presence of PZ were determined by the same methods except that the activation of fXa derivatives (0.5 nm) by ZPI (5–40 nm) was monitored in the presence of PZ derivatives (100 nm each) for 15 sec to 10 min under the same experimental conditions. The k2 values for the inhibition of APC derivatives were determined from the remaining activities after the incubation of proteases with 100 nm ZPI with 100 nm PZ for 166 min. All values are averages of at least three independent measurements ± S.D.
| Protease | -Cofactor | +rPZ | +PZ-fX/LC |
|---|---|---|---|
| fXa | (8.8 ± 0.4) × 102 | (1.8 ± 0.2) × 106 | (3.0 ± 0.2) × 105 |
| fXa-APC/Gla | (8.6 ± 0.8) × 102 | (6.5 ± 0.3) × 105 | —a |
| fXa-desEGF1 | (7.9 ± 0.2) × 102 | (4.6 ± 0.4) × 105 | — |
| APC-fX/Glab | NDc | (1.2 ± 0.3) × 102 | — |
| APC-fX/Gla/EGF1d | ND | (0.9 ± 0.1) × 102 | — |
(—), not determined.
APC-fX/Gla, activated protein C mutant in which the Gla domain has been replaced with the corresponding domain of factor X.
ND, no decline in the amidolytic activity of APC derivatives was detected in the absence of PZ under these conditions.
APC-fX/Gla/EGF1, activated protein C mutant in which both the Gla and EGF1 domains have been replaced with the corresponding domains of factor X.
Noting that the reactivity of fXa with ZPI was minimally affected by PC/PS vesicles in the absence of PZ, the 6-fold cofactor effect of GD-PZ in the ZPI inhibition of the protease may thus be attributed to a conformational change in the reactive center loop of the serpin. Alternatively, the cofactor function of PZ may involve structural changes in a secondary binding site of ZPI, thus optimizing its interaction with an exosite of fXa, as we have previously identified Arg-143 as a critical residue on the autolysis loop of fXa which interacts with an unidentified site of ZPI (15), by a mechanism similar to that observed with pentasaccharide activation of antithrombin (19). However, further studies will be required to validate these hypotheses.
Previous fluorescence resonance energy transfer studies have indicated that the binding of the Gla domain of fXa to PC/PS vesicles positions the active site of the protease some 60–70 Å above the membrane surface (20). Thus, it is possible that a key cofactor role for PZ may involve maintaining the reactive center loop of ZPI at an appropriate height above the same membrane surface on which fXa is bound. In the context of this model, the first EGF domain of ZPI could play a spacer function to properly accommodate ZPI into the active site pocket of the membrane-bound enzyme without making any specific interaction with fXa in the ternary complex. A similar role for the EGF1 domain of fXa has been postulated in the prothrombinase complex (14). Thus, the loss of the spacer function of EGF1 most likely accounts for ∼4-fold lower second-order rate constant observed for the PZ-mediated ZPI inhibition of fXa-desEGF1 on PC/PS vesicles. Further support for lack of an interactive site for EGF1 of PZ is provided by the observation that all fXa derivatives including E2-fXa lacking both the Gla and EGF1 domains reacted with similar k2 values with PZ and GD-PZ in solution in the absence of a membrane surface.
In summary, we have shown in this study that the ZPI interactive site of PZ is located within the C-terminal heavy chain homologue of the cofactor. PZ binds to ZPI a KD of ∼7 nm in the direct binding assay and a KD(app) of ∼1 nm on PC/PS vesicles in the inhibition assay. The Gla domain of PZ also makes a specific interaction with the Gla domain of fXa on the negatively charged membrane surface. The Gla-Gla interaction contributes ∼6-fold to the cofactor effect of PZ in the fXa inhibition reaction. A further ∼6-fold cofactor effect of PZ appears to be mediated through the PZ altering the conformation of ZPI, and the remaining cofactor activity of PZ in the promotion of fXa inhibition can be attributed to a Gla and Ca2+-dependent assembly of fXa and PZ-ZPI complex on PC/PS vesicles, which can be recapitulated by the Gla domains of other vitamin K-dependent coagulation proteins.
Acknowledgments
We thank Audrey Rezaie for proofreading the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants HL 62565 and HL 68571 (to A. R. R.) from the NHLBI. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement”in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: PZ, protein Z; pPZ, human plasma-derived PZ; rPZ, recombinant PZ; Gla, γ-carboxyglutamic acid; EGF, epidermal growth factor; GD-PZ, Gla-domainless PZ; ZPI, protein Z-dependent protease inhibitor; fXa, activated factor X; APC, activated protein C; BSA, bovine serum albumin; ELISA, enzyme-linked immunoassay.
References
- 1.Ichinose, A., Takeya, H., Espling, E., Iwanaga, S., Kisiel, W., and Davie, E. W. (1990) Biochem. Biophys. Res. Commun. 172 1139–1144 [DOI] [PubMed] [Google Scholar]
- 2.Sejima, H., Hayashi, T., Deyashiki, Y., Nishioka, J., and Suzuki, K. (1990) Biochem. Biophys. Res. Commun. 171 661–668 [DOI] [PubMed] [Google Scholar]
- 3.Broze, G. J., Jr., and Miletich, J. P. (1984) J. Clin. Investig. 73 933–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Han, X., Fiehler, R., and Broze, G. J., Jr. (2000) Blood 96 3049–3055 [PubMed] [Google Scholar]
- 5.Han, X., Fiehler, R., and Broze, G. J., Jr. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 9250–9255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Han, X., Huang, Z.-F., Fiehler, R., and Broze, G. J., Jr. (1999) Biochemistry 38 11073–11078 [DOI] [PubMed] [Google Scholar]
- 7.Tabatabai, A., Fiehler, R., and Broze, G. J., Jr. (2001) Thromb. Haemostas. 85 655–660 [PubMed] [Google Scholar]
- 8.Rezaie, A. R., Sun, M. F., and Gailani, D. (2006) Biochemistry 45 9427–9433 [DOI] [PubMed] [Google Scholar]
- 9.Heeb, M. J., Cabral, K. M., and Ruan, L. (2005) J. Biol. Chem. 280 33819–33825 [DOI] [PubMed] [Google Scholar]
- 10.Water, N., Tan, T., Ashton, F., O'Grady, A., Day, T., Browett, P., Ockelford, P., and Harper, P. (2005) Br. J. Haematol. 129 561–562 [DOI] [PubMed] [Google Scholar]
- 11.Kemkes-Matthes, B., Nees, M., Kuhnel, G., Matzdorff, A., and Matthes, K. J. (2002) Thromb. Res. 106 183–185 [DOI] [PubMed] [Google Scholar]
- 12.Rezaie, A. R., and Esmon, C. T. (1992) J. Biol. Chem. 267 26104–26109 [PubMed] [Google Scholar]
- 13.Chen, L., Manithody, C., Yang, L., and Rezaie, A. R. (2004) Protein Sci. 13 431–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qureshi, S. H., Yang, L., Yegneswaran, S., and Rezaie, A. R. (2007) Biochem. J. 407 427–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rezaie, A. R., Manithody, C., and Yang, L. (2005) J. Biol. Chem. 280 32722–32728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kittur, F. S., Manithody, C., and Rezaie, A. R. (2004) J. Biol. Chem. 279 24189–24196 [DOI] [PubMed] [Google Scholar]
- 17.Yang, L., and Rezaie, A. R. (2007) Thromb. Haemostas. 97 899–906 [DOI] [PubMed] [Google Scholar]
- 18.Smirnov, M. D., and Esmon, C. T. (1994) J. Biol. Chem. 269 816–819 [PubMed] [Google Scholar]
- 19.Olson, S. T., Björk, I., Sheffer, R., Craig, P. A., Shore, J. D., and Choay, J. (1992) J. Biol. Chem. 267 12528–12538 [PubMed] [Google Scholar]
- 20.Husten, E. J., Esmon, C. T., and Johnson, A. E. (1987) J. Biol. Chem. 262 12953–12962 [PubMed] [Google Scholar]