Abstract
Human Goodpasture antigen-binding protein (GPBP) is an atypical protein kinase that phosphorylates the Goodpasture auto-antigen, the α3 chain of collagen IV. The COL4A3BP gene is alternatively spliced producing two protein isoforms: GPBP and GPBPΔ26. The latter lacks a serine-rich domain composed of 26 amino acid residues. Both isoforms also function as ceramide transfer proteins (CERT). Here, we explored the function of Gpbp and GpbpΔ26/CERT during embryogenesis in zebrafish. We cloned both splice variants of the zebrafish gene and found that they are differentially expressed during development. We used antisense oligonucleotide-mediated loss-of-function and synthetic mRNA-based gain-of-function approaches. Our results show that the loss-of-function phenotype is linked to cell death, evident primarily in the muscle of the somites, extensive loss of myelinated tracks, and brain edema. These results indicate that disruption of the nonvesicular ceramide transport is detrimental to normal embryonic development of somites and brain because of increased apoptosis. Moreover, this phenotype is mediated by Gpbp but not GpbpΔ26/CERT, suggesting that Gpbp is an important factor for normal skeletal muscle and brain development.
The Goodpasture antigen-binding protein or GPBP (coded by the COL4A3BP gene) was originally identified in a screen for proteins expressed from a HeLa cDNA library for its capacity to bind the Goodpasture auto-antigen, the noncollagenous (NC1) domain of the α3 chain of human collagen (IV) (1). The protein is a nonconventional protein kinase that phosphorylates the auto-antigen. The gene is alternatively spliced and produces two protein isoforms: the full-length GPBP and GPBPΔ26. The latter lacks a serine-rich domain, composed of 26 amino acid residues, that is encoded by exon 11. The short isoform has less binding capacity to the Goodpasture auto-antigen and weaker kinase activity. GPBP can play a role in autoimmune responses, because it is overexpressed in many autoimmune conditions (2).
A recent study, using cell culture, has revealed a second function of both GPBP and GPBPΔ26, as ceramide transfer proteins (CERT) (3). The two isoforms share in common an amino-terminal pleckstrin homology (PH)3 domain and a serine-rich (SR) domain, a middle FFAT motif (two phenylalanines in acidic tract), and a carboxyl-terminal START domain. The PH domain and the FFAT domain permit the localization of the protein to the Golgi apparatus and the endoplasmic reticulum (ER), respectively, whereas the START domain binds and transfers ceramide between lipid membranes. A serine-rich motif in CERT undergoes phosphorylation, which down-regulates the ER to Golgi transport of ceramide. A recent study in Drosophila has shown that loss of function of a GPBP/CERT-like protein leads to enhanced oxidative damage that reduces lifespan (4).
To understand the physiological function of vertebrate GPBP and its shorter isoform, GPBPΔ26/CERT, we cloned the zebrafish col4a3bp gene and explored the function of the two splice variants during embryonic development. We found that both isoforms are dynamically expressed during early development and, when depleted, lead to apoptosis in selective tissues. Moreover, our results show that GPBP but not CERT carries the anti-apoptotic activity during early embryogenesis and that GPBP is an important factor for normal skeletal muscle and brain development.
EXPERIMENTAL PROCEDURES
Materials—The preparation of monoclonal antibody against human GPBP (Mab14) was previously described (1). Primary antibodies were used at the following dilutions: anti-FLAG M2 antibody (Sigma) at 1:1000. The secondary antibodies were anti-mouse IgG horseradish peroxidase-conjugated (Sigma) at 1:20,000, anti-mouse IgG1 anti-mouse biotin-conjugated antibody, and avidin-horseradish peroxidase (Vector, Burlingame, CA).
Synthetic Polymers—The following oligonucleotides were synthesized (Midland, Midland, TX): ZF-1F, 5′-GCAGGAGACGTGCAGTGTTGAGG-3′; ZF-2F, 5′-ATGTCAGACTGCAGTTCCTCGGG-3′; ZFE2-R, 5′-CCAGCGGTCCTGCCAGCCATGAATG-3′; ZF-2R, 5′-TCAGAAGAGGATGGCCTCACTGC-3′; ZF-3F, 5′-GCAGGCAGGTGGACACACTGC-3′; ZF-3R, 5′-CTTTGTCCCTGTGGAGCTCATC-3′; ZF-4F, 5′-GTTGAGGAGATGGTGCACAGTCAC-3′; ZF-4R, 5′-GCCGTTCTCCTCCACCTCTCC-3′; E-11F, 5′-CCTCACAGTCACACGTCCTCCT-3′; E-11R, 5′-CTGAGCACTGAACCTGTGCAC-3′; xba2F, 5′-AAATCTAGACATGTCAGACTGCAGTTCCTCGGG-3′; Sac2R, 5′-AAAACCGCGGTCAGAAGAGGATGGCCTCACTGC-3′; EcoMFLAGF, 5′-AAAGAATTCATGGACTACAAGGACGACGATGAC-3′; Eco2R, 5′-AAAGAATTCTCAGAAGAGGATGGCCTCACTGC-3′; ZFE10F, 5′-CCCAGGTGGAAAGGAGCAGGGCAT-3′; ZFE12R, 5′-CTCCTTCTTCAACAACCAGTTGCC-3′; LYAm-F, 5′-GAGTACGGCTGTAGAGAGTCCATCTGTCTCAGC-3′; LYAm-R, 5′-GCTGAGACAGATGGACTCTCTACAGCCGTACTC-3′; ZFGAPDH-F, 5′-CCTCCTGCACCACCAACTGCCTGG-3′; and ZFGAPDHR, 5′-CGGCAATCCCCATTGAAGTCAGTGG-3′.
Cloning of gpbp and gpbpΔ26—Zebrafish gpbp and gpbpΔ26 were amplified by PCR using as template a 24 h post-fertilization (hpf) embryonic cDNA library, which was cloned in uni-ZAP XR (Stratagene, La Jolla, CA). gpbpΔ26 was amplified as a single fragment using primers ZF-2F and ZF-2R and Pfu polymerase (Stratagene); the PCR product was cloned in the SmaI site of the pBluescript SK(-) vector (Stratagene) to form the pBczfGPBPΔ26 construct. gpbp was amplified in two pieces, which were then cloned independently in the HincII site of pBluescript SK(-). Thus, two constructs were formed, pBcR with the PCR fragment between primers ZF-2F and E11-R and pBcF with the fragment between primers E11-F and ZF-2R. The pBcF construct was digested with AflIII and XhoI and inserted into the pBcR construct to produce pBczfGPBP. The pcDNA3-FLAG-zfGPBP and pcDNA3-FLAG-zfGPBPΔ26 were cloned using a PCR approach. To this end, the pBluescript constructs served as templates, and PCR products were generated with primers Xba2F and Sac2R and digested with XbaI and SacII, and the digested product inserted in the NheI-SacII sites of the pRCX vector (5) producing pRCX-zfGPBP and pRCX-zfGPBPD26 sequences. The constructs are in frame with a FLAG sequence tag present in the vector. Using the pRCX vectors as templates and the primers EcoMFLAGF and Eco2R, the isoforms were reamplified and digested with EcoRI. Subsequently, both PCR products were inserted in the EcoRI site of pcDNA3.1 expression vector (Invitrogen).
For in vitro RNA synthesis, the gpbp fragment was subcloned in the pCS2+ vector without a FLAG sequence by digesting pBczfGPBP with EcoRI and XhoI and inserting in the polylinker of the pCS2+ vector producing the construct pCS2+zfGPBP. The pCS2+zfGPBP construct was digested with BamHI, and the resulting fragment from pBczfGPBPΔ26 was inserted to produce pCS2+zfGPBPΔ26. All of the constructs were verified by restriction mapping and nucleotide sequencing.
Site-directed Mutagenesis—For mutagenesis of gpbp into gpbpG67E (3), we used the QuikChange II site-directed mutagenesis kit (Stratagene) with the mutagenic primers LYAm-F and LYAmR, using pCS2+zfGPBP as template according to the manufacturer's instructions.
Cell Culture and DNA Transient Transfections—HEK293 cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin. Transient transfections were performed using the calcium phosphate precipitation method of the Profection mammalian system (Promega, Madison, WI).
Immunohistochemistry—Embryos were anesthetized and fixed overnight in 4% phosphate-buffered paraformaldehye in PBS at 4 °C, washed, dehydrated to methanol, and stored at -20 °C. After rehydration, the embryos were washed in PTT (0.3% Triton X-100, 0.1% Tween 20 in PBS) and bathed in blocking solution (1% Me2SO, 0.3% Triton X-100, 2% goat serum, 2 mg/ml bovine serum albumin) for 1 h. Anti-GPBP antibody (Mab14) was prepared in blocking solution at 1:200 dilutions of ascitic fluids and incubated for 1 h at room temperature or overnight at 4 °C. After extensive washes in PTT, the embryos were incubated with biotinylated secondary antibodies (Vector) at 1:200 dilution in blocking solution for 1 h at room temperature. The color reaction was developed using the Vectastain ABC kit with horseradish peroxidase and 3,3′ diaminobenzidine as chromogen (Vector). After staining, the embryos were cleared and stored in 80% glycerol. Monoclonal F59 antibody (anti-slow myosin heavy chain, a generous gift from F. E. Stockdale, Stanford University) (6) and anti-acetylated tubulin antibody (Sigma) were used at 1:100 and 1:200 dilutions, respectively. After extensive washes with PBT (0.1% Tween 20 in PBS), the embryos were incubated with secondary anti-mouse Alexa 488, and Alexa 555 IgG antibodies at 1:400 dilution (Molecular Probes). The F-actin in muscle cells was visualized with Alexa 555-conjugated phalloidin dye at 1:50 dilution in PBT (Molecular Probes). The embryos were washed, mounted on coverslips using ProLong gold medium, and photographed using LMS 510 META inverted confocal microscope (Zeiss). For plastic sections, staged embryos were anesthetized and fixed overnight in 4% phosphate-buffered paraformaldehyde at 4 °C, dehydrated to methanol, and stored at -20 °C until used. The samples were embedded in epoxy solution (Polysciences Inc., Warrington, PA) and cut in 5-micron-thick sections. The sections were stained with toluidine blue and photographed.
Reverse Transcription (RT)-PCR—Total RNA was extracted from staged embryos using TRIzol (Invitrogen) and retro-transcribed (1 μg) with Superscript II (Invitrogen). The subsequent cDNAs were subjected to PCR using primers ZF-E10F and ZF-E12R with Amplitaq polymerase (Applied Biosystems, Foster City, CA). The PCR products were separated by electrophoresis in 2% agarose gels and photographed.
Fish Maintenance and Breeding—Fish were maintained and kept under standard laboratory conditions at 28.5 °C (7). The embryos were staged and fixed at specific hours post-fertilization as described (8).
Protein Assays, SDS-PAGE, and Western Blotting—The proteins were extracted after transient transfection from cell plates scraping the cells in lysis buffer (50 mm Tris-HCl, 150 mm NaCl 0.5% Triton X-100 supplemented with 1 mm phenylmethylsulfonyl fluoride) and incubated on ice for 10 min. The lysates were clarified by centrifugation at 15,000 × g for 10 min at 4 °C, and the protein concentration was determined by the BCA method (Pierce) using known bovine serum albumin dilutions to construct a standard curve. For protein expression in morpholino-injected embryos, total proteins were extracted using TRIzol (Invitrogen) according to the manufacturer's instructions. SDS-PAGE and Western blotting were performed under reduced conditions, and the proteins were transferred to Immobilon P membranes (Millipore, Bilerica, MA). The presence of specific proteins was detected using anti-GPBP/GPBPΔ26 (Mab14) or FLAG antibodies and immunoperoxidaseor alkaline phosphatase-conjugated secondary antibodies.
In Vitro Translation and Synthetic mRNA—5 μg of pCS2+ vector plasmid containing the desired insert were digested with NotI, phenol/chlorophorm/isoamyl alcohol was extracted, and the DNA was precipitated and dissolved in nuclease-free water. One μg of the digested plasmid was transcribed using the mMESSAGE mMACHINE SP6 kit (Ambion, Austin, TX) for 2 h at 37 °C. The transcription reactions were treated with DNase I, synthetic RNA (cRNA) was purified using MEGAclear (Ambion), and the RNA concentration was calculated by UV absorbance. The RNA was stored frozen in aliquots at -80 °C (the samples were subjected to only one freezing-thawing cycle). The integrity of the synthetic cRNA was assayed in an in vitro translation assay with the Retic Lysate IVT™ (Ambion) kit according to the manufacturer's instructions using [35S]methionine (>1000 Ci/mmol, 10 mCi/ml) as radiolabeled amino acid (Amersham Biosciences).
Preparation and Injection of Morpholinos—Morpholino (MO) antisense oligonucleotides (Gene Tools, Corvallis, OR) were designed to complement the sense transcript of gpbp and gpbpΔ26 at the 5′-UTR (MO5′UTR, 5′-GGAAAACTCCGCGATAGTCGTGTTC-3′). A second morpholino was designed to complement the sense nuclear pre-mRNA affecting the 3′ splicing site in front of exon 11. Thus, this morpholino deletes the exon present only in gpbp (MOgpbp-SA, 5′-GACTGTGAGGCTGAACCCAAGAGC-3′). The morpholinos were solubilized in nuclease-free water, and the concentration was determined by UV absorbance.
Cell Death Assays—TUNEL and Acridine orange staining were used for apoptosis assays. For TUNEL analysis, the embryos were staged and fixed overnight in 4% phosphate-buffered paraformaldehyde in PBS at 4 °C. After washing, the embryos were dehydrated and stored in methanol. Rehydrated embryos were permeabilized by proteinase K digestion, washed in PBT, and assayed by TUNEL using the in situ cell death detection kit POD (Roche Applied Science) according to the manufacturer's instructions. The presence of positive cells was analyzed under a fluorescence microscope and photographed (Zeiss, Thornwood, NY). Live embryos were stained for apoptotic cells with the vital dye Acridine orange that permeates inside acidic lysosomal vesicles and becomes fluorescent, thus staining apoptotic cells. A stock solution of 5 mg/ml in egg water was diluted 300 times in egg water, and dechorionated live embryos were bathed in this solution for 20 min in the dark, extensively washed in egg water, analyzed under a fluorescence microscope, and photographed.
RESULTS
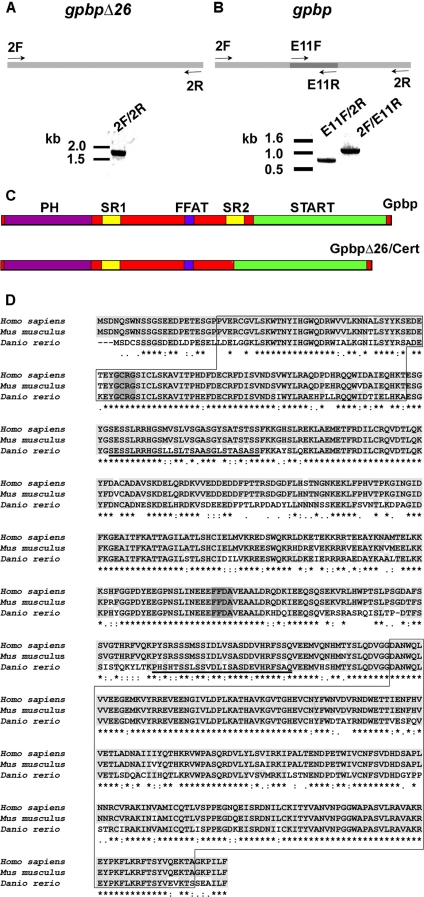
Identification and Cloning of Zebrafish gpbp and gpbpΔ26 cDNA—To identify zebrafish gpbp, we searched expressed sequence tag databases using human GPBP cDNA as bait (Gen-Bank™ accession number AF136450). The search picked two clones with homologous sequences, each containing the start (AL910168) and the stop codon (BM859835). To obtain full-length coding sequences, we used PCR primers located within the expressed sequence tags and amplified a single product from a zebrafish 24 hpf cDNA library (Fig. 1A). The PCR product was sequenced revealing a 1,785-bp transcript representing gpbpΔ26, the splicing variant of gpbp that codes for a 594-amino acid polypeptide. The BM890535 expressed sequence tag showed an extra 78-bp sequence not present in gpbpΔ26, thus suggesting that it encoded the unspliced variant of gpbp. Based on this sequence, we designed two primer pairs and amplified the putative gpbp in two fragments using the 24-hpf cDNA library as a template (Fig. 1B). Both PCR fragments shared the 78-bp sequence and contained either the 5′ (2F/E11R; 1,176 bp long) or the 3′ (E11F/2R; 765 bp) parts of the cDNA. The two fragments were combined to produce the complete gpbp cDNA of 1,863 bp encoding the 620-amino acid polypeptide. The gpbp sequences were deposited in Gen-Bank™ with the accession numbers EU000165 (gpbp) and EU000166 (gpbpΔ26).
FIGURE 1.
Molecular cloning of gpbp and gpbpΔ26. A and B depict schematically the cloning strategy of gpbpΔ26 and gpbp, respectively. GpbpΔ26 was cloned in one-step RT-PCR (one band), whereas gpbp in a two-step manner (two bands). C, schematic representation of the secondary protein structures of Gpbp and GpbpΔ26. The PH, SR1, FFAT, SR2, and START domains are marked. D, sequence alignment of Gpbp from zebrafish (D. rerio) with human (Homo sapiens) and mouse (Mus musculus) GPBP with accession numbers AF136450 for human, AF232932 for mouse, and EU000165 for zebrafish. Identical amino acids are highlighted in light gray and marked with an asterisk below. Conserved substitutions are marked by dots. Closed boxes mark the PH (top) and START (bottom) domains; the SR1 and SR2 domains are underlined; dark gray marks the GCRG and FFDA motifs.
The alignment of the zebrafish Gpbp sequence with human and mouse proteins using ClustalW shows high conservation at the amino acid level (Fig. 1D). The zebrafish protein shares a 75% identity with the human, whereas the human and mouse proteins are 96% identical. The secondary structure of the protein includes a PH domain and a serine-rich domain in the amino terminus, a FFAT motif and a second serine-rich domain in the middle part of the protein, which is not present in gpbpΔ26, and a START domain in the carboxyl terminus (Fig. 1C).
The zebrafish col4a3bp gene that codes for the gpbp and gpbpΔ26 mRNAs is located on chromosome 5 and spans 51 kb of the zebrafish genome. Alignment of the genomic and cDNA sequences revealed 18 highly conserved exons consistent with the GT-AG rule (detailed in supplemental Table S1). Exon 11 is not present in gpbpΔ26, and exon 18 contains the 3′-UTR. The same number of coding exons and a similar arrangement of splicing junctions are found in the human and mouse genes (data not shown).
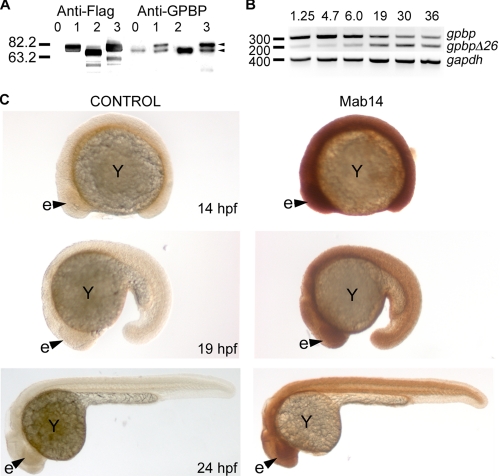
Recombinant Expression of Zebrafish Gpbp and GpbpΔ26—We have produced recombinant FLAG-tagged human GPBP and zebrafish Gpbp and GpbpΔ26 proteins in HEK293 cells, and the expression levels in the lysates were assayed by immunoblotting using the anti-FLAG M2 antibody. We observed a single band for each of the splice variants with higher mobility for GpbpΔ26, similar to the previously described human homologs (Fig. 2A); (2). The monoclonal antibody Mab14, which was raised against the human GPBP protein (1), is cross-reacting with both zebrafish isoforms, corroborating the anti-FLAG antibody results (Fig. 2A).
FIGURE 2.
Protein characterization and spatio-temporal expression of gpbp and gpbpΔ26 during zebrafish development. A, to characterize the zebrafish proteins, HEK293 cells were transiently transfected using FLAG-tagged constructs representing no insert (lanes 0), human GPBP (lanes 1), zebrafish GpbpΔ26 (lanes 2), and zebrafish Gpbp (lanes 3). The cell lysates were separated by reducing SDS-PAGE in 10% acrylamide gels, transferred to polyvinylidene difluoride membranes, and blotted with the M2 Anti-FLAG antibody. Similarly, Western blots, prepared as described for anti-FLAG labeled blots, were also stained with a monoclonal antibody against human GPBP (Mab14, -anti-GPBP). The extra band that is present in lanes 0, 1, and 3, is likely representing GPBPΔ26 from human origin of the HEK293 cells and is masked in lane 2 by recombinant GpbpΔ26. B, total RNA (1 μg) isolated at the indicated developmental stages in hpf was reversed transcribed and used as template in PCRs with primers surrounding exon 11, and thus able to distinguish between gpbp and gpbpΔ26 transcripts (forward primer in exon 10, reverse primer in exon 12). We also used primers for the house keeping gene gapdh for normalization purposes. The PCR products were resolved in a 2% TAE-agarose gel and stained with ethidium bromide. C, lateral views of whole mount embryos at the indicated stages stained with the Mab14 antibody or secondary antibodies alone as control. e, eye (arrowhead); Y, embryonic yolk.
Expression Pattern of gpbp—To investigate the temporal expression of gpbp during development, we performed RT-PCR using RNA from zebrafish embryos at different developmental stages. The full-length gpbp is maternally deposited (expressed before mid-blastula transition at 3 hpf), and its level gradually subsides as development progresses. The short isoform, gpbpΔ26, is barely detectable during the first 12 h post-fertilization and increases by the second day of development (Fig. 2B). Therefore, although the gene is expressed at all of the tested developmental stages, gpbp is more abundant at early stages as compared with gpbpΔ26. To gain insight into the spatial distribution of the protein, we analyzed the expression pattern of Gpbp in whole embryos using the Mab14 monoclonal antibody. We found that Gpbp is widely expressed during embryogenesis (Fig. 2C). Because the Mab14 antibody recognizes both splice variants, the staining pattern illustrates the combined expression domains of Gpbp and GpbpΔ26.
Knockdown of gpbp and gpbpΔ26 Reveals Specific Brain and Somite Phenotypes—To investigate the function of Gpbp in zebrafish development, we first designed a strategy to knock down gene function by using a morpholino-modified antisense oligonucleotide directed against the 5′-UTR sequence (MO5′UTR). This morpholino blocks protein translation, but it does not affect pre-mRNA processing, e.g. splicing (Fig. 3A). Because the targeted 5′-UTR area is common in both gpbp and gpbpΔ26 transcripts, MO5′UTR is expected to knock down Gpbp and GpbpΔ26 simultaneously.
FIGURE 3.
Phenotypes of morpholino-mediated knockdown of gpbp and gpbpΔ26. A, the position of the target sequence for the translation-blocking morpholino is depicted. The arrow marks the start ATG codon. B, uninjected (wild type, WT) or MO5′UTR injected embryos at 24 hpf were lysed, and the protein extracts were resolved in reducing 10% acrylamide gels, blotted, and stained with the Mab14 antibody. On the left is the molecular mass expressed in kDa. Arrowheads point to isoforms of Gpbp and GpbpΔ26. C, lateral views of embryos, injected with 5 ng of morpholino (MO5′UTR) or uninjected controls (WT), were photographed at the indicated times. Arrowheads point to brain edema and to the jaw region. D, wild-type (WT) and MO5′UTR-injected embryos were fixed at 48 hpf and embedded in plastic resin, sectioned, and stained with toluidine blue. The top four panels show sagittal (left) and cross-sections (right) through the fourth ventricle region of the brain. The arrowhead points to myelinated axonal tracts, and the asterisk marks large brain edema and loss of neuronal tissues. The bottom two panels depict sagittal sections through the mid-trunk region of wild-type and morphant embryos at 48 hpf. The morphant somites are smaller and contain large numbers of dying cells and cellular debris (arrows).
To determine the specificity and effectiveness of the morpholino, we injected increasing amounts of MO5′UTR into one- to four-cell stage embryos. The embryos were then scored for the presence of unspecific necrosis to select morpholino dosage with no apparent toxic effects. To gauge the effectiveness of the morpholino, the proteins were extracted from MO5′UTR-injected embryos and analyzed by Western blotting to test for possible residual Gpbp activity. Staining with the Mab14 antibody showed a significant reduction in the intensity of the Gpbp band, demonstrating that the 5′-UTR morpholino effectively reduces but does not ablate the amount of Gpbp protein (Fig. 3B).
MO5′UTR did not interfere with gastrulation and early somitogenesis. Instead, we observed the first phenotypic effects of the morpholino knockdown at 24 hpf. At this stage, the injected embryos exhibited tissue loss in the head region, edema in the fourth brain ventricle, and small eyes. This phenotype persisted at later stages resulting in small head and eyes at 48 hpf (Fig. 3C).
To further analyze this phenotype, we prepared histological, plastic sections of the 48-hpf morphants and counterstained them with toluidine blue to reveal tissue and cell morphology. We found that the brain was the most affected organ, with a clear reduction of the myelinated tracts and hydrocephaly of the fourth ventricle (Fig. 3D). We also observed severe tissue damage in the somites with extended loss of muscle fibers (Fig. 3D). To further analyze the neuronal defects, we examined patterning of axonal tracts in the brain using an anti-acetylated tubulin monoclonal antibody. We found that in wild-type embryos the tracts of postotic commissure, posterior commissure, and medial longitudinal fasciculus were established at 24 hpf (9). However, in knockdown animals, we observed a single axonal tract posterior to the eye (Fig. 4).
FIGURE 4.
Immunohistochemistry analysis of brain and embryonic muscles. Labeling with an anti-acetylated tubulin antibody shows axonal tracts in wild-type (WT) uninjected controls (arrowheads), and in morphant embryo where the staining is greatly reduced detecting only a single thin axonal tract. The somitic phenotype is revealed by myosin heavy chain labeling with the F59 antibody that shows shortening of slow muscle fibers, disorganization, and focal loss of tissue in morphant embryos at 24 and 48 hpf (blue arrowheads). Falloidin labeling of F-actin filaments shows all muscle fibers (slow and fast). This staining reveals that muscle fibers in morphants are not parallel to each other as in wild types and appear as tangled bundles (yellow arrowheads). e, eye; m, midbrain; c, cerebellum; ne, neuro-ectoderm; n, notochord; v, ventral tissues; pc, posterior commissure; tpoc, tract of the postoptic commissure; mlf, medial longitudinal fasciculus; isb, intersomitic boundary; my, myotome.
Histological analysis of the embryonic trunk muscles showed disorganized muscle fibers that were peppered with cell debris (Fig. 3D). To determine whether the defect is limited to slow or fast muscle fibers, we performed immunohistochemical studies using the F59 antibody that recognizes myosin heavy chain in slow muscle fibers (10) and found that these fibers are present but greatly disorganized, shorter, and with areas of tissue debris between them. Labeling with fluorophore-conjugated phalloidin dye that marks F-actin in slow and fast muscle fibrils showed similar defects, which were first observed at 24 hpf and significantly worsened at 48 hpf (Fig. 4). Thus, it appears that all muscle fibrils are dependent on Gpbp and Cert for normal development and function.
Taken together, these data suggest that impairment of Gpbp and GpbpΔ26 function leads to specific brain and muscle defects, whereas gastrulation and early segmentation stages are not affected at these morpholino doses.
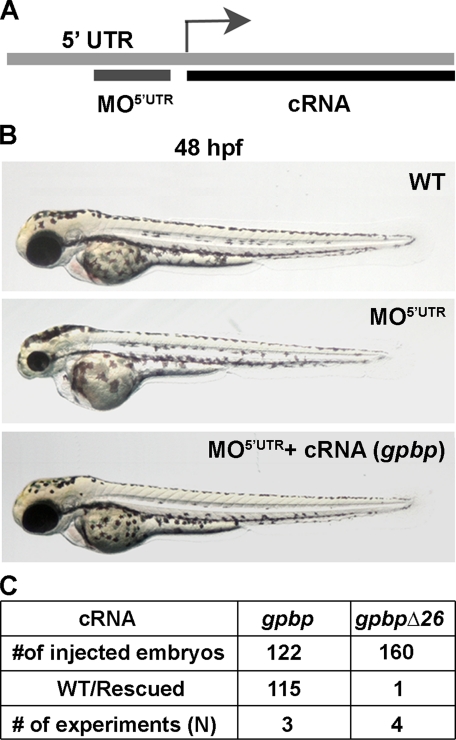
Suppression of Loss-of-Function Phenotype by mRNA Overexpression—To test the specificity of the morpholino-induced phenotype and to further exclude potential toxic or mistargeting effects, we designed a rescue experiment of the MO5′UTR morphants with synthetic cRNAs encoding gpbp variants (Fig. 5A). The injection of cRNA alone, gpbp, or gpbpΔ26 did not produce any phenotype, and the embryos developed normally (data not shown). When we co-injected 5 ng of 5′-UTR morpholino with 50 pg of gpbp mRNA at one-cell stage embryos, we observed a total suppression of the morpholino-induced phenotype at 24 and 48 hpf in 94% of the injected embryos (Fig. 5, B and C). In contrast, when we co-injected 5′-UTR morpholino and gpbpΔ26 mRNA, we did not observe suppression of the loss-of-function phenotype (Fig. 5C). These results suggest that the long splice variant of Gpbp exerts critical function in neural and muscle development, and its loss leads to severe developmental deficits. Gpbp most likely mediates these effects, because the short variant does not rescue the knockdown phenotype.
FIGURE 5.
Suppression of MO5 ′UTR phenotype by gpbp mRNA. A, schematic outline of the strategy used for the phenotypic rescue of MO5′UTR. The target sequence for the morpholino is not present in the recombinant capped RNA expression constructs marked as cRNA. B, fertilized eggs were co-injected at the one cell stage with 5 ng morpholino alone (middle panel) or together with 50 pg of gpbp cRNA (bottom panel) and imaged at 48 hpf. Wild-type (WT) uninjected embryos served as a control (top panel). C, Summary of the experimental animals and rescue outcomes.
Selective Knockdown of the Full-length Gpbp Isoform—The rescue experiments suggested that the visible morpholino phenotype might be due predominantly to the loss of gpbp and not gpbpΔ26. To test this notion and to further corroborate the specificity and strength of the MO5′UTR-caused defects, we selectively knocked down the long isoform of gpbp in live embryos. To this end, we designed a morpholino, MOgpbp-SA, to target the 3′ splice site sequence in intron 10 blocking the inclusion of exon 11 in the col4a3bp transcript. This way, the full-length variant of gpbp could be removed without affecting the expression of gpbpΔ26 (Fig. 6A).
FIGURE 6.
Knockdown of gpbp by splicing blocking morpholino, preserving intact gpbpΔ26, produces a similar phenotype to MO5 ′UTR. A, schematic drawing of the location of the splicing-blocking morpholino in the gpbp transcript. The morpholino interferes with the 3′ splicing sequence at the boundary between intron 10 and exon 11 producing only gpbpΔ26 mRNA. B, groups of uninjected controls (lane 1) and embryos injected with the indicated amounts of MOgpbp-SA (lane 2) were monitored for the presence of gpbp mRNA by RT-PCR and gel electrophoresis at 6 hpf. The gel images indicate that MOgpbp-SA leads to a specific loss of the long splice variant leaving the short form undisturbed. C, lateral views of uninjected controls (WT) and embryos injected with 4 ng of MOgpbp-SA at 30 and 48 hpf. An arrowhead points to brain edema. D, the table presents a summary of experimental outcomes based on number of injected embryos. The term Phenotype in the table represents number of embryos that showed mutant phenotype as in C.
To test the efficacy of this approach, we injected increasing amounts of MOgpbp-SA into one- to four-cell embryos, isolated RNA, and amplified cDNA fragments spanning exon 11 from injected and uninjected embryos. cDNA samples from control embryos gave rise to two bands of the expected size corresponding to gpbp and gpbpΔ26 transcripts. Conversely, we found that injection of MOgpbp-SA effectively and specifically eliminated the high MW band of the gpbp transcript leaving gpbpΔ26 intact (Fig. 6B). Sequencing of the low molecular weight band confirmed that it is gpbpΔ26, demonstrating that MOgpbp-SA effectively removed exon 11 (data not shown).
To investigate the effects of the specific gpbp loss-of-function phenotype, we injected 4 ng of MOgpbp-SA into one- to four-cell embryos. Morphological analysis indicated that the MOgpbp-SA-caused defects at 30 hpf strongly resembled the phenotype observed with MO5′UTR, including extensive cell death in the head region and edema in the fourth brain ventricle (Fig. 6C). At 48 hpf, the phenotype became more severe with strong signs of microphthalmia, microcephaly, and pericardial edema. Of note, the mortality of the splice site morpholino-injected embryos was higher (40%) as compared with 5′-UTR morphants (10%).
These data indicate that knockdown of both Gpbp and GpbpΔ26 isoforms or elimination of the long Gpbp form alone results in almost identical defects. Thus, it appears that the brain and muscle dysmorphologies are mediated by loss of function of the full-length Gpbp. These data are also in agreement with the fact that gpbp but not gpbpΔ26 RNA could rescue the morpholino defects.
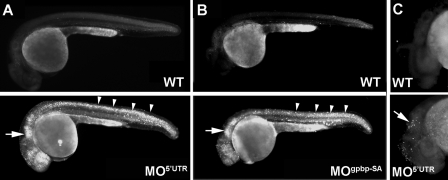
Gpbp Regulates Apoptosis—To investigate whether cell death might be causing the tissue damage after the gpbp knockdown, we assayed apoptosis in living embryos by Acridine orange staining. Acridine orange is a vital dye that binds to DNA and RNA and is routinely used to discriminate between apoptosis and necrosis (11). When MO5′UTR- or MOgpbp-SA-injected embryos and uninjected controls were stained with Acridine orange, morpholino-injected embryos showed more intense staining than controls. Moreover, staining was confined to the areas affected by the gpbp knockdown. Specifically, the highest count of Acridine orange-positive cells were in the hindbrain and the fourth ventricle, telencephalon, diencephalon, and the neural tube, somites, and paraxial mesoderm of the tail (Fig. 7, A and B). The results were further confirmed by TUNEL assay, which detects end stage apoptotic DNA fragmentation, in MO5′UTR-injected and uninjected control embryos. We found an abundance of positive cells in the morpholino-injected embryos at the areas with morphological defects, consistent with the live phenotype and the Acridine orange staining pattern (Fig. 7C). These results indicate that the depletion of the long gpbp splice variant leads to massive programmed cell death predominantly in brain and muscle tissues.
FIGURE 7.
Knockdown of Gpbp leads to increased levels of apoptosis. A and B, lateral views of Acridine orange-stained, live control (wild type, WT), or morpholino-injected embryos as indicated: MO5′UTR at 24 hpf (A) and MOgpbp-SA at 27 hpf (B). Arrows (brain region) and arrowheads (somites) point to apoptotic cells. C, TUNEL assay in wild-type and MO5′UTR-injected embryos at 30 hpf in the brain region (arrow).
Ceramide Accumulation in the Gpbp Knockdown Embryos—It has been previously demonstrated that a single amino acid substitution in the CERT PH domain abolishes ceramide transfer between the endoplasmic reticulum and the Golgi complex by preventing interaction of CERT with phosphatidylinositol 4-phosphate in the Golgi membrane (3). To test whether aberrant ceramide transport is behind the observed Gpbp knockdown defects, we engineered a G64E mutation in zebrafish gpbp (the equivalent of hamster G67E) and assayed the capacity of this mutant protein to rescue the morpholino-induced phenotype. When one-cell stage embryos were injected with a mix containing 4 ng of MO5′UTR and 50 pg of the mutant gpbp mRNA, the mutant phenotype was not suppressed (data not shown). This result implies that the observed apoptosis in muscle and brain is most likely caused by the loss of function of the PH domain of Gpbp, which has been demonstrated to play a key role in intracellular ceramide distribution (3). It is likely that disruption of intercompartmental ceramide flow in the morphant cells results in apoptosis.
DISCUSSION
The Goodpasture antigen-binding protein (GPBP) was isolated during the pursuit of two independent biological questions. The initial discovery was made in the attempt to understand the molecular complexity of Goodpasture syndrome (1), where Gpbp was shown to bind and phosphorylate the Goodpasture antigen. Subsequently, CERT, a spliced variant of GPBP, was identified in a genetic screen of sphingomyelin-deficient cells for proteins able to restore intracellular lipid transport (3). These two strikingly different isolation strategies might reflect the complex functions of the COL4A3BP (collagen type IV α3-binding protein) gene. Intrigued by the diversity of the potential biological activities of Gpbp, we set out to study the fundamental characteristics of the gene and its protein products by analyzing the genomic structure and the protein function of Gpbp/Cert during development in the teleost, zebrafish (Danio rerio).
Our results show that zebrafish col4a3bp is located on chromosome 5 and spans 51 kb of genomic sequence, a more compact gene structure compared with COL4A3BP, which is located on human chromosome 5q13.3 and spans 130 kb. However, both orthologs retain a highly conserved intron-exon structure over 18 exons, with exon 18 coding for the 3′-UTR. The occurrence of two splice variants (Gpbp and GpbpΔ26/Cert) has been also conserved during evolution, suggesting that the two isoforms likely perform distinct functions. At the protein level, both variants are 75% identical between zebrafish and human orthologs and over 95% among mammals. The high conservation at the amino acid level correlates well with the presence of a number of functional domains, which can be divided into three parts: the amino-terminal pleckstrin homology domain, which has been shown to interact with phosphatidylinositol 4-phosphate and localizes the protein in the Golgi membranes (12, 13); the central region containing the serine-rich and a FFAT domain that localizes the protein to the ER by interacting with VAP (14); and the carboxyl-terminal steroidogenic acute regulatory protein-related domain (START) (15) that extracts and transports ceramide (3). The remarkable similarities in protein structural elements, RNA processing, and genomic organization imply that the basic biological functions of COL4A3BP have been highly conserved between lower vertebrates and mammals.
Surveillance of adult human RNA samples using Northern blots showed that high levels of COL4A3BP are expressed in the striated muscles (heart and skeletal) and in the brain, whereas pancreas, kidney, placenta, and lung express lower levels of both splice variants (1). Further immunohistochemistry analysis in human samples revealed that both GPBP isoforms are present in the plasma membrane of kidney (epithelia of tubules and mesenchymal cells), lung (pneumocytes), and prostate (epithelial) cells; the axonal tracts, but not the neurons, in the central nervous system; the nuclei of spermatogonia in testis; and the extracellular matrix of lung alveoli (1). However, the expression of COL4A3BP was never analyzed during vertebrate development. Here, we present evidence that col4a3bp is dynamically expressed during zebrafish embryogenesis. RT-PCR based experiments showed that the expression patterns of the two splice variants behave in opposite fashion. The gpbp is maternally deposited and highly expressed at gastrulation and early somitogenesis. As development progresses, the levels of gpbp transcripts decrease, whereas the levels of gpbpΔ26/cert that are initially very low gradually increase at later developmental stages. In whole mount antibody labeling, we observed initially widespread, high protein expression that later concentrated in the brain and the embryonic muscle (somites). These data correlate well with the expression in adult human tissues, suggesting that both the growth and homeostasis of brain and muscle might critically dependent on col4a3bp.
In support of this idea, we found that knockdown of Gpbp leads to loss of myelinated tracks in the central nervous system and to extensive apoptosis and tissue loss in the brain and somites. To tease out which of the two splice variants mediate the observed phenotype, we conducted a set of rescue experiments using full-length gpbp and gpbpΔ26 mRNA transcripts (summarized in Table 1). We found that the recombinant gpbp mRNA is able to rescue the morpholino-induced phenotypes, whereas gpbpΔ26 is not. Furthermore, the splice site morpholino knockdown, which cleanly deletes exon 11 effectively converting endogenous gpbp to gpbpΔ26, presented an identical phenotype as the 5′-UTR MO knockdown of both variants. Our results suggest that the observed phenotype is a consequence of depletion of the full-length Gpbp splice isoform, selectively inducing apoptosis in muscle and brain during early development.
TABLE 1.
Summary of morpholino knockdown (KD), cRNA overexpression (OE), and cRNA rescue (R) experiments
| Injection | Experiment | Absent | Present | Phenotype |
|---|---|---|---|---|
| gpbp cRNA | OE | high gpbp; gpbpΔ26 | Wild type | |
| gpbpΔ26 cRNA | OE | gpbp; high gpbpΔ26 | Wild type | |
| MO5′UTR | KD | gpbp; gpbpΔ26 | Apoptosis | |
| MOgpbp-SA | KD | gpbp | gpbpΔ26 | Apoptosis |
| MO5′UTT and gpbp cRNA | R | gpbpΔ26 | gpbp | Wild type |
| MO5′UTR and gpbpG64E cRNA | R | gpbp; gpbpΔ26 | gpbpG64E | Apoptosis |
| MO5′UTR and gpbpΔ26 cRNA | R | gpbp | gpbpΔ26 | Apoptosis |
The accumulation of ceramide in cellular membranes in the absence of Gpbp could explain the morphant phenotypes because free ceramide and its derived products are known to act as second messengers and to regulate apoptotic pathways (16-19). Also, it has been demonstrated that the imbalance in enzymatic activities controlling ceramide levels, sphingomyelinase or ceramidase, results in pathologies such as pulmonary edema in mice (20) or impaired development in zebrafish (21). Moreover, our results suggest that this anti-apoptotic activity is carried primarily by the long splice variant. Interestingly, the single difference between the short and long splice variants that could account for this outcome is the second serine-rich domain (SR2). Recent elegant biochemical analysis of the adjacent SR1 domain showed that phosphorylation of 7-9 Ser/Thr residues in the SR1 domain reduces ceramide transport from ER to Golgi, and dephosphorylation of the same residues increases ceramide transport by relaying conformational changes to the PH and START domains (22). The phosphorylation of the SR1 domain appears to be regulated by the levels of sphingomyelin and cholesterol in the lipid rafts of the plasma membrane. Although to date there are no biochemical data available for the function of SR2, we postulate that it might also have a crucial role in ceramide trafficking, because we observed ceramide accumulation in zebrafish morphants lacking specifically the SR2 domain of Gpbp (data not shown).
Previous analysis revealed that the chemically induced Chinese hamster ovary mutant cell line, LY-A, which is defective in sphingomyelin metabolism, harbors a point mutation, G67E, in CERT. Glycine 67 is conserved among multicellular organisms, and its conversion to glutamic acid ablates the interaction of the PH domain with phosphatidylinositol 4-phosphate in Golgi membranes, leaving other functional domains undisturbed (3). Thus, the Chinese hamster ovary mutant cells most likely express a hypomorphic allele of CERT. Alternatively, it is conceivable that because the remaining domains are fully functional, the mutant might act as a neomorph or exerts a dominant-negative effect, for example by picking up ceramide in the ER but being unable to transfer it to the Golgi apparatus. The fact that we were unable to rescue the gpbp knockdown phenotype by overexpressing the zebrafish equivalent G64E mutant form further supports the idea that a defect in ceramide transport is a plausible cause of the apoptotic defects in gpbp morphants.
It is likely that the anti-apoptotic activity of Gpbp might be beneficial to the gastrulating embryo, which is fast growing generating large numbers of organ-specific progenitor cells. As the organism reaches maturity, the physiological ratio of the short to the long splice variant is ∼9:1, shifting in favor of the short isoform (2). However, in several autoimmune conditions, the long splice variant accumulates, reducing the differential levels between the two proteins. Presently, it is unclear whether the variable balance and differences in activities between the two isoforms have similar roles in development, tumorigenesis, and autoimmune conditions. It is possible that the short, highly abundant GpbpΔ26/Cert might serve as a basic ceramide transporter between the ER and Golgi cellular compartments, whereas Gpbp might be playing an anti-apoptotic role during embryogenesis and under pathophysiological conditions. It is interesting that in invertebrates as Drosophila melanogaster the col4a3bp gene produces only one isoform. The phenotype associated with the loss of function of Dcert is reduced lifespan because of oxidative stress rather than increased apoptosis (4).
As mentioned above, GPBP was initially isolated as the Goodpasture antigen-binding protein, which has been linked to an autoimmune disease with kidney defects called Goodpasture progressive glomerulonephritis. Further expression analysis in human tissues revealed that organs expressing GPBP are also associated with other autoimmune disorders (e.g. Lupus erythromatosus, multiple sclerosis, myasthenia gravis, Addison disease, male infertility, type I diabetes, etc.). Although we did not observe a kidney phenotype in live embryos and histological sections, we further analyzed the ultrastructure of the kidney glomeruli by electron microscopy. We reasoned that because morphants die early in development, the kidney phenotype might not be severe enough by this stage of development that we can easily observe it. However, electron microscopy analysis showed normal kidney podocytes and glomerular basement membrane in morphants as in controls (supplemental Fig. S1). This result could be explained in a number of ways. For example, the embryos may die before kidney defects appear, and thus we cannot address this question in the current experimental paradigm, or the zebrafish protein could have a more ancestral function that does not include a part in kidney morphogenesis and physiology. Alternatively, Gpbp may play a yet unknown role in the ER quality control system that is linked to the autoimmune conditions. This would be consistent with a recent study showing that GPBP interacts with the ER stress response pathway (23). Localization at the ER would place GPBP in the same cellular compartment as the synthesis of Col(IV)α3 chain, which could explain previous observations of phosphorylation of the Col4α3 NC1 domain by GPBP. It remains to be determined whether this is the effect or the cause of the disease. Our results, as well as the novel set of genetic and developmental tools described here might contribute to further analysis of the full gamut of biological functions of Gpbp/Cert in physiological and pathological conditions.
Supplementary Material
Acknowledgments
We thank Antonis Hatzopoulos, Todd Graham, David Melville, and Fernando Revert for critical reading of the manuscript and helpful discussions, Lila Solnica-Krezel for reagents, F. Stockdale for the F59 antibody, and Mercedes Montero-Balaguer for assistance in the initial stages of the project.
This work was supported, in whole or in part, by National Institutes of Health Grants DK065123 (to B. G. H. and D. A.), DK18381 (to B. G. H.), DK70929-01A11 (to A. S.), and DE018477 (to E. W. K.) and National Institutes of Health Grants CA68485, DK20593, DK58404, HD15052, DK59637, and EY08126 (to the Vanderbilt University Medical Center Cell Imaging Shared Resource). This work was also supported by Grant SAF2006-12520-C02-01 from Plan Nacional I+D del MEC of Spain (to J. S.) and the Vanderbilt University Academic Venture Capital Fund (to E. W. K.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1 and Table S1.
Footnotes
The abbreviations used are: PH, pleckstrin homology; ER, endoplasmic reticulum; START, steridogenic acute regulatory protein-related lipid transfer; SR, serine-rich; MO, morpholino; RT, reverse transcription; PBS, phosphate-buffered saline; UTR, untranslated region; TUNEL, terminal transferase-mediated dUTP nick end labeling; hpf, hour(s) post-fertilization; cRNA, capped RNA; FFAT, two phenylalanines in acidic tract.
References
- 1.Raya, A., Revert, F., Navarro, S., and Saus, J. (1999) J. Biol. Chem. 274 12642-12649 [DOI] [PubMed] [Google Scholar]
- 2.Raya, A., Revert-Ros, F., Martinez-Martinez, P., Navarro, S., Roselló, E., Vieites, B., Granero, F., Forteza, J., and Saus J. (2000) J. Biol. Chem. 275 40392-40399 [DOI] [PubMed] [Google Scholar]
- 3.Hanada, K., Kumagai, K., Yasuda, S., Miura, Y., Kawano, M., Fukasawa, M., and Nishijima, M. (2003) Nature 426 803-809 [DOI] [PubMed] [Google Scholar]
- 4.Rao, R. P., Yuan, C., Allegood, J. C., Rawat, S. S., Edwards, M. B., Wang, X., Merrill, A. H., Jr., Acharya, U., and Acharya, J. K. (2007) Proc. Natl. Acad. Sci. U. S. A. 11364-11369 [DOI] [PMC free article] [PubMed]
- 5.Leinonen, A., Netzer, K., Boutad, A., Gunwar, S., and Hudson, B. G. (1999) Kidney Int. 55 926-935 [DOI] [PubMed] [Google Scholar]
- 6.Crow, M. T., and Stockdale, F. E. (1986) Dev. Biol. 113 238-254 [DOI] [PubMed] [Google Scholar]
- 7.Westerfield, M. (1995) The Zebrafish Book, University of Oregon Press, Eugene, OR
- 8.Kimmel, C. B., Ballard, W. W., Kimmel, S. R., Ullmann, B., and Schilling, T. F. (1995) Dev. Dyn. 203 253-310 [DOI] [PubMed] [Google Scholar]
- 9.Wilson S. W., Ross L. S., Parrett, T., and Easter, S. S., Jr. (1990) Development 108 121-145 [DOI] [PubMed] [Google Scholar]
- 10.Henry, C. A., and Amacher, S. L. (2004) Dev. Cell. 7 917-923 [DOI] [PubMed] [Google Scholar]
- 11.Abrams, J., White, K., Fessler, L. I., and Steller, H., (1993) Development 117 29-43 [DOI] [PubMed] [Google Scholar]
- 12.Levine P. T., and Munro, S. (1998) Curr. Biol. 8 729-739 [DOI] [PubMed] [Google Scholar]
- 13.De Matteis, M. A., and Godi, A. (2004) Nat. Cell Biol. 6 487-498 [DOI] [PubMed] [Google Scholar]
- 14.Loewen, C. J. R., Roy, A., and Levine, T. P. (2003) EMBO J. 22 2025-2035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kallen, C. B., Billheimer, J. T., Summers, S. A., Stayrook, S. E., Lewis, M., and Strauss, J. F., III (1998) J. Biol. Chem. 273 26285-26288 [DOI] [PubMed] [Google Scholar]
- 16.Fugmann, T., Hausser, A., Schöffler, P., Schmid, S., Pfizenmaier, K., and Olayioye, M. A. (2007) J. Cell Biol. 178 15-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hannun, Y. A. (1994) J. Biol. Chem. 269 3125-3128 [PubMed] [Google Scholar]
- 18.Osawa, Y., Uchinami, H., Bielawski, J., Schwabe, R. F., Hannun, Y. A., and Brenner, D. A. (2005) J. Biol. Chem. 280 27879-27887 [DOI] [PubMed] [Google Scholar]
- 19.Gómez-Muñoz, A. (2006) Biochim. Biophys. Acta 1758 2049-2056 [DOI] [PubMed] [Google Scholar]
- 20.Göggel, R., Winoto-Morbach, S., Vielhaber, G., Imai, Y., Lindner, K., Brade, L., Brade, H., Ehlers, S., Slutsky, A. S., Schütze, S., Gulbins, E., and Uhlig, S. (2004) Nat. Med. 10 155-160 [DOI] [PubMed] [Google Scholar]
- 21.Yoshimura, Y., Tani, M., Okino, N., Iida, H., and Ito, M. (2004) J. Biol. Chem. 279 44012-44022 [DOI] [PubMed] [Google Scholar]
- 22.Kumagai, K., Kawano, M., Shinkai-Ouchi, F., Nishijima, M., and Hanada, K. (2007) J. Biol. Chem. 282 17758-17766 [DOI] [PubMed] [Google Scholar]
- 23.Swanton, C., Marani, M., Pardo, O., Warne, P. H., Kelly, G., Sahai, E., Elustondo, F., Chang, J., Ahmed, A. A., Brenton, J. D., Downward, J., and Nicke, B. (2007) Cancer Cell 11 498-512 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.