Abstract
Selenium is an essential dietary element with antioxidant roles in immune regulation, but there is little understanding of how this element acts at the molecular level in host defense and inflammatory disease. Selenium is incorporated into the amino acid selenocysteine (Sec), which in turn is inserted into selenoproteins in a manner dependent on Sec tRNA[Ser]Sec. To investigate the molecular mechanism that links selenium to T cell immunity, we generated mice with selenoprotein-less T cells by cell type-specific ablation of the Sec tRNA[Ser]Sec gene (trsp). Herein, we show that these mutant mice exhibit decreased pools of mature T cells and a defect in T cell-dependent antibody responses. We also demonstrate that selenoprotein deficiency leads to oxidant hyperproduction in T cells and thereby suppresses T cell proliferation in response to T cell receptor stimulation. These findings offer novel insights into immune function of selenium and physiological antioxidants.
Numerous health benefits have been attributed to selenium, including roles in preventing cancer, metabolic, and inflammatory disorders, and in inhibiting viral expression and pathogenesis (1). Overall, little is known about the molecular action of this element in effecting these many health benefits. This is certainly true in the case of the role of selenium in immune function (2–4).
Many of the physiological roles of selenium are thought to be derived from selenium-containing proteins (selenoproteins) (1). Selenium makes its way into protein as selenocysteine (Sec),3 the 21st amino acid in the genetic code, which is decoded by UGA (5, 6). Although numerous studies have elucidated various aspects of the incorporation of Sec into protein (5–7), the complete biosynthesis of this amino acid on its tRNA in eukaryotes and archaea was only determined recently (8).
One of the principal activities of selenoproteins is related to scavenging physiological oxidants and thereby protecting redox-sensitive biological processes (9). Reactive oxygen species (ROS) and other forms of oxidants have long been considered deleterious byproducts of mitochondrial and endosomal metabolic activities. However, their regulatory roles in cellular signaling have begun to be unveiled in the study of growth factor-stimulated cells (10, 11). The effects of selenium on immunity and other aspects of human health may be achieved through selenoprotein action in ROS regulation. In this paper, we describe our study focused on the function of selenoproteins in T cell development and activation and propose a model wherein their antioxidant activity links selenium to T cell immunity.
EXPERIMENTAL PROCEDURES
Generation of ΔtrspT Mice—A C57BL/6 mouse line carrying floxed trsp (trspfl/fl, designated as the control) was described previously (12). A transgenic C57BL/6 line carrying the LckCre transgene (13) was from The Jackson Laboratory. These lines were mated to obtain ΔtrspT mice. The maintenance and care of all mice were in accordance with the National Institutes of Health institutional guidelines.
75S Labeling and Analysis of Selenoproteins—Mice were injected with 50 μCi of 75Se/g and sacrificed after 24 h. T cells were isolated (see below) and lysed; the resulting protein extractions were electrophoresed on gels; and the gels were stained with Coomassie Blue R-250, vacuum-dried, and exposed to a PhosphorImager as described (12, 14).
Isolation and Analysis of T Cells—CD3+ T cells were purified by negative selection using the Mouse T Cell Negative Isolation Kit (Dynal). Cell purity and marker expression (CD3, CD4, CD8, and IL-2Rα) were determined by flow cytometry using fluorescence-conjugated antibodies and a FACScan (BD Biosciences).
T Cell Activation—1 × 106 T cells were placed into 96-well plates in triplicate and stimulated with 10 μg/ml each anti-mouse CD3ε (145-2C11) and anti-CD28 (37.51) antibodies (BD Biosciences). Culture supernatants were assayed for IL-2 production using the Searchlight Assay (Pierce). To measure cell proliferation, cells were pulsed with 1 μCi/well of [3H]thymidine at various time points after T cell receptor (TCR) stimulation, and the amount of [3H]thymidine incorporation was measured in a scintillation counter. Cells were preincubated with NAC (Sigma) in some experiments as described under “Results”.
Immunoblotting and ROS Measurement—ROS were measured by flow cytometry using the oxidation-sensitive dye DCFDA by modifying the previous procedure (15–17). The level of ERK activation was determined by immunoblotting with antibodies specific to total and phosphorylated ERK proteins (Cell Signaling Technology).
Antibody Response in Vivo—Mice were immunized with 100 μg of NP-OVA (Biosearch Technologies). NP-specific immunoglobulin levels were determined in all serum samples (diluted 1:100) by standard enzyme-linked immunosorbent assay on NP-bovine serum albumin-coated plates.
RESULTS
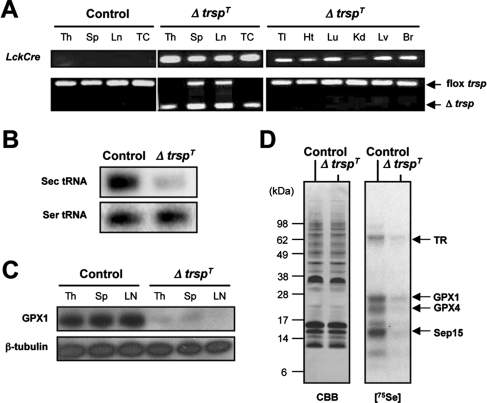
Generation of Mice Lacking Selenoproteins in T Cells—Gene expression profiling shows that specific sets of selenoprotein mRNAs are expressed in T cells from various lymphoid organs (supplemental Fig. S1). To generate mice whose T cells are devoid of selenoproteins, we targeted the removal of the floxed Sec tRNA[Ser]Sec gene (trsp) in mice (12) by Cre recombinase expressed under the control of the Lck promoter (13). The T cell-restricted expression of LckCre was confirmed previously (18). The lymph nodes, splenocytes, and thymocytes, as well as purified T cells of trspfl/fl-LckCre mice (hereafter designated ΔtrspT), exhibited trsp deletion, whereas other non-lymphoid tissues did not (Fig. 1A). Northern blot analysis of Sec RNA[Ser]Sec in purified T cells showed that trsp ablation resulted in the loss of this tRNA (Fig. 1B), whereas Western blot analysis of glutathione peroxidase 1, which is among the selenoproteins highly expressed in T cells (see below), showed that this selenoprotein was virtually absent in T cells of ΔtrspT mice (Fig. 1C). To further examine selenoprotein expression in T cells, control (genotype trspfl/fl) and ΔtrspT mice were labeled with 75Se, the T cells were isolated from thymus, and labeled selenoproteins were examined by gel electrophoresis. The level of 75Se-labeled selenoproteins was substantially reduced in T cells from the ΔtrspT mouse as compared with the corresponding control cells (Fig. 1D). The pattern of labeling is similar to that observed in human T cells wherein the various selenoproteins were identified (14).
FIGURE 1.
Generation and characterization of ΔtrspT mice. A, the genotypes of tissues and T cells obtained from control and ΔtrspT mice are shown. Th, thymus; Sp, spleen; Ln, lymph nodes; TC, T cells; Tl, tail; Ht, heart; Lu, lung; Kd, kidney, Lv, liver; Br, brain; flox, floxed. B, the levels of tRNA[Ser]Sec and tRNASer in thymic T cells isolated from control and ΔtrspT mice as determined by Northern blotting are shown. C, the protein levels of glutathione peroxidase 1 (GPX1) andβ-tubulin in T cells obtained from tissues as indicated of control and ΔtrspT mice as determined by immunoblotting are shown. D, 75Se-labeled selenoproteins are visualized by autoradiography after SDS electrophoresis. The identities of the major labeled selenoproteins (14) are designated to the right. GPX4, glutathione peroxidase 4; TR, thioredoxin reductase; CBB, Coomassie Brilliant Blue.
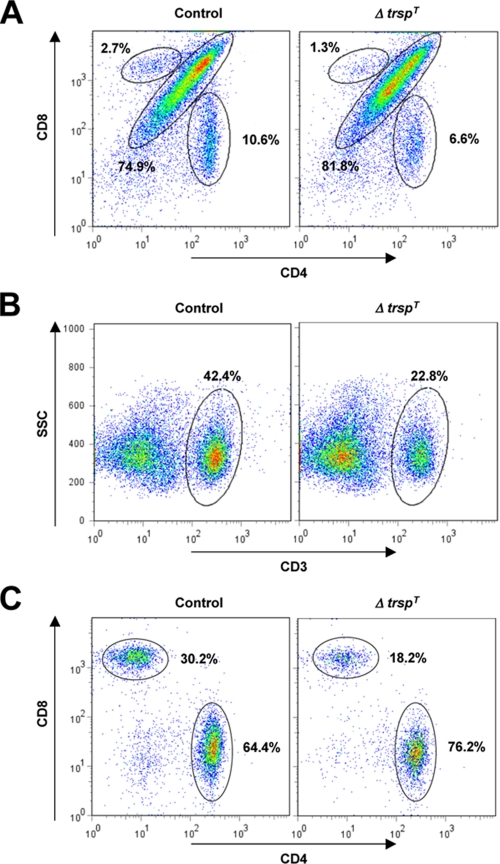
Analysis of Lymphoid Organs and T Cell Differentiation in ΔtrspT Mice—We examined thymus, spleen, and lymph nodes of ΔtrspT mice and found that they exhibited moderate to severe atrophy compared with their control counterparts; the mass and cellularity of the thymus, spleen, and lymph nodes were decreased to varying extents ranging between 50 and 80% of those of control organs. We further investigated the T cell population of ΔtrspT lymphoid organs by flow cytometry. The level of CD4/CD8 double positive cells was slightly higher, and those of single positive CD4+ and CD8+ cells were significantly less in the ΔtrspT thymocytes relative to control thymocytes (Fig. 2A). CD4- and CD8- thymocytes were unaffected by selenoprotein loss. We next examined splenic T cells in control and ΔtrspT mice. The CD3+ population in ΔtrspT splenocytes was about 50% as compared with that in control splenocytes (Fig. 2B), suggesting that trsp deficiency brings about a reduction in the mature and functional pool of T cells in lymphoid tissues. The relative fraction of CD4+ cells among the CD3+ population in ΔtrspT spleen was similar to that in control spleen (Fig. 2C). However, it should be noted that the absolute numbers of splenic CD3+ and CD4+ cells in ΔtrspT mice are lower than in control mice at ratios similar to those of thymic CD4+ cells (Fig. 2A). The CD8+ T cell subpopulation was more substantially reduced in the same ΔtrspT CD3+ splenocytes. The partial loss of functional T cells, particularly CD8+ T cells, that results from T cell-specific selenoprotein deficiency may affect the responsiveness of the immune system to external as well as self antigens.
FIGURE 2.
Analysis of lymphoid organs and T cell differentiation in ΔtrspT mice. T cells isolated from lymphoid organs of control and ΔtrspT mice were analyzed by flow cytometry. The results shown are representative of six different experiments (n = 6). A, CD4 and CD8 expression in thymocytes were analyzed. The subpopulations corresponding to CD4/CD8 DP, as well as CD4+ and CD8+ SP cells, are indicated by circles along with their percent levels. B and C, total splenocytes were gated into CD3+ cells and further analyzed for CD4 and CD8 expression. CD3+ cells CD4/CD8 DP (B) and CD4+ and CD8+ SP cells (C) are indicated as in A. SSC, side scatter.
It is unclear at present how T cell-specific trsp deficiency leads to defects in the development and/or maintenance of mature T cell populations. Nonetheless, the decrease in the CD8+ T cell population in ΔtrspT mice may result in inadequate cytotoxic T cell immunity. Of note, several studies showed that selenium deficiency in diet, a condition that also causes loss or reduction of selenoprotein function, exacerbates viral pathogenesis and impairs antiviral immunity (19, 20).
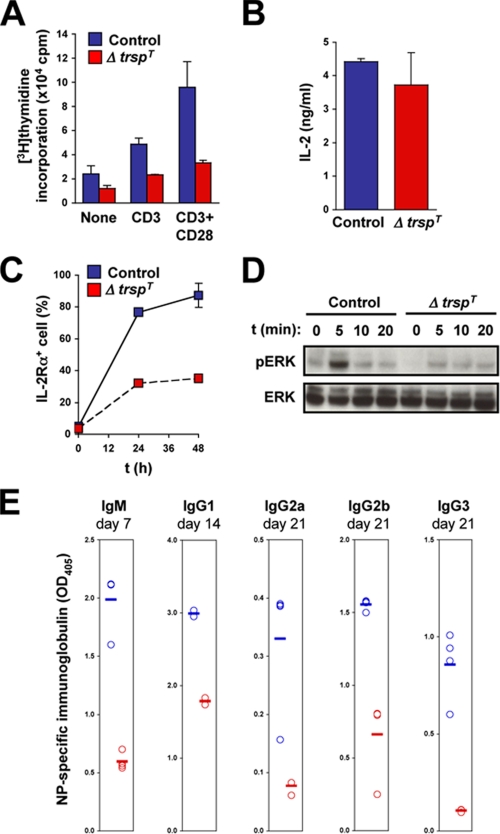
Activation of Selenoprotein-deficient T Cells by T Cell Receptor Signaling—Antigenic stimulation of the TCR-CD3 complex induces proliferation and functional activation of T cells. These responses are greatly potentiated by concomitant activation of costimulatory receptors such as CD28. To test whether TCR-triggered responses require selenoprotein function, T cells were purified from control and ΔtrspT lymph nodes and were cultured in the presence of anti-CD3 and anti-CD28 antibodies, a condition that mimics TCR and costimulatory receptor activation. We first investigated the ability of the T cells to proliferate in response to TCR activation by measuring the rate of [3H]thymidine incorporation. Control T cells that were incubated with anti-CD3 antibody alone or together with anti-CD28 antibody showed greatly enhanced levels of proliferation compared with unstimulated cells. The TCR-induced proliferation of T cells from ΔtrspT lymph nodes was dramatically lower under the same conditions (Fig. 3A). We also examined splenic T cells from control and ΔtrspT mice and obtained similar results (data not shown; also see Fig. 4B). Therefore, selenoprotein expression in T cells appears to be crucial for their ability to proliferate in response to TCR stimulation.
FIGURE 3.
Activation of selenoprotein-deficient T cells by TCR stimulation. A, the level of proliferation of purified lymph node T cells is shown as the amounts of [3H]thymidine incorporation after 60 h of incubation in the absence or presence of anti-CD3/CD28 or anti-CD3 alone. Results are the averages of triplicate readings from six control and ΔtrspT mice, respectively. B and C, the levels of IL-2 production (B) and cell surface expression of IL-2Rα (C) in CD3/CD28-stimulated T cells are shown. D, ERK activation in lymph node T cells stimulated with anti-CD3 and anti-CD28 was analyzed at the time intervals indicated. Both phosphorylated (pERK) and total (ERK) proteins are shown. E, serum levels of major immunoglobulins at the indicated time points were determined by enzyme-linked immunosorbent assay in control and ΔtrspT mice following immunization with NP-OVA. Circles represent immunoglobulin levels of each animal. Error bars represent the mean values.
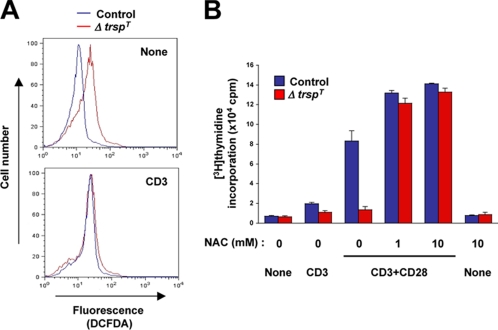
FIGURE 4.
ROS production in TCR-stimulated T cells and its role in the impaired proliferation of selenoprotein-deficient T cells. A, ROS production was analyzed by flow cytometry using DCFDA as described under “Experimental Procedures.” T cells, either unstimulated or stimulated with anti-CD3 antibody for 12 min, were used. B, T cells were either unstimulated (None) or stimulated as indicated in the presence of various concentrations of NAC. T cell proliferation was analyzed as in Fig. 3A. Values represent the average and S.E. of three separate experiments.
Activated T cells produce IL-2, which exerts an autocrine, feed-forward effect on T cell proliferation. In parallel, TCR activation also leads to higher expression of the IL-2 receptor (IL-2R) complex on T cells. We analyzed IL-2 production and IL-2R expression in T cells from control and ΔtrspT mice following CD3/CD28 stimulation. Both groups of T cells produced similar amounts of IL-2 in response to TCR activation (Fig. 3B). The IL-2R induction, on the other hand, occurred to a much lesser extent in T cells lacking trsp (Fig. 3C). Therefore, we expect that selenoproteins in activated T cells function to enhance their IL-2 responsiveness. TCR is coupled to multiple intracellular signaling modules. It was shown that the mitogen-activated protein kinase ERK links TCR activation to T cell proliferation (21). We therefore analyzed ERK activation in T cells in response to CD3/CD28 stimulation. T cells from control but not ΔtrspT mice showed rapid appearance of the active (phosphorylated) forms of ERK following treatment with anti-CD3 and anti-CD28 antibodies (Fig. 3D).
We found that elimination of tRNA[Ser]Sec and resultant selenoprotein deficiency in T cells cause defects in the development of functionally mature T cells and their TCR-dependent activation. It is highly likely that these multiple anomalies collectively contribute to impairing T cell-mediated immune responses. To verify this reasoning, we immunized control and ΔtrspT mice with NP-OVA, an antigen that elicits antibody production in a T cell-dependent fashion. At different time points after immunization, the serum levels of antigen-specific immunoglobulins were determined by enzyme-linked immunosorbent assay. Regardless of the immunoglobulin classes, the serum levels of antigen-specific antibodies were poorly raised in ΔtrspT mice (Fig. 3E). This in vivo finding reveals a link between selenoprotein loss and defective T cell immunity in mice.
A Role for Reactive Oxygen Species in the Impaired Proliferation of Selenoprotein-deficient T Cells—Many selenoproteins serve as antioxidant enzymes and possess Sec residues at their catalytic center. Such selenoenzymes eliminate ROS within and outside the cell and thereby protect ROS-sensitive molecular events against oxidative inactivation. It was shown that TCR activation leads to rapid production of ROS (15) and also that an NADPH oxidase is involved in this process (16). Most importantly, ROS production in TCR-stimulated cells was found to inhibit ERK signaling in T cells (17). These pieces of information prompted us to test whether selenoprotein-deficient T cells produce higher ROS than control cells and, if so, how this relates to T cell defects in ΔtrspT mice.
We first determined the level of basal and TCR-stimulated ROS generation by labeling cells with cell-permeant, oxidation-sensitive DCFDA dye. Intriguingly, DCFDA oxidation due to ROS generation occurred at higher rates in T cells from ΔtrspT mice than in control cells even without TCR stimulation (Fig. 4A). TCR stimulation resulted in increased ROS production in control T cells up to a level comparable with that of basal ROS production in trsp-deficient T cells (Fig. 4A). T cells from ΔtrspT mice did not show further increases in ROS production upon TCR stimulation. These data suggest that selenoproteins are indeed important for suppressing ROS production in T cells. Therefore, loss of selenoprotein function may render T cells defective in TCR-induced and ROS-sensitive responses.
The action of endogenously produced ROS can be blocked by pharmacological antioxidants such as NAC. We tested whether NAC can reverse the effect of selenoprotein deficiency in T cells. Incubation of trsp-deficient T cells with NAC restored the ability of the cell to proliferate in response to TCR stimulation (Fig. 4B). NAC also increases the proliferation response of control T cells. However, NAC alone did not induce T cell proliferation (Fig. 4B). Taken together, these findings indicate that selenoproteins are critically required for limiting ROS production in T cells and thus preventing ROS-mediated suppression of T cell activation.
DISCUSSION
Chronically elevated ROS levels may perturb a variety of cellular functions mediated by ROS-sensitive signaling pathways. We found that selenoproteins play a crucial role in T cell proliferation in response to TCR stimulation. The finding that the antioxidant NAC can compensate for selenoprotein deficiency in T cells and reverse their defect of TCR-induced proliferation strongly argues for a function of selenoproteins as antioxidants. Exact molecular targets in T cells that selenoproteins protect against ROS effects remain to be identified. ERK signaling is one of the likely candidates, as it is essential for T cell proliferation (21), and its activation is inhibited by endogenous ROS produced upon TCR stimulation (17). However, we do not rule out possible contributions of other molecules in this selenoprotein-ROS regulatory network.
It was previously shown that dietary selenium deprivation in mice led to reductions in T cell marker expression in muscle (22) and T cell proliferation (23). Intriguingly, replenishment of selenium reversed those changes in both studies. Furthermore, selenium supplementation has been found to be an effective treatment in delaying the progression of viral replication and boosting host defense in immunocompromised patients (19, 20). The data presented in this report provide first-hand molecular evidence that selenoproteins indeed have essential and non-redundant functions in T cell immunity. Our ΔtrspT mouse model can open up new horizons of research in immune regulation by dietary trace elements and antioxidants.
Supplementary Material
This work was supported, in whole or in part, by the National Institutes of Health NCI Intramural Research Program and the Center for Cancer Research; by a National Institutes of Health grant from the Office of Dietary Supplements, NCI, and the Cutaneous Biology Research Center through the Massachusetts General Hospital/Shiseido Co. Ltd. Agreement (to J. M. P.); and by National Institutes of Health Grants GM065204 and CA080946 (to V. N. G.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures,” Fig. S1, Table 1, and references.
Footnotes
The abbreviations used are: Sec, selenocysteine; ROS, reactive oxygen species; IL, interleukin; TCR, T cell receptor; ERK, extracellular signal-regulated kinase; DCFDA, dichlorodihydrofluorescein diacetate; NAC, N-acetyl cysteine; NP, 4-hydroxy-3-nitrophenyl; OVA, ovalbumin.
References
- 1.Hatfield, D. L., Berry, M. L., and Gladyshev, V. N. (2006) Selenium: Its Molecular Biology and Role in Human Health, 2nd Ed., pp. 1-419, Springer, New York, NY
- 2.Hoffmann, P. R. (2007) Arch. Immunol. Ther. Exp. 55 289-297 [DOI] [PubMed] [Google Scholar]
- 3.Hoffmann, P. R., Jourdan-Le Saux C., Hoffman, F. W., Chang, P. S., Bollt, O., He, Q., Tam, E. K., and Berry, M. J. (2007) J. Immunol. 179 3258-3267 [DOI] [PubMed] [Google Scholar]
- 4.Kiremidjian-Schumacher, L., Roy, M., Wishe, H. I., Cohen, M. W., and Stotzky, G. (1992) Biol. Trace Elem. Res. 33 23-35 [DOI] [PubMed] [Google Scholar]
- 5.Driscoll, D. M., and Copleand, P. R. (2003) Annu. Rev. Nutr. 23 17-40 [DOI] [PubMed] [Google Scholar]
- 6.Hatfield, D. L., Carlson, B. A., Xu, X. M., Mix, H., and Gladyshev, V. N. (2006) Prog. Nucleic Acid Res. Mol. Biol. 81 97-142 [DOI] [PubMed] [Google Scholar]
- 7.Copeland, P. R. (2005) Genome Biol. 6 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu, X. M., Carlson, B. A., Mix, H., Zhang, Y., Saira, K., Glass, R. S., Berry, M. J., Gladyshev, V. N., and Hatfield, D. L. (2006) PLoS Biol. 5 96-105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Papp, L. V., Lu, J., Holmgren, A., and Khanna, K. K. (2007) Antioxid. Redox Signal. 9 775-806 [DOI] [PubMed] [Google Scholar]
- 10.Rhee, S. G. (2006) Science 312 1882-1883 [DOI] [PubMed] [Google Scholar]
- 11.D'Autréaux, B., and Toledano, M. B. (2007) Nat. Rev. Mol. Cell Biol. 8 813-824 [DOI] [PubMed] [Google Scholar]
- 12.Kumaraswamy, E., Carlson, B. A., Morgan, F., Miyoshi, K., Robinson, G. W., Su, D., Wang, S., Southon, E., Tessarollo, L., Lee, B. J., Gladyshev, V. N., Hennighausen, L., and Hatfield, D. L. (2003) Mol. Cell. Biol. 23 1477-1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Orban, P. C., Chui, D., and Marth, J. D. (1992) Proc. Natl. Acad. Sci. U. S. A. 89 6861-6865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gladyshev, V. N., Stadtman, T. C., Hatfield, D. L., Jeang, K. T. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 835-839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Devadas, S., Zaritskaya, L., Rhee, S. G., Oberley, L., and Williams, M. S. (2002) J. Exp. Med. 195 59-70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jackson, S. H., Devadas, S., Kwon, J., Pinto, L. A., and Williams M. S. (2004) Nat. Immunol. 5 818-827 [DOI] [PubMed] [Google Scholar]
- 17.Kwon, J., Devdas, S., and Williams, M. S. (2003) Free Radic. Biol. Med. 35 406-417 [DOI] [PubMed] [Google Scholar]
- 18.Mao, X., Fujiwara, Y., and Orkin, S. H. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 5037-5042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beck, M. A. (2007) J. Nutr. 137 1338-1340 [DOI] [PubMed] [Google Scholar]
- 20.Hurwitz, B. E., Klaus, J. R., Llabre, M. M., Gonzalez, A., Lawrence, P. J., Maher, K. J., Greeson, J. M., Baum, M. K., Shor-Posner, G., Skyler, J. S., and Schneiderman, N. (2007) Arch. Intern. Med. 167 148-154 [DOI] [PubMed] [Google Scholar]
- 21.Pagès, G., Guérin, S., Grall, D., Bonino, F., Smith, A., Anjuere, F., Auberger, P., and Pouysségur, J. (1999) Science 286 1374-1377 [DOI] [PubMed] [Google Scholar]
- 22.Hooven, L. A., Butler, J., Ream, L.W., and Whanger, P. D. (2006) Biol. Trace Elem. Res. 109 173-179 [DOI] [PubMed] [Google Scholar]
- 23.Ueno, H., Hasegawa, G., Ido, R., Okuno, T., and Nakamuro, K. (2008) J. Trace Elem. Med. Biol. 22 9-16 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.